Abstract
Adipocyte differentiation and function relies on a network of transcription factors, which is disrupted in obesity-associated low grade, chronic inflammation leading to adipose tissue dysfunction. In this context, there is a need for a thorough understanding of the transcriptional regulatory network involved in adipose tissue pathophysiology. Recent advances in the functional annotation of the genome has highlighted the role of non-coding RNAs in cellular differentiation processes in coordination with transcription factors. Using an unbiased genome-wide approach, we identified and characterized a novel long intergenic non-coding RNA (lincRNA) strongly induced during adipocyte differentiation. This lincRNA favors adipocyte differentiation and coactivates the master adipogenic regulator peroxisome proliferator-activated receptor gamma (PPARγ) through interaction with the paraspeckle component and hnRNP-like RNA binding protein 14 (RBM14/NCoAA), and was therefore called PPARγ-activator RBM14-associated lncRNA (Paral1). Paral1 expression is restricted to adipocytes and decreased in humans with increasing body mass index. A decreased expression was also observed in diet-induced or genetic mouse models of obesity and this down-regulation was mimicked in vitro by TNF treatment. In conclusion, we have identified a novel component of the adipogenic transcriptional regulatory network defining the lincRNA Paral1 as an obesity-sensitive regulator of adipocyte differentiation and function.
Similar content being viewed by others
Introduction
White adipose tissue (WAT) is a dynamic organ responding to dietary intakes by a rapid morphological remodeling whose kinetics depends on WAT localization within the body1. Expanding WAT mass stores energy in periods of plenty and is a safeguard against lipid accumulation in peripheral tissues, a major contributor to insulin resistance and associated co-morbidities such as type 2 diabetes (T2D)2. Indeed, increased fat deposition in WAT may be protective and metabolic health thus relies in part on WAT expandability, which depends on WAT hyperplasia and adipocyte hypertrophy3. In the context of obesity, hypertrophied adipocytes are prone to cell death4, hence triggering macrophage infiltration and TNF-induced PPARγ downregulation among other processes5. Furthermore, adipocyte size positively correlates with insulin resistance and T2D and is thus pathologically meaningful6. In contrast, WAT hyperplasia is metabolically more beneficial than hypertrophy7.
De novo adipogenesis, leading to WAT hyperplasia, is thus required for WAT to cope with a positive energy balance. Adipogenesis is a highly complex mechanism relying on the sequential activation or repression of transcriptional regulators leading to a mature lipid-storing adipocyte phenotype. The core of the terminal differentiation signaling pathway is constituted by the transcription factor CCAATT enhancer-binding protein β (C/EBPβ) which regulates the expression of PPARγ8 and of C/EBPα9. The coordinated interplay of these 2 transcription factors triggers complex epigenomic remodeling to achieve adipocyte maturation8,10,11,12.
Pervasive transcriptional events throughout the genome generate numerous RNA transcripts without protein coding potential [non-coding (nc) RNAs] and covering ~60% of the genome. Among those, long non-coding RNAs (lncRNAs, > 200 nt) play a role in diverse biological processes such as cellular differentiation13,14. LncRNAs are expressed in a highly tissue-specific manner and display a wide array of functions in the cytoplasm and/or the nucleus often related to transcriptional and post-transcriptional gene regulation, as well as to organization of chromosome and nucleus topology15,16. Considering their generally low abundance and cell-specific expression, lncRNAs have also been proposed to be mere by-products of transcription which is a nuclear structure-regulatory event per se17.
Several lncRNAs (Neat1, Adinr and lnc-U90926) interfere with terminal adipocyte differentiation by modulating PPARγ or C/EBPα expression18,19,20. The exact molecular mechanisms involved in lncRNA-mediated control of adipogenesis remain however poorly defined. As no investigation of their possible contribution to WAT physiopathology has been reported, a case-by-case investigation remains necessary to decipher the mechanism of action of lncRNAs.
In this study, we characterized a novel adipocyte-specific lincRNA potentiating the adipogenic function of PPARγ through interaction with RNA Binding Motif Protein 14 (RBM14/CoAA), hereafter called Paral1 for PPARγ-activator RBM14-associated lncRNA. Loss-of-function experiments demonstrated its positive contribution to adipocyte differentiation. Expression studies in obese mice and humans showed a similarly decreased expression of Paral1 in obese WAT, thereby identifying a novel adipogenic pathway dysregulated in obesity.
Results
Paral1 is a long intergenic non-coding RNA specifically expressed in mature white adipocytes
To identify lincRNA(s) expressed in adipose tissue and regulated during adipogenesis, we mined the NONCODE v3.0 database (http://www.noncode.org) containing 36,991 lncRNAs, from which 9,364 lincRNAs could be identified by filtering out transcripts overlapping with RefSeq genes. Using NGS data from differentiating 3T3-L1 cells21, a well-established model for adipocyte differentiation, 406 lincRNAs from the NONCODE database displaying an increased density in H3K4me3 and H3K27ac ChIP-seq signals within +/− 2.5 kb from the TSS upon differentiation were identified (Supplemental Table 2, Fig. 1A). Additional filtering using PPARγ ChIP-Seq signals narrowed this list down to 3 lincRNAs, amongst which BC034902, hereafter termed Paral1 (PPARγ-activator RBM14-associated lincRNA 1), displayed the strongest levels of transcriptional activation marks (Fig. 1A, lower inset, and Fig. 1B). This 2.4 kb transcript is devoid of strong coding potential (Supplemental Table 3) and may occur as 2 isoforms in 3T3-L1 cells, of which isoform 1 is predominantly expressed (Fig. 1B, Supplemental Fig. 1). The 2 flanking protein-coding genes Pak7 and Ankrd5 genes display no histone activating marks neither in 3T3-L1 cells (Supplemental Fig. 2A) nor in primary adipocytes (Supplemental Fig. 2B) and are poorly activated during 3T3-L1 differentiation (Fig. 1C). This suggests that Paral1 is an autonomous transcription unit not stemming from spurious read-through processes. In contrast, Paral1 expression was potently induced during 3T3-L1 [fold change (FC = 70)], Fig. 1C) and 3T3-F442A differentiation (FC = 25, Supplemental Fig. 3). Murine mesenchymal stem cell (MSC) differentiation toward the adipocyte lineage was equally accompanied by a strong upregulation of Paral1 (FC = 250), in contrast to osteoblastic differentiation during which Paral1 expression was not modified compared to osteoblastic markers (Runx2, Osteocalcin) [Fig. 1D, Supplemental Fig. 4]. Paral1 expression was restricted to mouse white adipose tissue (WAT) (Fig. 1E). Paral1 was almost exclusively detected in mature adipocytes (AF) but not in the stromal vascular fraction (SVF) (Fig. 1F,G), in line with the data on in vitro differentiated adipocytes (Fig. 1C) and with the specific marking of the Paral1 promoter with H3K4me3 and H3K27ac in isolated adipocytes (Supplemental Fig. 2B). Paral1 expression is therefore markedly restricted to adipocytes and increases during adipocyte differentiation.
Identification of Paral1, an adipocyte-specific lincRNA. (A) Identifying long intergenic non-coding (linc) RNA promoter regions activated during 3T3-L1 adipogenesis. LincRNA promoter regions were scanned for increasing ChIP-seq signals for H3K4me3 (red) or H3K27Ac (blue) upon 3T3-L1 cell differentiation as well as PPARγ binding in adipocytes (yellow). The resulting Venn diagram shows signal overlaps between the identified lincRNA promoters. Numbers indicate the number of transcripts potentially regulated by identified promoters. Lower inset: H3K4me3 profiles in 3T3-L1 adipocytes for the 3 identified lincRNAs (Noncode v.3). (B) ChIP-seq profiles at the Paral1 locus. PPARγ-, H3K4me3- and H3K27-enriched sequences were visualized using the IGV browser, as well as the RNA-seq profile allowing the identification of Paral1 exons (lower panel). (C) Paral1 transcript level in differentiating 3T3-L1 cells. Paral1 expression was monitored at indicated times by RT-qPCR and normalized against the reference Rplp0 housekeeping gene expression level. The expression of neighboring genes (5’: Pak7, 3′: Ankrd5) was assayed similarly. Results are expressed as the mean ± S.E.M. (n = 3) relative to Paral1 RNA level in preadipocytes arbitrarily set to 1. The statistical significance of differences was established using a 1-way ANOVA followed by a Dunnett post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. (D) Paral1 expression in mouse mesenchymal stem cells (MSCs). MSCs were differentiated for 14 days and Paral1 expression was assessed by RT-qPCR as in (C). (E) Paral1 expression in mouse tissues. Paral1 transcript levels in indicated mouse tissues were assayed by RT-qPCR as above. The statistical significance of differences was established using a 1-way ANOVA followed by a Tukey post hoc test. Comparing to eWAT control: *p < 0.05; **p < 0.01; ***p < 0.001. Comparing to iWAT control: $p < 0.05; $$p < 0.01; $$$p < 0.001. eWAT: epididymal white adipose tissue, iWAT: inguinal white adipose tissue, BAT: brown adipose tissue, SKM: skeletal muscle. (F,G) Paral1 expression level in fractionated adipose tissues. Paral1 expression was measured in the stromal vascular fraction (SVF) and adipocyte fraction (AF) from epididymal (eWAT, F) and inguinal (iWAT, G) white adipose tissues (WAT).
Paral1 is required for adipocyte differentiation
In vitro loss-of-function experiments (Fig. 2A) were carried out in 3T3-L1 cells by transfecting a siRNA targeting the Paral1 transcript (Paral1/1-siRNA). A decreased expression (>70%, Fig. 2B) corresponded to a significantly blunted lipid storage (Fig. 2C) and decreased adipocyte-specific gene expression (Supplemental Fig. 5). Transfection with other siRNA targeting Paral1 (Paral1/2-siRNA) similarly showed an impact on adipogenesis (Supplemental Fig. 6). The most efficient siRNA (Paral1/1-siRNA) was selected for further experiments. Silencing of the neighbouring gene Ankrd5, whose expression is modestly increased during differentiation (Fig. 1C), did not impact on lipid storage (Supplemental Fig. 7), indicating that Paral1 does not act through regulation of this flanking gene. To identify regulatory pathway(s) controlled by Paral1, we compared gene expression patterns in differentiating adipocytes (D2) depleted or not of Paral1. This allowed the identification of ~500 genes deregulated upon Paral1 silencing (Fig. 2D). Importantly, the expression of genes associated with white adipogenesis (AdipoQ, aP2) was decreased, in contrast to genes associated with brown adipogenesis (Ebf2, Prdm16, Ucp1) which remained unchanged (Supplemental Table 4). Gene set enrichment analysis (GSEA) revealed that many down-regulated genes are associated with oxidative metabolism and PPAR signalling, whereas up-regulated genes are functionally related to protein synthesis and cytokine signalling (Fig. 2E,F). Additional term enrichment analysis based on the Gene Ontology “Biological Process” functional annotation table (GO BP FAT) indicated that down-regulated genes are involved in lipid homeostasis and metabolism (Fig. 2G). This is in line with a role for Paral1 in the acquisition of the mature adipocyte phenotype, as indicated by the altered lipid-storing capacity of Paral1-depleted 3T3-L1 cells (Fig. 2C). A significant and selective down-regulation of the adipogenic master genes Pparγ and C/ebpα was observed (Fig. 2G and Supplemental Table 5). Paral1 knockdown using an unrelated LNA gapmer generated a similar phenotype, confirming the role of Paral1 in adipogenesis (Supplemental Fig. 8). However, inducible Paral1 overexpression did not promote 3T3-L1 cell differentiation on its own, in contrast to Pparγ overexpression (Supplemental Fig. 9). Taken as a whole, our data show that Paral1 is necessary, but not sufficient in our conditions, for terminal adipocyte differentiation.
Paral1 expression is required for adipocyte differentiation. (A) 3T3-L1 differentiation and transfection protocol. B) Validation of Paral1 expression knockdown. 3T3-L1 cells were transfected with control (Ctrl-siRNA) or Paral1-targeting siRNA (Paral1/1-siRNA) at D0 and differentiation was initiated at the same time. Paral1 transcripts were assayed by RT-qPCR at D2 and results are expressed as the mean ± S.E.M. (n = 3) relative to the Paral1 RNA level in Ctrl-siRNA transfected preadipocytes arbitrarily set to 1. Values were compared using a 1-way ANOVA followed by a Dunnett post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. NT: non transfected preadipocytes. (C) Lipid accumulation in differentiated adipocytes. Oil Red O staining of 3T3L1 cells was performed at D8. Numbers indicate intracellular ORO stain and intracellular triglycerides quantification relative to non-transfected (NT) cells for a representative experiment. (D) Knockdown of Paral1 expression alters the expression of a specific subset of genes. The 3T3-L1 cell transcriptome was characterized by DNA microarray analysis as described in the Materials & Methods section. GSEA was performed against the KEGG database. NES: normalized enrichment score, FDR: false discovery rate), red: up-regulated after Paral1/1-siRNA treatment, green: down-regulated after Paral1/1-siRNA treatment. (E) GSEA enrichment plots of oxidative phosphorylation, TCA cycle, PPAR signaling pathway and cytokine-cytokine receptor signature genes in differentiating 3T3-L1 cells. Green curves depict the enrichment score for each gene (vertical black line) ranked along a heatmap (red to green, up- to down-regulated genes) indicating the observed fold change in Paral1/1-siRNA treated cells. NES: normalized enrichment score, FDR: false discovery rate. (F) Top-ranking genes in Paral1/1-siRNA treated cells. (G) Expression level variation of major contributors to the adipogenic program and top ranking biological theme (using the gene ontology biological process functional annotation table, GO BP FAT) of down-regulated genes in Paral1-depleted cells.
Paral1 contributes to adipocyte phenotype maintenance
We next assessed whether Paral1 is required to maintain a fully mature adipocyte phenotype. Silencing of Paral1 in differentiated 3T3-L1 adipocytes at D7 (Supplemental Fig. 10A,B) perturbed the expression of a minor fraction of transcripts (∼100 genes, FC > 1.5, p < 0.05). A gene set enrichment analysis of expression data emphasized the down-regulation of genes involved in functions similar to those previously identified upon Paral1 silencing in differentiating cells (D2) and notably including oxidative phosphorylation (Supplemental Fig. 10C). A gene-by-gene analysis showed Paral1 depletion affected neither Pparγ nor C/ebpα expression (Supplemental Fig. 10E) but reduced that of genes participating to glycerolipid and cholesterol synthesis (Agpat2, Gpam, Lpin1, Cyp51a1, Lss…). These biosynthetic pathways are not only under the tight control of PPARγ but are also coordinately regulated by SREBP1c, CHREBP and/or LXR. Paral1 depletion affected mostly PPARγ target genes (Supplemental Fig. 10E). However, the expression of several other bona-fide direct PPARγ target genes such as aP2/Fabp4, Cd36 or Glut4 was left unchanged in these conditions. Whether this results from distinct regulatory mechanisms or from a differential sensitivity to Paral1 depletion remains to be established. Taken together, these data however suggest that Paral1 sustains at least in part PPARγ transcriptional activity in mature adipocytes.
Paral1 localization is mainly nuclear and interacts with the transcriptional coactivator and paraspeckle component RBM14
As lincRNA functions are highly dependent on their subcellular localization22, we quantified Paral1 transcripts in the chromatin (nuclear insoluble), nuclear (nuclear soluble) and cytosolic fractions from differentiated 3T3-L1 cells. Ribosomal Rplp0 RNA was used as a cytosolic RNA control and Neat1 as a chromatin-associated lincRNA18. Paral1 was mainly nuclear (~70%) and predominantly detected in the chromatin fraction (~47%) (Fig. 3A). A RNA pull-down was performed using biotinylated Paral1 as a bait (Fig. 3B). This assay identified by mass spectrometry (Fig. 3C) known components of paraspeckles [Splicing Factor Proline And Glutamine Rich (SFPQ), Non-POU Domain Containing, Octamer-Binding (NONO), Paraspeckle Component 1 (PSPC1) and RNA Binding Motif Protein 14 (RBM14)]23. Among them, RBM14/ Coactivator Activator (COAA) is not only an RNA-binding protein, but a secondary coactivator of several nuclear receptors24,25. The presence of RBM14 in RNA pulldown eluates from cellular extracts from differentiating (D2) and differentiated (D7) was validated by western blotting (Fig. 3D,E, Supplemental Fig. 11). Like Paral1, RBM14 was located in the chromatin fraction (Fig. 3F).
Paral1 interacts with chromatin-bound RBM14. (A) Subcellular localization of Paral1. RNA from 3T3-L1 cells was fractionated into nuclear insoluble, nuclear soluble and cytosolic fractions at D5 and analysed for their content in Paral1, Rplp0 and Neat1. Rplp0 and Neat1 RNA were used as cytosolic and nuclear RNA controls, respectively. (B) Proteins from RNA pull-down eluates (no RNA, Paral1 and Input) were visualized by silver staining after SDS-polyacrylamide gel electrophoresis (PAGE). (C) Top hits of identified proteins by Q-Exactive nano-LC tandem mass spectrometry specifically interacting with Paral1 in the RNA pulldown assay. (D) RNA pull-down eluates were analysed by western blotting for their content in RBM14 at D2. (E) RNA pull-down eluates were analysed by western blotting for their content in RBM14 at D8. (F) 3T3-L1 whole cell extracts (D5) were separated into nuclear insoluble, nuclear soluble and cytosolic fractions. Proteins were resolved by SDS-PAGE and identified by western blotting. β-actin and histone H3 were used as cytosolic and nuclear controls respectively. RNA pull-down eluates were analysed by western blotting for their content in RBM14. (G) RNA immunoprecipitation using an anti-RBM14 antibody. Total extracts were immunoprecipitated and Paral1 level was measured by RT-qPCR experiments. Rplp0 and Neat1 RNA were used as a negative and positive control respectively.
Immunoprecipitation using an antibody against RBM14 followed by RT-qPCR (RIP-qPCR) (Fig. 3G) showed that Paral1 was specifically enriched (x5) in RBM14-containing complexes, confirming that RBM14 interacts specifically with Paral1. The Rplp0 cytosolic RNA did not interact with RBM14, whereas the paraspeckle-associated Neat1 lncRNA16 was enriched (x 3.4) in RBM14 immunoprecipitates. The interaction of RBM14 with Paral1 was mapped to the 5′ half of Paral1 (Supplemental Fig. 12), whose secondary structure prediction did not reveal any peculiar features (Supplemental Fig. 13).
RBM14 cooperates with Paral1 in regulation of adipocyte differentiation
RBM14 protein expression increases during 3T3-L1 differentiation similar to Paral1 (Fig. 4A). Rbm14 mRNA expression however did not strictly match protein expression (Fig. 4A), suggesting that post-transcriptional processes may influence RBM14 protein stability. We investigated whether RBM14 is also required for 3T3-L1 adipogenesis by loss-of-function experiments (Fig. 4B and Supplemental Fig. 14). RBM14-depleted 3T3-L1 cells accumulated less lipid at D8 (Fig. 4C), and adiponectin and Pparγ gene expression was decreased (Fig. 4D). Paral1 and RBM14 are thus both necessary for adipocyte differentiation. A similar loss-of-function study was performed for the other Paral1-associated paraspeckles components (Supplemental Fig. 15). Both Pspc1 and Sfpq knockdowns interfered with adipogenesis, whereas the contribution of NONO was not significant. Paral1 may thus belong to a large functional protein complex comprising several paraspeckle components.
RBM14 protein expression increases during adipogenesis and is required for the adipogenic process. (A) RBM14 RNA level (upper panel) and protein level (lower panel) were assayed by RT-QPCR and western blotting respectively. RT-QPCR results were expressed as the mean ± S.E.M. (n = 3) relative to the indicated control in non-treated 3T3-L1 preadipocytes arbitrarily set to 1. Values were compared using ANOVA followed by a Tukey’s post hoc test. *p < 0.05; **p < 0.01. RBM14 protein was visualized in 3T3-L1 cells lysates (70 µg) at D0, D2, D5 and D8 by western blot using a specific anti-RBM14 polyclonal antibody. (B) Rbm14 knockdown by LNA gapmer transfection. Control (Ctrl-LNA) or LNA gapmers targeting Rbm14 (Rbm14-LNA, RBM14/2-LNA) were transfected at D0 and Rbm14 RNA and protein levels were assayed by RT-qPCR and western blotting as above at D8. (C) ORO staining of 3T3-L1 cells at D8. (D) AdipoQ expression in RBM14-depleted cells. Pparγ, Paral1 and AdipoQ transcript abundance was measured by RT-qPCR in RNA extracted from 3T3-L1 cells at D2. Results are expressed as the mean ± S.E.M. (n = 3) relative to the indicated control in non-treated 3T3-L1 preadipocytes arbitrarily set to 1. Values were compared using a t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
Paral1 potentiates RBM14 coactivation of PPARγ transcriptional activity
As our data pointed to a regulatory role of the Paral1:RBM14 complex in PPARγ–driven events, we investigated its potential contribution to PPARγ transcriptional activity. Using a 1-hybrid assay in which Gal4 DBD-fused PPARγ activity was monitored in the presence or not of overexpressed Paral1 and/or Rbm14, we observed that, in contrast to RBM14 which increased PPARγ transcriptional activity, Paral1 had no effect on its own in this assay (Fig. 5A). However, Paral1 potentiated RBM14 coactivation of PPARγ. This combination had no effect on PGC1α-regulated transactivation (Fig. 5B).
Paral1 potentiates RBM14 coactivation of PPARγ. (A,B) One-hybrid transactivation assay monitoring the transcriptional potential of PPARγ (A) or PGC1α (B). HEK cells were transfected using the indicated combination of reporter (pUAS-tk Luc) and expression (pGal4-Pparγ, pcDNA3-Rbm14, pcDNA3-Paral1) vectors. Results are expressed as fold change relative to luciferase level detected in cells transfected without the indicated transcription factor. Results are expressed as means ± S.E.M (n = 3–5). The statistical significance of differences was analyzed by ANOVA followed by a Tukey’s post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. (C) Structure and expression of RMB14. The structure of wild type RBM14 is depicted with numbers indicating aminoacid sequence positions. Right panel: western blot analysis of RBM 14 derivatives when expressed in HEK cells (NT: non transfected). (D) PPRE-dependent transactivation assay. HEK cells were transfected with a PPRE-driven reporter vector and the indicated combination of expression vectors. Results are expressed as fold change relative to luciferase level detected in cells transfected without the indicated transcription factor. Results are expressed as means ± S.E.M (n = 3). The statistical significance of differences was analyzed by ANOVA followed by a Tukey’s post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001.
RBM14/NCoAA comprises 2 main functional domains including at its N-terminus 2 RNA recognition motifs (RRMs) and a large TRBP/AIB3 interacting domain (TRBP-ID), that mediates its interaction with nuclear receptor coactivators (Fig. 5C 24). We monitored PPARγ transcriptional activity using a PPARγ response element (PPRE)-driven reporter gene and an expression vector coding for RXRα, PPARγ’s obligate heterodimerization partner, in the presence of wild type RBM14 or of a RBM14 N-terminally truncated mutant (Fig. 5C,D). In this system, the RXRα/PPARγ dimer induced a 15-fold increase over control (no RXRα/PPARγ) of the reporter gene activity in the presence of rosiglitazone (Rosi), which was not altered by the overexpression of Paral1. In contrast, wild type RBM14 significantly increased both basal (FC = 15) and Rosi-induced (FC = 240) luciferase activity. The RBM14 deletion mutant was inactive in similar conditions. Overexpression of Paral1 did not affect the ability of RBM14 to potentiate the basal activity level of the system, but dramatically increased its activity in the presence of rosiglitazone (FC = 33), confirming the functional synergy between Paral1 and RBM14 on PPARγ-mediated transcription. In sharp contrast, the RRM-truncated RBM14 mutant was unable to convey such a potentiation. We also observed that a TRBP-ID deleted RBM14 was devoid of any activity in this system (data not shown). While requiring further investigation to reach a definitive conclusion, this suggests that the RBM14 RRM domain is required for such a functional interaction to occur.
Adipose tissue inflammation lowers Paral1 expression in murine models of obesity
Paral1 expression was therefore assessed in 2 mouse models of obesity. Epididymal (e)WAT from both leptin-deficient (ob/ob) and high-fat diet-fed (HFD) mice displayed clear signs of increased expression of inflammation-related pathways (Fig. 6A,B, Supplemental Fig. 16), in agreement with our previous studies26. GSEA comparing gene expression patterns of Paral1-depleted 3T3-L1 cells at D7 to that of ob/ob and HFD mouse eWAT revealed that pathways related to oxidative phosphorylation were commonly dysregulated (Fig. 6A,B and Supplemental Fig. 10). Epididymal WAT from obese mice displayed a markedly decreased expression of Paral1, in line with our data showing that depletion of Paral1 yields dysfunctional 3T3-L1 cells, (Fig. 6C,D). PPARγ gene expression was also decreased in eWAT from obese mice, but not RBM14 (Supplemental Fig. 17A,C and B,D respectively). Increased pro-inflammatory cytokines production is a hallmark of obese WAT. TNF stimulation of differentiated 3T3-L1 cells, mimicking part of the M1 macrophage-induced inflammatory response, decreased Paral1 expression, suggesting that obesity-induced inflammation may regulate Paral1 expression (Supplemental Fig. 18). IRF5 is a key driver of the pro-inflammatory response in WAT and Irf5 gene knockout protects from metabolic damages caused by diet-induced obesity, notably through impaired IL-1β and TNF release27. eWAT from HFD-fed Irf5 +/+ mice displayed decreased Paral1 expression, which paralleled Pparγ expression, when compared to chow diet (CD) fed mice (Fig. 6E,F). In sharp contrast, eWAT from HFD fed Irf5 −/− mice displayed Paral1 and Pparγ expression levels comparable to CD-fed Irf5 −/− mice (Fig. 6E,F). Thus Paral1 expression is impacted by TNF in vitro and by a chronic pro-inflammatory background in an Irf5-dependent manner in vivo.
Paral1 expression is decreased in obese eWAT in an Irf5-dependent manner. (A) GSEA was performed using the KEGG pathway gene sets. Red: up-regulated in obese (ob/ob) eWAT, green: down-regulated in obese (ob/ob) eWAT. (B) GSEA was performed using the KEGG pathway gene sets. Red: up-regulated in obese (HFD) eWAT, green: down-regulated in obese (HFD) eWAT. Framed: pathways common to both obese eWAT and Paral1-depleted 3T3-L1 cells at D7. NES: normalized enrichment score, FDR: false discovery rate. (C) Paral1 expression in eWAT. eWAT RNA from wild type (wt) C57Bl6/J and from ob/ob mice were analysed for their content in Paral1 transcripts by RT-qPCR. Results are expressed as the mean ± S.E.M. (n = 6–8) relative to the wild type level arbitrarily set to 1. Values were compared using a t-test. *p < 0.05; **p < 0.01; ***p < 0.001. (D) Paral1 expression in eWAT. eWAT RNA from wild type (wt) C57Bl6/J fed either a chow diet (CD) or a high fat diet (HFD) were analysed for their content in Paral1 transcripts by RT-qPCR. Results are expressed as the mean ± S.E.M. (n = 6–8) relative to the wild type level arbitrarily set to 1. Values were compared using a t-test. *p < 0.05; **p < 0.01; ***p < 0.001. (E) Paral1 and Pparγ expression in eWAT. eWAT RNA from wild type (Irf5 +/+) or from Irf5-deficent-mice (Irf5 −/−) fed either a chow diet (CD) or a high fat diet (HFD) were analysed by RT-qPCR. Results are expressed as the mean ± S.E.M. (n = 5–8) relative to the wild type level arbitrarily set to 1. Values were compared using a t-test. * p < 0.05; ** p < 0.01; ***p < 0.001.
Identification of a human Paral1 homolog with decreased expression in obesity
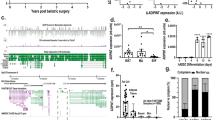
The most recent compendium of human lncRNAs28 suggest that functional lncRNAs are most conserved across species. Sequence alignment across multiple species showed that mouse Paral1 displays significant similarities in the transcribed region with a human homolog (Fig. 7A). Identified as ENSG00000243961.2 in the human lncRNA repertoire (http://fantom.gsc.riken.jp/cat/v1/#/genes/ENSG00000243961.2), hPARAL1 is also flanked by PAK7 and ANKRD5 and displays a conserved promoter, indicating a potential functionality28. Like its murine counterpart, it harbors no significant coding potential and is induced during adipocyte differentiation28, a feature confirmed by the analysis of epigenetic marks around the TSS in undifferentiated and differentiated human adipose-derived stem cells [hASC21], (Fig. 7B). Expression atlas data (Supplemental Fig. 19) and RT-qPCR assays (Fig. 7C) show that hPARAL1 expression is highest in lung and breast and significantly detected in subcutaneous (sc) and omental (vis)WAT.
Identification of a human Paral1 homolog. (A) Multiz alignments of vertebrate homologs to mouse Paral1 in different species visualized with the UCSC Genome browser59. (B) Gene tracks visualized in IGV show the PPARγ ChIP-seq signal [from differentiated human adipose stem cells (HASC), yellow], the H3K4me3 ChIP-seq signal (from undifferentiated and differentiated HASC; same scale; red) and the H3K27ac ChIP-Seq signal (from undifferentiated and differentiated HASC; same scale; blue) at the human PARAL1 locus. (C) hParal1 expression level was measured by RT-qPCR in total subcutaneous adipose tissue (scWAT), omental adipose tissue (visWAT) and liver. Results are expressed as the mean ± S.E.M. (n = 6) relative to the wild type level arbitrarily set to 1. The statistical significance of differences were assessed by ANOVA and Tukey’s post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. (D) hPARAL1 expression level was measured in subcutaneous adipose tissue (scWAT) by RT-qPCR. Results are expressed as the mean ± S.E.M. (n = 5) relative to lean control level arbitrarily set to 1 (not shown). The statistical significance of differences were assessed by ANOVA and Dunnett’s post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. (E) hParal1 expression level was measured in visceral adipose tissue (visWAT) by RT-qPCR. Results are expressed as the mean ± S.E.M. (n = 5) relative to lean control level arbitrarily set to 1. The statistical significance of differences was assessed by ANOVA and Dunnett’s post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001. (F) Correlation between hPARAL1 expression level in scWAT and BMI (n = 20). G) Correlation between hParal1 expression level in visWAT and BMI (n = 20).
Quantification of hPARAL1 transcripts in subcutaneous (sc)WAT and visceral (vis)WAT from lean, obese, glucose-intolerant obese and obese diabetic patients showed decreased hPARAL1 expression in obesity (Fig. 7D,E). In line with mouse data, altered pathways in human obese visWAT again pertained to oxidative phosphorylation and the PPAR signaling pathway in a pro-inflammatory context, both being biological processes affected upon Paral1 depletion (Supplemental Fig. 20). hPARAL1 expression was inversely correlated to BMI (Fig. 7F,G) but not to other biometric or biochemical parameters (Supplemental Table 6), suggesting an exclusive link between hPARAL1 expression and WAT dysfunctions in human WAT.
Discussion
Adipocyte differentiation and function relies on an intricate network of interconnected transcription factors centered on PPARγ29. We report here the identification and characterization of the lincRNA Paral1 as a novel member of this adipogenic master regulatory network. Paral1, whose expression is a feature of mature adipocytes, is required for PPARγ expression and activity during adipogenesis and sustains adipocyte functions. In line, an unbiased search for potential functions of Paral1 based on tissue specific co-expression studies30 predicted a role for hPARAL1 in triglyceride and pyruvate metabolisms. Interestingly, Paral1 expression is decreased in an obesogenic context in both humans and mice and is sensitive to pro-inflammatory signals suggesting that loss of Paral1 expression may contribute to WAT dysfunction in obese individuals with chronic inflammation. Our data also show that Paral1 expression parallels that of PPARγ, as both transcripts increase during adipocyte differentiation. The occurrence of a PPARγ binding site upstream of the Paral1 TSS suggests that PPARγ may drive the expression of a coactivating RNA molecule. This would establish a positive feedforward loop on PPARγ expression (Fig. 8), raising the question of the impact of PPARγ agonism on Paral1 expression. In mature 3T3-L1 adipocytes, acute treatment by insulin-sensitizing thiazolidinediones (TZD such as rosiglitazone or pioglitazone) increases the expression of many genes induced during differentiation, with a few notable exceptions including PPARγ itself whose expression is decreased upon TZD treatment31. We could replicate this finding and, as expected from our data, Paral1 expression followed that of PPARγ (data not shown). While the mechanistic basis of this repression is still elusive31,32, this demonstrates that Paral1 expression is exquisitely dependent on PPARγ expression level, a feature that we also confirmed in WAT from ob/ob mice treated for 5 days by rosiglitazone (3mpk/day,26). In this setting, TZD treatment increased PPARγ expression and that of several bona-fide target genes such as adiponectin, and also induced Paral1 expression (data not shown and26).
Paral1 mechanism of action. PPARγ expression is activated during adipogenesis (a) creating an heterodimer complex with RXR (b) in order to regulate adipogenic factors such as Paral1 (c) necessary for adipogenesis. The Paral1 RNA transcript (d) interacts with RBM14 (e) potentiating its coactivating function (f). The interaction between RBM14-Paral1 and PPARγ-RXR complexes (f) promotes PPARγ activity leading to positive feedback in the upregulation of adipogenic factors (g).
The molecular basis of Paral1 pro-adipogenic activity stems from its ability to interact with and potentiates the co-activating potential of RBM14, whose expression parallels that of Paral1 during adipocyte differentiation. This newly identified property of RBM14 is probably due to its ability to act as an indirect coactivator, through synergistic interactions with nuclear receptor coactivators (hence its alias CoActivator Activator CoAA)24,33. Our work extends its role, initially reported for estrogen and glucocorticoid receptor-dependent transcriptional activation34, to the pro-adipogenic nuclear receptor PPARγ. RBM14, a hnRNP-like protein, encompasses 2 RNA binding modules (RRM), which are likely to interact with RNA molecules such as Paral1, as described for hnRNP U and Xist35. In the chimeric transcription assay used here, we could indeed show that Paral1 function is dependent on the RRM domains.
We also note that RBM14/CoAA inhibits the transcriptional activity of the osteoblastic Runx2 transcription factor, a differentiation pathway that opposes commitment to the adipocyte lineage36. Together with other studies37, our work indicates that lincRNAs act as regulators of expression/activities of key developmental transcription factors. Another way by which lncRNAs regulate gene expression and ensuing biological processes is to act in cis at neighbouring protein-coding loci38. Paral1 is flanked by Ankrd5 and Pak7, none of these genes being reportedly involved in adipogenesis and dysregulated upon Paral1 expression modulation, thereby excluding a possible contribution of Paral1 through such a mechanism.
As noted above, RBM14 possesses RNA binding activity and is a component of paraspeckles, which assemble on the lincRNA Neat1. Interestingly, we found that Paral1 also interacts with paraspeckle components NONO, PSPC1 and SFPQ which are three multifunctional nuclear factors and bind Neat1. This suggested that RBM14-Paral1 could be involved in splicing events. However, our exon microarray analysis did not reveal major changes in the splicing of adipogenic genes (data not shown). This however does not rule out other paraspeckle-associated roles for this RNA-protein complex which remain to be formally investigated, such as the nuclear retention of edited RNA molecules known to play a role in cellular differentiation39.
About 30% of total Paral1 is localized in the cytosol. Although our approach did not allow to appreciate potential roles of Paral1 as a miRNA sponge, we note that Paral1 could hybridize to mir27a, a post-transcriptional regulator of PPARγ40 through its sequence ACUGUGA.
Paral1 interacts also with Eukaryotic Translation Elongation Factor 1 Alpha 1 (EEF1A1) and with Tubulin beta (Tubb), a component of the centriole cytoskeleton with which RBM14 interacts41, hinting at a possible role in mRNA translation or cell division. The basis for this subcellular repartition is unknown. A recent report demonstrated that PSPC1 is involved in the nuclear export of adipogenic RNAs, including PPARγ42. These observations raise the possibility that Paral1 may be part of this RNA shuttling complex, since the RNA shuttling protein DDX3X interacts with Paral1 (Fig. 3C) and PSPC142.
In conclusion, we have identified on the basis of an unbiased genome-wide approach a novel lincRNA with functions in adipocyte physiology, whose expression is decreased in obese rodents and humans. At least part of its mechanism of action can be attributed to co-activating properties of PPARγ, thereby impacting on genes involved in metabolic regulations. This adds a new piece to the puzzle of lncRNA contribution to adipogenesis. In this context, the structural versatility of RNA molecules make them attractive druggable entities43 and a complete inventory of anti- or pro-adipogenic lncRNAs may expand the therapeutic repertoire to combat obesity.
Experimental procedures
Chemicals
Dulbecco’s Modified Eagle’s Medium, alpha Modified Eagle’s Medium, Cosmic Calf Serum and Fetal Bovine Serum (FBS) were from Life Technologies. GW4064 was from Tocris. Ascorbic acid, β-glycerophosphate, bovine insulin, IBMX, dexamethasone, indomethacine, pioglitazone and rosiglitazone were from Sigma.
Cell culture
3T3-L1 and 3T3-442A cells were routinely grown and differentiated as described26. Mesenchymal stem cells (MSC) were isolated from adult mouse bone marrow and maintained in αMEM (Life Technologies) as described44.
Oil Red O staining
Oil red O (ORO) staining was performed at D8 as described8.
Triglycerides quantification
Triglycerides quantification were performed at D8 as described45.
RNA extraction and RT-qPCR
Total RNA was extracted, analyzed by RT-qPCR and transcript level were quantified as described26.
Microarray analysis
Total RNA (100–300ng) from 3T3-L1 cells was processed for labelling, purification and hybridized to Affymetrix Genechip Mouse Genome 430 2.0 or to Mouse Transcriptome Array 1.0 according to the manufacturer’s protocol. Raw data (available on the GEO website under the accession number GSE 97241) were pre-processed using the GCCN and SST algorithms (Expression Console, v1.4.1. Affymetrix). RMA background correction and gene-level probe set summarization were performed with the Partek Genomics Suite software (v6.6, Partek Inc.). Microarray analysis of leptin-deficient (ob/ob) mouse WAT has been described elsewhere26. Microarray data from mouse WAT fed either a chow or a high fat diet were from the GEO dataset GSE21069 46. Human WAT RNAs were analysed as described26.
Human gene symbols were attributed to each murine gene using the Orthologue Conversion software (https://biodbnet-abcc.ncifcrf.gov/db/dbOrtho.php)47 and resulting files were analysed with the GSEA software [Broad Institute, v2.2.248]. Pathway-enrichment scores were calculated with the GSEA pre-ranking tool and the KEGG Pathway gene set. Default parameters were used except for the permutation number (10,000) and the enrichment score statistic (weighted). The GEO dataset is available under the number GSE97241.
Data mining and bioinformatics
Long non-coding RNA identification
Murine lncRNA sequences were extracted from the NONCODE database v.3 (http://www.noncode.org/NONCODERv3/; mouse genome reference: mm9).
Gene Expression Omnibus (GEO) datasets were: GSE84888 for differentiating human adipose stromal (hASC) and murine 3T3-L1 cells21 and GSE92590 for isolated primary mouse adipocytes from WAT49. RNA-Seq data were from the GEO dataset GSE3572450.
ChIP-Seq data were analyzed using the Galaxy Cistrome platform11. RNA-Seq and ChIP-Seq data were visualized using the Integrative Genomics Viewer software (IGV, v2.3.14, Broad Institute).
Coding potential analysis and proteomic databases
Three softwares were used with default parameters to determine lincRNAs’ coding potential (CPC, http://cpc.cbi.pku.edu.cn/)51; CPAT, http://lilab.research.bcm.edu/cpat/index.php)52 and GenView2 (http://bioinfo.itb.cnr.it/~webgene/wwwgene.html). GAPDH and XIST RNA were used as representative of mRNA and lincRNA respectively. Potential ORFs were converted into protein sequences and matching peptides were searched in four protein databases [UniProtKB (SwissProt and TEMBL; http://www.uniprot.org/); PDB (http://www.rcsb.org/pdb/home/home.do); Ensembl (www.ensembl.org/)].
Total protein extraction, western blotting and antibodies
Proteins (30–100 µg) were extracted and analysed by western blotting as described53. Primary antibodies used in this study were: anti-RBM14 (Euromedex, ref. 10196-1-AP), anti-PPARγ (Santa Cruz, ref. sc-7196), anti-TFIIB (Santa Cruz, ref. sc-225), anti-β-actin (Santa Cruz, ref. sc-1616), anti-H3 (Abcam, ref. ab1791) and control IgG (Merck-Millipore, ref. 17–658). Secondary antibodies were anti-rabbit IgG-peroxidase antibody (Sigma, ref. A0545) or anti-goat IgG-peroxidase antibody (Sigma, ref. A5420).
Subcellular fractionation
3T3-L1 cells were plated in P100 dishes and differentiated as described above. At indicated times, cells were washed twice using ice-cold 1x PBS and lysed in 500 µL lysis Buffer 1 [10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 40 U/mL RNasin and protease inhibitors (Roche)] supplemented with 0.1% Triton-X100 and 1 mM DTT upon use. Lysates were centrifuged to separate cytosolic and nuclear fractions (1,300 G, 5′, 4 °C). After removal of the upper lipid phase, supernatants (cytosolic fraction) were centrifuged (16,000 G, 5′, 4 °C) and used for RNA and protein characterization. Nuclei were washed once into 400 µL lysis buffer 1 supplemented with 1 mM DTT extemporarily and incubated in 100 µL lysis buffer 2 (10 mM HEPES pH 7.9, 3 mM EDTA pH8, 0.2 mM EGTA pH8, 40 U/mL RNasin, 1 mM DTT and protease inhibitors) for 30 min. on ice. Insoluble and soluble fractions were obtained by centrifugation (1,300 G, 5′, 4 °C). The insoluble fraction was digested with benzonase [50 mM Tris-HCl, pH7.5, 1 mM MgCl2, 3 mM EDTA pH8, 0.2 mM EGTA pH8 supplemented with 25U benzonase (Millipore) and 1 mM DTT extemporarily] for 20 min. on ice.
Oligonucleotide design and synthesis
LNA gapmers complementary to mouse Rbm14 mRNA (Rbm14-LNA) were designed using the online Exiqon software (https://www.exiqon.com/ls/Pages/GDTSequenceInput.aspx): Rbm14-LNA: ATGACTGAGTGCGGTA; Paral1-LNA: AGGAGCATAATGAATA. Exiqon LNA gapmers were synthesized by Exiqon (Qiagen) with a phosphorothioate backbone and purified by a standard desalting method.
Plasmids
The pcDNA3-Myc-Rbm14 vector and derivatives were from D. Monté (Univ. Lille, France)54. Other plasmids used in this study were the pUAS-tk-Luc vector55, pSG5-hRXRα55, pcDNA3-Flag-Pparγ 56, pGal4-PGC1α vector57. pGal4-empty vectors was from Addgene. The pGL4.25 vector was from Promega. The pcDNA3.1 vector was from Life Technologies.
The Paral1 RNA was reverse-transcribed and amplified by PCR with primers containing BamHI and NotI restriction sites (Supplemental Table 1) then ligated into the pCR blunt II TOPO vector (TOPO TA Cloning Kit, Life Technologies). The cDNA insert was then excised as a BamHI/ fragment and inserted into pcDNA3.1 to generate the pcDNA3-Paral1 plasmid.
pRetroX-Paral1 and pRetroX-Pparγ vectors were generated by ligating inserts into the pRetrox-Tight-Pur plasmid (ClonTech) as BamHI/NotI or NotI/XbaI inserts.
The pGL4-3xPPRE Luc reporter vector was generated by inserting an oligonucleotide containing 3 consensus PPRE sequences (in bold) (5′-AAGCTTGACAGGGGACCAGGACAAAGGTCACGTTCGGGAAGCTTGTCGACAGGGGACCAGGACAAAGGTCACGTTCGGGAAGCTTG TCGACAGGGGACCAGGACAAAGGTCACGTTCGGGAAGCTT-3′) into the pGL4.25 [luc2CP/minP] backbone (Promega).
Biotinylated RNA synthesis and purification
pcDNA3-Paral1 was linearized by NotI, and biotinylated RNA transcripts were synthesized with the MEGAscript® T7 Transcription Kit (Ambion) and biotin-CTP (Enzo-Life Sciences). RNA was purified as suggested by the manufacturer (Ambion). RNA integrity and biotinylation efficiency were checked by agarose gel analysis and dot blotting, respectively.
RNA pulldown assay
3T3-L1 cell lysates were pre-cleared [1 mg protein for 40 µL streptavidin-coupled Dynabeads M-280 (Invitrogen)] for 3 hours. Beads were removed by magnetic separation. Biotinylated Paral1 RNA was incubated at 65 °C for 5 minutes then at room temperature for 10 min. Paral1 RNA (60 µg) was coupled to streptavidin-coupled Dynabeads M-280 (1 µL/µg RNA) in water containing RNasin (40 U/mL) for 30 min. at 20 °C. The supernatant was removed by magnetic separation. Pre-cleared 3T3-L1 cell lysates (1 mg total protein) were incubated with RNA-coupled beads for 60 min. After 3 washes with washing buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 40 U/mL and protease inhibitors), beads were incubated in 100 µL 1.5x Laemmli buffer and supernatants were processed for further analysis.
RNA immunoprecipitation
3T3-L1 cells were washed in ice-cold 1x PBS and incubated in 1x PBS-1% formaldehyde for 10 min. The crosslinking reaction was quenched with 0.25 M glycine for 5 min. Cells were washed once with 1x PBS and lysed in 25 mM Tris-HCl pH 7.4, 500 mM NaCl, 1 mM EDTA, 40 U/mL RNasin, 5% glycerol and protease inhibitors. Lysates were sonicated using a Bioruptor UCD-200 and centrifuged (16,000 G, 10 min. at 4 °C).Supernatants were brought to 150 mM NaCl using 25 mM Tris-HCl pH 7.4, 1 mM, 40 U/mL RNasin, 5% glycerol and protease inhibitors. Protein concentration was measured using the DC Protein Assay (Biorad).
Crosslinked lysates (1 mg) were incubated with 5 µL Protein G-coupled Dynabeads® (Invitrogen) for 1 hour at 4 °C. Beads were removed by magnetic separation and 1 µg of primary antibody against RBM14 or control IgG were added and incubated overnight. Magnetic Protein G-Dynabeads were coated overnight in 5% BSA, 100 µg/mL yeast tRNA and 40 U/mL RNasin, added to the pre-cleared lysate/antibody mix and incubated for 3 hours at 4 °C. Dynabeads were washed three times with Washing Buffer and incubated with a Reverse-Crosslinking Solution (100 mM Tris-HCl pH 7.5, 5 mM EDTA, 1% SDS, 10 mM DTT and 40 U/mL RNasin) during 15 min. at room temperature. Supernatants were digested with 20 µg proteinase K and incubated at 65 °C for 120 min. Immunoprecipitated RNAs were extracted with TRIzol according to the manufacturer’s instructions.
siRNA transfection
3T3-L1 cells were transfected by siRNAs (100 nM) using INTERFERin (Polyplus Transfection) as described26. siRNAs were: control siRNA (Dharmacon, ON-TARGETplus non-targeting, ref. D-001810-10-10,), Pparγ-siRNA (Dharmacon, ON-TARGETplus, ref. L-040712-00-0010n), Paral1/1-siRNA (Silencer Select ref. 101240, Ambion) or Paral1/2-siRNA (Silencer Select ref. 101167, Ambion).
Transient transfections and reporter gene assay
HEK293T cells (7.5 × 104) were plated 1 day prior to transfection in 6-well plates. Plasmids were transfected using jetPEI according to the manufacturer’s instructions. A combination of the following vectors was used as indicated in the figure legends: pUAS-tk-Luc (1 µg), pGL4-3PPRE (1 µg), Gal4-Pparγ (100ng), Gal4-PGC1α (100ng), pcDNA3-Myc-Rbm14 (250ng), pcDNA3-Paral1 (1 µg), pcDNA3-Flag-Pparγ (500 pg) and pSG5-RXRα Rxrα (500 pg). The next day, HEK293T cells were trypsinized and transferred into 96-wells plates. Twenty four hours later, cells were incubated with either vehicle (DMSO) or indicated compounds [Rosiglitazone (2 µM), GW4064 (1 µM)] overnight. Cells were lysed and luciferase activities were measured as described53.
Retroviral infection and clone selection
Phoenix cells (106) were transfected using Lipofectamine 2000 (Life Technologies) and indicated plasmids according to the manufacturer’s instructions. Transfected plasmids were the pRetroX-Tight-Pur plasmids (pRetroX-empty, pRetroX-Pparγ or pRetrox-Paral1) and the pRetroX-Tet-On vector. After a 5-hour incubation in serum-free medium, cells were washed twice with 1x PBS and grown in complete medium supplemented with 10% FBS. Retroviral supernatants were collected and stored at −80 °C until use.
Stable cell line production: 3T3-L1 cells (2.5 × 104) were plated into 12-wells plates and transduced overnight with the above retroviral supernatants (500 µL pRetroX-Tet-On supernatant and 500 µL pRetroX-Tight-Pur). Clones were selected with 10 µg/mL puromycin and 800 µg/mL G418 for 7-10 days. Doxycycline (5 µg/mL) was used as the Tet-ON system inducer.
Electrophoresis, gel staining and mass spectrometry
Protein separation, in-gel trypsin cleavage and mass spectrometry analysis were carried out as described58. Peptide separation was performed using an EASY-nLC 1000 UHPLC (Thermo Scientific) equipped with a 75 µmX 2 cm Acclaim PepMap 100 pre-column with nanoViper fittings and a 50 µm I.D. × 500mm Acclaim PepMap RSLC analytical column (Thermo Scientific). Peptides were eluted using a 5%-30% acetonitrile gradient for 60 min. at a flow rate of 300 nL/min. The Q-Exactive instrument acquisition mode was set to the top 10 MS2 method. The survey scans were taken at 70,000FWHM (at m/z 400) resolving power in positive mode and using a target of 1e6 and default charge state of + 2. Unassigned and + 1 charge states were rejected, and dynamic exclusion was enabled for 30 sec. The scan range was set to m/z 300–1600 m/z. For MS/MS, a microscan was obtained at 17,500FWHM and with an isolation window of 3.0 m/z, using a scan range between m/z 200–2000 m/z. Tandem mass spectra were processed with the Thermo Scientific Proteome Discoverer software v 1.3. Spectra were searched against UniprotKB/Swiss-Prot mouse databases (version 09/2015) using the SEQUEST HT algorithm (v1.3.0.339). The search was performed choosing trypsin as the cleaving enzyme with one missed cleavage site allowed. Precursor mass tolerance was 10 ppm, and fragment mass tolerance was 0.1 Da. N-terminal acetylation, cysteine carbamidomethylation and methionine oxidation were set as variable modifications. Peptide identification was performed with the Percolator algorithm by selecting only peptides with a q-value < 0.01, which corresponds to a false discovery rate (FDR) of 1%.
Animal experimentation
All animal experiments were approved by the ethical committee for animal experimentation of Institut Pasteur de Lille, Pierre and Marie Curie University and French Research Council guidelines. All methods were performed in accordance with the relevant French and European guidelines and regulations. Detailed procedures can be found elsewhere8,27.
Human biopsies
Human tissue samples were provided by Dr P. Gélé and the Centre d’Investigations Cliniques (C.H.R.U. Lille, France). Human WAT samples were collected from patients undergoing abdominal surgery by laparoscopy or coelioscopy after informed consent was obtained. All procedures were approved by the C.H.R.U. Lille Ethical committee and were compliant to the French National Ethics Committee guidelines. Tissue samples from female patients (35–59 year-old) and are from the ABOS cohort (ClinicalTrials.gov identifier: NCT01129297). More details can be found elsewhere26.
Statistical analysis
Statistical analysis was performed using Prism 6.0 (GraphPad Software, La Jolla, CA). Values are expressed as the mean +/− SEM. Statistical significance was evaluated using either a two-tailed t test or by one-way ANOVA followed by Tukey’s or Dunnett’s post hoc tests. *p < 0.05; **p < 0.01; ***p < 0.001.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Affymetrix raw files are available on the GEO web site.
References
Wang, Q. A., Tao, C., Gupta, R. K. & Scherer, P. E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med 19, 1338–1344 (2013).
Pellegrinelli, V., Carobbio, S. & Vidal-Puig, A. Adipose tissue plasticity: how fat depots respond differently to pathophysiological cues. Diabetologia 59, 1075–1088, https://doi.org/10.1007/s00125-016-3933-4 (2016).
Virtue, S. & Vidal-Puig, A. Adipose tissue expandability, lipotoxicity and the Metabolic Syndrome–an allostatic perspective. Biochim. Biophys. Acta 1801, 338–349 (2010).
Nishimura, S. et al. In vivo imaging in mice reveals local cell dynamics and inflammation in obese adipose tissue. J Clin Invest 118, 710–721, https://doi.org/10.1172/JCI33328 (2008).
Sun, K., Kusminski, C. M. & Scherer, P. E. Adipose tissue remodeling and obesity. J Clin Invest 121, 2094–2101, https://doi.org/10.1172/JCI45887 (2011).
Salans, L. B., Knittle, J. L. & Hirsch, J. The role of adipose cell size and adipose tissue insulin sensitivity in the carbohydrate intolerance of human obesity. J. Clin. Invest 47, 153–165 (1968).
Lee, M. J., Wu, Y. & Fried, S. K. Adipose tissue remodeling in pathophysiology of obesity. Curr. Opin. Clin. Nutr. Metab Care 13, 371–376 (2010).
Oger, F. et al. Peroxisome proliferator-activated receptor gamma regulates genes involved in insulin/insulin-like growth factor signaling and lipid metabolism during adipogenesis through functionally distinct enhancer classes. J Biol Chem 289, 708–722, https://doi.org/10.1074/jbc.M113.526996 (2014).
Tang, Q. Q. & Lane, M. D. Adipogenesis: from stem cell to adipocyte. Annu Rev Biochem 81, 715–736, https://doi.org/10.1146/annurev-biochem-052110-115718 (2012).
Siersbaek, R. et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep 7, 1443–1455, https://doi.org/10.1016/j.celrep.2014.04.042 (2014).
Dubois-Chevalier, J. et al. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res 42, 10943–10959, https://doi.org/10.1093/nar/gku780 (2014).
Serandour, A. A. et al. Dynamic hydroxymethylation of DNA marks differentiation-driven enhancers. Nucl. Acid Res 40, 8255–8265 (2012).
Fatica, A. & Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15, 7–21, https://doi.org/10.1038/nrg3606 (2014).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81, 145–166, https://doi.org/10.1146/annurev-biochem-051410-092902 (2012).
Hacisuleyman, E. et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21, 198–206, https://doi.org/10.1038/nsmb.2764 (2014).
Clemson, C. M. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33, 717–726, https://doi.org/10.1016/j.molcel.2009.01.026 (2009).
Mele, M. & Rinn, J. L. “Cat’s Cradling” the 3D Genome by the Act of LncRNA Transcription. Mol Cell 62, 657–664, https://doi.org/10.1016/j.molcel.2016.05.011 (2016).
Cooper, D. R. et al. Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARgamma2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes (Basel) 5, 1050–1063, https://doi.org/10.3390/genes5041050 (2014).
Xiao, T. et al. Long Noncoding RNA ADINR Regulates Adipogenesis by Transcriptionally Activating C/EBPalpha. Stem Cell Reports 5, 856–865, https://doi.org/10.1016/j.stemcr.2015.09.007 (2015).
Chen, J. et al. The role and possible mechanism of lncRNA U90926 in modulating 3T3-L1 preadipocyte differentiation. Int J Obes (Lond) 41, 299–308, https://doi.org/10.1038/ijo.2016.189 (2017).
Mikkelsen, T. S. et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell 143, 156–169, https://doi.org/10.1016/j.cell.2010.09.006 (2010).
Chen, L. L. L. L. & Noncoding, R. N. A. Localization and Function. Trends Biochem Sci 41, 761–772, https://doi.org/10.1016/j.tibs.2016.07.003 (2016).
Bond, C. S. & Fox, A. H. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 186, 637–644, https://doi.org/10.1083/jcb.200906113 (2009).
Iwasaki, T., Chin, W. W. & Ko, L. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J Biol Chem 276, 33375–33383, https://doi.org/10.1074/jbc.M101517200 (2001).
Auboeuf, D. et al. CoAA, a Nuclear Receptor Coactivator Protein at the Interface of Transcriptional Coactivation and RNA Splicing. Molecular and Cellular Biology 24, 442–453 (2004).
Lefebvre, B. et al. Proteasomal degradation of retinoid X receptor alpha reprograms transcriptional activity of PPARgamma in obese mice and humans. J Clin Invest 120, 1454–1468, https://doi.org/10.1172/JCI38606 (2010).
Dalmas, E. et al. Irf5 deficiency in macrophages promotes beneficial adipose tissue expansion and insulin sensitivity during obesity. Nat Med 21, 610–618, https://doi.org/10.1038/nm.3829 (2015).
Hon, C. C. et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. https://doi.org/10.1038/nature21374 (2017).
Eeckhoute, J., Oger, F., Staels, B. & Lefebvre, P. Coordinated Regulation of PPARgamma Expression and Activity through Control of Chromatin Structure in Adipogenesis and Obesity. PPAR Res 2012, 164140, https://doi.org/10.1155/2012/164140 (2012).
Perron, U., Provero, P. & Molineris, I. In silico prediction of lncRNA function using tissue specific and evolutionary conserved expression. BMC Bioinformatics 18, 144, https://doi.org/10.1186/s12859-017-1535-x (2017).
Haakonsson, A. K., Stahl Madsen, M., Nielsen, R., Sandelin, A. & Mandrup, S. Acute genome-wide effects of rosiglitazone on PPARgamma transcriptional networks in adipocytes. Mol Endocrinol 27, 1536–1549, https://doi.org/10.1210/me.2013-1080 (2013).
Vernochet, C. et al. C/EBPalpha and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor gamma agonists. Mol Cell Biol 29, 4714–4728, https://doi.org/10.1128/MCB.01899-08 (2009).
Auboeuf, D., Honig, A., Berget, S. M. & O’Malley, B. W. Coordinate Regulation of Transcription and Splicing by Steroid Receptor Coregulators. Science 298, 416–419 (2002).
Perani, M. et al. The proto-oncoprotein SYT interacts with SYT-interacting protein/co-activator activator (SIP/CoAA), a human nuclear receptor co-activator with similarity to EWS and TLS/FUS family of proteins. J Biol Chem 280, 42863–42876, https://doi.org/10.1074/jbc.M502963200 (2005).
Hasegawa, Y. et al. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19, 469–476, https://doi.org/10.1016/j.devcel.2010.08.006 (2010).
Li, X., Hoeppner, L. H., Jensen, E. D., Gopalakrishnan, R. & Westendorf, J. J. Co-activator activator (CoAA) prevents the transcriptional activity of Runt domain transcription factors. J Cell Biochem 108, 378–387, https://doi.org/10.1002/jcb.22263 (2009).
Herriges, M. J. et al. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev 28, 1363–1379, https://doi.org/10.1101/gad.238782.114 (2014).
Engreitz, J. M. et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455, https://doi.org/10.1038/nature20149 (2016).
Fox, A. H. & Lamond, A. I. Paraspeckles. Cold Spring Harbor Perspectives in Biology 2, https://doi.org/10.1101/cshperspect.a000687 (2010).
Kim, S. Y. et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun 392, 323–328, https://doi.org/10.1016/j.bbrc.2010.01.012 (2010).
Shiratsuchi, G., Takaoka, K., Ashikawa, T., Hamada, H. & Kitagawa, D. RBM14 prevents assembly of centriolar protein complexes and maintains mitotic spindle integrity. EMBO J 34, 97–114, https://doi.org/10.15252/embj.201488979 (2015).
Wang, J. et al. RNA-binding protein PSPC1 promotes the differentiation-dependent nuclear export of adipocyte RNAs. J Clin Invest 127, 987–1004, https://doi.org/10.1172/JCI89484 (2017).
Matsui, M. & Corey, D. R. Non-coding RNAs as drug targets. Nat Rev Drug Discov 16, 167–179, https://doi.org/10.1038/nrd.2016.117 (2017).
Ding, J. et al. TNF-alpha and IL-1beta inhibit RUNX2 and collagen expression but increase alkaline phosphatase activity and mineralization in human mesenchymal stem cells. Life Sci 84, 499–504, https://doi.org/10.1016/j.lfs.2009.01.013 (2009).
Le Guevel, R. et al. Inactivation of the Nuclear Orphan Receptor COUP-TFII by Small Chemicals. ACS Chem Biol, https://doi.org/10.1021/acschembio.6b00593 (2017).
Zhao, W. et al. Genome-wide expression profiling revealed peripheral effects of cannabinoid receptor 1 inverse agonists in improving insulin sensitivity and metabolic parameters. Mol Pharmacol 78, 350–359, https://doi.org/10.1124/mol.110.064980 (2010).
Mudunuri, U., Che, A., Yi, M. & Stephens, R. M. bioDBnet: the biological database network. Bioinformatics 25, 555–556, https://doi.org/10.1093/bioinformatics/btn654 (2009).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550, doi:0506580102 [pii];10.1073/pnas.0506580102 [doi] (2005).
Roh, H. C. et al. Simultaneous Transcriptional and Epigenomic Profiling from Specific Cell Types within Heterogeneous Tissues In Vivo. Cell Rep 18, 1048–1061, https://doi.org/10.1016/j.celrep.2016.12.087 (2017).
Lo, K. A. et al. Analysis of in vitro insulin-resistance models and their physiological relevance to in vivo diet-induced adipose insulin resistance. Cell Rep 5, 259–270, https://doi.org/10.1016/j.celrep.2013.08.039 (2013).
Kong, L. et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35, W345–349, https://doi.org/10.1093/nar/gkm391 (2007).
Wang, L. et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41, e74, https://doi.org/10.1093/nar/gkt006 (2013).
Caron, S. et al. Farnesoid X receptor inhibits the transcriptional activity of carbohydrate response element binding protein in human hepatocytes. Mol. Cell Biol 33, 2202–2211, MCB.01004-12 [pii];10.1128/MCB.01004-12 (2013).
Verreman, K. et al. The coactivator activator CoAA regulates PEA3 group member transcriptional activity. Biochem J 439, 469–477, https://doi.org/10.1042/BJ20110728 (2011).
Depoix, C., Delmotte, M. H., Formstecher, P. & Lefebvre, P. Control of retinoic acid receptor heterodimerization by ligand-induced structural transitions. a novel mechanism of action for retinoid antagonists. J. Biol. Chem 276, 9452–9459 (2001).
Hauser, S. et al. Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J. Biol. Chem 275, 18527–18533 (2000).
Fan, M. et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev 18, 278–289, https://doi.org/10.1101/gad.1152204 (2004).
Sacchetti, P., Carpentier, R., Segard, P., Olive-Cren, C. & Lefebvre, P. Multiple signaling pathways regulate the transcriptional activity of the orphan nuclear receptor NURR1. Nucleic Acids Res 34, 5515–5527, https://doi.org/10.1093/nar/gkl712 (2006).
Kuhn, R. M. et al. The UCSC genome browser database: update 2007. Nucleic Acids Res 35, D668–673, https://doi.org/10.1093/nar/gkl928 (2007).
Acknowledgements
We thank E. Bouchaert and O. Ghali (University of Lille, France) for their advices and help with the isolation and differentiation of MSC. We are very grateful to D. Monté (Univ. Lille 1) for providing us with the CoAA expression vectors. The authors also thank the Research Federation FRABio (Univ. Lille, CNRS, FR 3688, FRABio, Biochimie Structurale et Fonctionnelle des Assemblages Biomoléculaires) and the CLIC-Imaging Mass Spectrometry facility of the PRISM laboratory (INSERM U1192, Univ. Lille) for the NanoLC-Orbitrap experiments. We are also grateful to L. Casteilla, X. Maréchal, E. Arnaud, A. Carrière and S. Mandrup for helpful discussion and data sharing. This work has been supported by Fondation pour la Recherche Médicale (Equipe labellisée, DEQ. 20150331724), INSERM, University of Lille, and Agence Nationale pour la Recherche (EGID, ANR-10-LBEX-46). FFF received a PhD fellowship from the French Ministry of Research.
Author information
Authors and Affiliations
Contributions
Conceptualization: P.L., J.E., B.S.; Methodology: F.F., C.G., F.O., A.B., E.W., H.D., F.A., A.S.V.; Formal analysis and investigation: F.F., T.L., N.V., J.E., F.A., J.A., Q.D., B.D., M.P., C.M., P.L.; Writing - original draft preparation: J.E., P.L.; Writing - review and editing: F.F., T.L., N.V., J.E., B.S., P.L.; Funding acquisition: B.S., P.L.; Resources: T.L., N.V., J.D.C., F.P., J.E., P.L.; Supervision: B.S., J.E., P.L.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Firmin, F.F., Oger, F., Gheeraert, C. et al. The RBM14/CoAA-interacting, long intergenic non-coding RNA Paral1 regulates adipogenesis and coactivates the nuclear receptor PPARγ. Sci Rep 7, 14087 (2017). https://doi.org/10.1038/s41598-017-14570-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-14570-y
This article is cited by
-
The landscape of the long non-coding RNAs and circular RNAs of the abdominal fat tissues in the chicken lines divergently selected for fatness
BMC Genomics (2022)
-
Long non-coding RNAs in metabolic disorders: pathogenetic relevance and potential biomarkers and therapeutic targets
Journal of Endocrinological Investigation (2021)
-
Regulation of zebrafish dorsoventral patterning by phase separation of RNA-binding protein Rbm14
Cell Discovery (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.