Abstract
Xpert MTB/RIF (Xpert) is a widely-used test for tuberculosis (TB) and rifampicin-resistance. Second-line drug susceptibility testing (DST), which is recommended by policymakers, typically requires additional specimen collection that delays effective treatment initiation. We examined whether cartridge extract (CE) from used Xpert TB-positive cartridges was, without downstream DNA extraction or purification, suitable for both genotypic DST (MTBDRplus, MTBDRsl), which may permit patients to rapidly receive a XDR-TB diagnosis from a single specimen, and spoligotyping, which could facilitate routine genotyping. To determine the limit-of-detection and diagnostic accuracy, CEs from dilution series of drug-susceptible and -resistant bacilli were tested (MTBDRplus, MTBDRsl). Xpert TB-positive patient sputa CEs (n = 85) were tested (56 Xpert-rifampicin-susceptible, MTBDRplus and MTBDRsl; 29 Xpert-rifampicin-resistant, MTBDRsl). Spoligotyping was done on CEs from dilution series and patient sputa (n = 10). MTBDRplus had high non-valid result rates. MTBDRsl on CEs from dilutions ≥103CFU/ml (CT ≤ 24, >“low” Xpert semiquantitation category) was accurate, had low indeterminate rates and, on CE from sputa, highly concordant with MTBDRsl isolate results. CE spoligotyping results from dilutions ≥103CFU/ml and sputa were correct. MTBDRsl and spoligotyping on CE are thus highly feasible. These findings reduce the need for additional specimen collection and culture, for which capacity is limited in high-burden countries, and have implications for diagnostic laboratories and TB molecular epidemiology.
Similar content being viewed by others
Introduction
Of the 10.4 million individuals with active tuberculosis (TB) in 2015, 580 000 were rifampicin (RIF) resistant or multidrug-resistant (MDR), defined as resistance to isoniazid (INH) and RIF1. Only ~20% of MDR-TB cases were diagnosed and started on treatment, and only half started on treatment were cured1. Extensively drug-resistant (XDR)-TB, which is MDR with resistance to a fluoroquinolone (FQ) and a second-line injectable drug (SLID) comprises 10% of MDR-TB cases, yet is even more underdiagnosed than MDR-TB, very costly to treat, and represents an emerging public health emergency2,3,4,5,6.
Xpert MTB/RIF (Xpert) (Cepheid, United States) is a Food and Drug Administration and World Health Organization (WHO)-endorsed nucleic acid amplification test (NAAT) that rapidly detects Mycobacterium tuberculosis complex-DNA and RIF-resistance directly from sputa7,8,9. Over 25 million Xpert MTB/RIF cartridges have been consumed and over 30 000 test modules are installed worldwide10. The WHO and several national programmes recommend that if Xpert detects resistance, an additional sputum is collected for further drug susceptibility testing (DST) using line probe assays (LPAs), such as MTBDRplus (RIF and INH) and MTBDRsl (FQs and SLIDs), or phenotypic testing1,9,11,12.
Patients, however, often do not rapidly return to the clinic to give another sputum or receive DST results. For example, a study in South Africa found that, even after MTBDRplus roll-out, time-to-treatment since initial diagnosis was unacceptably long (~55 days), and that this was partly due to challenges with patient loss-to-follow-up13. Furthermore, many patients do not produce sufficient sputum of adequate quality, especially in settings with high rates of HIV14,15,16,17.
MTBDRplus and MTBDRsl have suboptimal sensitivity on specimens, and culture is often required prior to DNA extraction and further genotypic testing. Not only can this cause diagnostic delay, but many high burden countries lack the necessary biosafety and laboratory infrastructure for mycobacterial culture and DNA extraction18,19,20,21. Furthermore, culture can result in the loss of potentially clinically-meaningful resistance22. There is hence a need to reduce delays in the diagnosis of drug-resistant TB and use rapid methods that minimise reliance on culture through the direct testing of specimens23.
Poor adherence to diagnostic algorithms using MTBDRplus and MTBDRsl has been reported5,24,25. For example, in South Africa, 34% of Xpert RIF-resistant patients failed to receive MTBDRplus and, of those confirmed to have MDR-TB, 28% did not receive second-line DST with MTBDRsl – despite both LPAs being mandated by the national programme21. Novel approaches to reduce this gap in the TB care cascade, which is worsened by the requirement for extra patient visits and additional specimen collection, is a major research priority26,27. If TB-testing and first- and second-line DST were possible on the first available specimen, fewer patients would potentially be lost and patients could be diagnosed earlier. This could result in earlier effective treatment initiation, fewer patient- and health systems-costs, and better long-term clinical outcomes.
We therefore conducted a proof-of-concept evaluation on whether M. tuberculosis-complex genomic DNA in the PCR-reaction mix from used Xpert cartridges (cartridge extract; CE) - that would otherwise be discarded - was detectable in an accurate manner using MTBDRplus and MTBDRsl. The feasibility of genotyping on CE by spoligotyping was also tested as this would potentially be useful for research laboratories and programmes seeking to implement routine strain surveillance. We explored the feasibility of Sanger sequencing on CE, as this may be useful for additional genotypic DST. Critically, we evaluated CE for all tests without additional downstream DNA extraction or purification, as not only would extraction require equipment not readily available in routine diagnostic laboratories in high burden settings, but it would complicate laboratory workflows and reduce the attractiveness of our approach. If the CE approach was feasible, it would mean that many laboratories would already have instrumentation available for mycobacterial genomic DNA extraction in the form of GeneXpert10 and not need to procure new equipment.
Material and Methods
Ethics statement
Methods and protocols were carried out in accordance with relevant guidelines and regulations. The study was approved by the Health Research Ethics Committee of Stellenbosch University (N09/11/296) and the City of Cape Town (#10570). Permission was granted to use anonymised residual specimens collected as part of routine diagnostic practice and thus informed consent was waived.
Xpert MTB/RIF on dilution series of drug-susceptible- and drug-resistant bacilli
A triplicate tenfold dilution series was made using phenotypically-confirmed drug-susceptible (DS)-TB, MDR-TB and XDR-TB clinical isolates (0–106 CFU/ml) in phosphate buffer (33 mM Na2HPO4, 33 mM KH2PO4; pH 6.8) with 0.025% Tween80 (Sigma-Aldrich, United States). Colony counts were done by plating on 7H11 Middlebrook agar (BD Biosciences, United States). Dilutions containing bacilli (1 ml aliquots) were tested by Xpert (54 in total: six dilutions ranging from 101–106 CFU/ml in triplicate for three strains and hence 18 dilutions each for the DS, MDR, and XDR strains) as well as 0 CFU/ml controls in triplicate, according to the manufacturer’s instructions9. Used cartridges were stored at 4 °C prior to CE extraction within 24 h and freezing of the CE at −20 °C.
Xpert MTB/RIF on clinical specimens
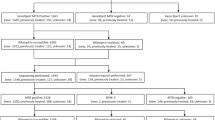
Used Xpert-TB-positive cartridges done on sputa from people with symptoms suggestive of TB tested as part of the South African national TB diagnostic algorithm were collected between February 2016 and November 2016 from the National Health Laboratory Services (NHLS), a South African National Accreditation System-accredited, quality-assured diagnostics laboratory in Cape Town, South Africa11. Cartridges were stored at 4 °C prior to CE extraction within 5 days. Fifty-six Xpert TB-positive, RIF-susceptible cartridges and 29 Xpert-TB-positve RIF-resistant cartridges were collected. When the NHLS did a MGIT 960 liquid culture on sputum from RIF-resistant patients, we collected the isolate [20/29 (69%) had available isolates]. Isolates were not available from Xpert TB-positive, RIF-susceptible specimens as culture is not routinely done in these patients11,28.
Recovery of mycobacterial genomic DNA from used Xpert MTB/RIF cartridges
The transparent diamond-shaped reaction chamber on the back of the cartridge was punctured with a sterile fixed-needle insulin syringe (1 ml; 29 G) (Fig. 1) in a biosafety level 2 cabinet. The full CE volume, typically ~15 µl, was withdrawn and stored in sterile, safe-lock micro-centrifuge tubes at −20 °C prior to analysis. Each cartridge and the surrounding surface was wiped down thoroughly with 1% sodium hypochlorite and 70% EtOH before and after extraction and UV sterilization was done after each batch of extraction. Used needles were discarded in a sharps container containing 1% sodium hypochlorite. Before and after each cartridge extraction session, hood surface area was decontaminated with sodium hypochlorite and EtOH and UV sterilised. No DNA extraction or purification steps were done on CE.
Cartridge extract extraction procedure. (a) The arrow indicates the diamond-shaped reaction chamber where the PCR amplification takes place and contains cartridge extract with mycobacterial genomic DNA. The needle is placed at the top of the diamond and the film is slowly and carefully pierced. (b) The needle is then slowly inserted deeper into the pocket and cartridge extract mix drawn out without piercing the other side.
Line probe assays on cartridge extract
MTBDRplus and MTBDRsl (both version 2.0) were done according to the manufacturer’s instructions29,30 except for Xpert TB-positive, RIF-susceptible clinical specimens CE (n = 56), 7.5 µl CE was used as imput volume into MTBDRplus and MTBDRsl. For the Xpert TB-positive, RIF-resistant clinical specimen CEs (n = 29) and the dilution series, 5 µl (the recommended imput volume) CE was used in order to have enough CE remaining for Sanger sequencing. MTBDRplus and MTBDRsl results were reported as susceptible or resistant (RIF and INH for MTBDRplus; FQ and SLID for MTBDRsl), indeterminate [M. tuberculosis complex DNA-positive (reported by the test as TUB-positive) but no gene loci control bands] or TUB-band negative. LPA strips were read by two independent, experienced readers blinded to each other’s calls and Xpert results (and, for dilution series, the strain used).
Spoligotyping on cartridge extract
Spoligotyping was done as described31,32 on 2 µl CE from the MDR-TB dilution series. A set of Xpert TB-positive, RIF-susceptible cartridges (n = 10) done on specimens and separate from those used for genotypic DST on CE were collected with paired culture isolates from an ongoing research study. To determine whether the correct spoligotype was obtained from CE, crude DNA extracted through heat inactivation from the corresponding culture isolates was spoligotyped. SITVIT was used to identify strain families33.
Targeted Sanger sequencing on cartridge extract
For dilution series, PCR clean-up and Sanger sequencing on 5 µl CE was done by the Stellenbosch University Central Analytical Facility using primers overlapping with LPA-binding sites (Supplementary Table 1). The gyrA and rrs regions in the DS-TB and XDR-TB strains were sequenced.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Results
Patient characteristics
A summary of the patient demographic and clinical data is in Table 1. For Xpert TB-positive, RIF-susceptible patients the median age (IQR) was 40 (31–49) years and for RIF-resistant specimens was 35 (23–42) years. 37/55 (67%) of RIF-susceptible patients and 12/20 (60%) of RIF-resistant patients were male.
Feasibility and diagnostic accuracy of MTBDRplus and MTBDRsl on dilution series Xpert TB-positive cartridge extract
Xpert detected M. tuberculosis-complex DNA in all dilutions ≥102 CFU/ml and correctly identified RIF-susceptibility and -resistance (Fig. 2). MTBDRplus showed poor overall sensitivity for M. tuberculosis-complex DNA [22% (12/54) TUB-band-positive] in CE from Xpert TB-positive cartridges. MTBDRplus had high rates of non-actionable (TUB-band negative or TUB-band positive but indeterminate) and false RIF-heteroresistant results (Figs 2 and 3).
Results of MTBDRplus and MTBDRsl on Xpert CE from a dilution series of DS-, MDR- and XDR-TB strains. MTBDRplus (irrespective of concentration and strain) had high TUB-band negativity and indeterminate rates. However, MTBDRsl had high sensitivity and specificity and low indeterminate rates. For each dilution, left bars are for rifampicin (MTBDRplus, top row) or fluoroquinolones (MTBDRsl, bottom row) and right bars are for isoniazid (MTBDRplus) or second-line injectables (MTBDRsl). Data from LPA on DS-TB, MDR-TB and XDR-TB strains are shown. The experiment was done in triplicate. Abbreviations: CFU – colony forming; DS-TB – drug susceptible TB; MDR-TB – multidrug resistant TB; XDR-TB – extensively drug resistant TB; units; Xpert - Xpert MTB/RIF.
Xpert MTB/RIF quantitative information [average cycle threshold (CT) values] (line graph, right y-axes) versus bacterial load (CFU/ml) in a triplicate dilution series for MTBDRplus (a) and MTBDRsl (b) done on CE. Left y-axes (bars) show the proportion of assays with non-valid results, disaggregated into non-actionable (TUB-band negative, indeterminate) and non-valid (false-susceptible, false-resistant). For each dilution, left bars are for rifampicin (MTBDRplus, top) or fluoroquinolones (MTBDRsl, bottom) and right bars are for isoniazid (MTBDRplus) or second-line injectables (MTBDRsl). Beyond 103 CFU/ml, there were no false resistance or susceptibility calls for MTBDRsl, which corresponds to CT ≤ 24. CT ≥ 38 (horizontal dashed line) correspond to a negative Xpert. Error bars show standard error (SE) of average CT. Right y-axes show CT corresponding to Xpert semiquantitation levels of very low (CT > 28), low (CT = 22–28), medium (CT = 16–22) and high (CT < 16). Pooled data from LPAs on DS-TB, MDR-TB and XDR-TB strains are shown. Abbreviations: CFU – colony forming; DS-TB – drug susceptible TB; MDR-TB – multidrug resistant TB; XDR-TB – extensively drug resistant TB; CFU – colony forming units; Xpert - Xpert MTB/RIF.
In contrast, MTBDRsl on CE had high sensitivity and specificity [87% (47/54) and 100% (9/9) respectively] for M. tuberculosis-complex DNA and a limit of detection of 103 CFU/ml. Susceptibility and resistance to FQs and SLIDs were correctly detected for all strains ≥103 CFU/ml, corresponding to CT ≤ 24 (the higher CT range of the Xpert “low” semiquantitation category) in all but one sample (one replicate of the MDR-TB strain was indeterminate for FQs; Fig. 3). Once non-actionable results were excluded, overall sensitivities and specificities of 87% (13/15) and 96% (25/26) for FQ-resistance and 94% (15/16) and 97% (30/31) for SLID-resistance, respectively were obtained. When the threshold of ≥103 CFU/ml (CT ≤ 24) was applied, the sensitivity and specificity were both 100% (12/12 and 23/23, respectively) for FQs and for SLIDs (12/12 and 24/24, respectively).
Diagnostic accuracy of MTBDRplus and MTBDRsl on clinical specimen Xpert TB-positve cartridge extract
Xpert MTB/RIF rifampicin-susceptible specimens
As with the dilution series, MTBDRplus had high rates of indeterminate and false-resistance results on clinical specimen CE (Table 2). However, most MTBDRsl results from Xpert TB-positive, RIF-susceptible clinical specimen CE were valid (TUB-positive, not indeterminate, and no false-susceptible or -resistant results): 53/56 (95%) for FQ (two TUB-band negative, one indeterminate) and 51/56 (91%) for SLID (two TUB-band negative, three indeterminate). The few CEs that yielded indeterminate MTBDRsl results corresponded to “low” or “very low” Xpert semiquantitation levels (CT > 24). The median (IQR) CT of indeterminate (26.3, 24.4–26.7) vs. determinate (17.62, 15.6–20.6) MTBDRsl results differed (p < 0.001), indicating that indeterminate results are likely a function of low DNA concentrations in CE. There was not enough CE volume remaining or a matching clinical isolate for confirmatory testing from the three MTBDRsl-detected SLIDs resistant patients.
Xpert MTB/RIF rifampicin-resistant specimens
MTBDRsl on Xpert TB-positive, RIF-resistant CE had 24/29 (83%) valid results. For FQs, 14/24 (58%) were susceptible and 10/24 (42%) were resistant. For SLIDs, 15/24 (63%) were susceptible and 9/24 (37%) resistant. The five non-valid results were TUB-band-negative [2/29 (7%)] or indeterminate for both FQs and SLIDs [3/29 (10%); Table 2]. All CEs corresponding to the higher CT ranges of the Xpert “low” semiquantitation category (CT ≤ 24) had valid results, whereas those that had indeterminate or TUB band-negative results corresponded to the lower semiquantitation levels (CT > 25.0). The median (IQR) CT of indeterminate (29.1, 26.5-31.1) vs. determinate (20.5, 16.−23.2) results differed significantly (p < 0.001).
MTBDRplus and MTBDRsl performance on Xpert MTB/RIF cartridge extract by smear status
MTBDRplus had high non-valid result rates irrespective of smear status. However, MTBDRsl on CE from smear-negative sputums had significantly higher rates of non-actionable results [5/23 (22%) vs. 1/43 (2%) for FQ, p = 0.01; 6/23 (23%) vs. 2/43 (5%) for SLIDs, p = 0.01] compared to smear-positive patients (Supplementary Table 2).
Concordance of MTBDRsl results on cartridge extract and culture isolates
Of the 29 Xpert TB-positive, RIF-resistant patients, 20 (69%) matched culture isolates were collected while the remaining nine had negative or contaminated cultures. The CEs and isolates showed 18/20 (90%) matching MTBDRsl FQ results and 17/20 (84%) matching SLID results. There were 2/20 (10%) discordant TUB-band MTBDRsl results on culture isolates (one TUB-positive and FQ and SLID sensitive, one TUB-positive and FQ and SLID resistant) where both CE results were TUB-band negative. There was also 1/20 (5%) discordant SLID result (CE showed resistance but the isolate showed susceptibility). Importantly, all three discordant results corresponded to a “very low” semiquantitation (CT > 28.0). All TUB-band, susceptibility and resistance calls were concordant at CT ≤ 24, indicating that the diagnostic accuracy of MTBDRsl on CE vs. isolates is likely comparable at this threshold.
Spoligotyping on cartridge extract
Dilution series
Spoligotyping resulted in a readable strain type for dilutions ≥103 CFU/ml, corresponding to the same threshold seen for MTBDRsl.
Clinical specimens
Spoligotyping on specimen CE and crude DNA from matched culture isolates were highly concordant 10/10 (100%) at the threshold defined by the dilution series (Table 3). A variety of strain families were observed with Beijing as the predominant family type [6/10 (60%)] as well as 2/10 (20%) LAM and 2/10 (20%) T1 family type.
Targeted sequencing on extract from used Xpert MTB/RIF cartridges
Dilution Series
Targeted Sanger sequencing was done on dilution series CE. For the rrs PCR on CE, sequence shorter than the expected length was observed. PCR of gyrA from CE from dilutions 103–104 CFU/ml resulted in sequence expected length, however high background noise occurred and the sequence did not align to H37Rv [NC_000962]. gyrA on CE from dilutions 105–106 CFU/ml aligned to the reference genome, however, several SNPs known to be present in the resistance determining regions (identified by sequencing of the corresponding isolate) were not detected. Due to the relatively poor limit of detection and accuracy of Sanger sequencing on dilution series CE, we did not do sequencing on clinical specimen CEs.
Discussion
Our key findings are: (1) MTBDRsl on CE enabled genotypic drug-susceptibility testing for FQs and SLIDs with high accuracy and low indeterminate rates when the Xpert semiquantitation category was at least “medium” or CT ≤ 24 (corresponding to ≥ 103 CFU/ml), (2) spoligotyping was feasible and accurate on CE at the same threshold, (3) MTBDRplus was not feasible or accurate on CE and (4) neither was Sanger sequencing. These data have implications for the routine diagnosis of drug-resistant TB, researchers, and test developers.
Xpert is one of the most widely used tests for TB and drug-resistance9,34 and although it is a significant advancement, time-to-treatment – especially for MDR- and XDR-TB - is still very long35,36,37,38. Our results show that accurate second-line drug testing using MTBDRsl is possible on CE from Xpert cartridges that would otherwise be discarded. This potentially allows for a rapid, single-specimen diagnosis of XDR-TB without additional specimen collection. Importantly, we defined a threshold at which this approach is feasible, meaning that MTBDRsl assays do not need to be wasted on CE unlikely to give a valid result. Using this threshold, we showed that on clinical specimen CEs, susceptibility and resistance calls were concordant with those from the isolate19,39. Furthermore, we showed that it is possible to do spoligotyping on CE at this threshold, which will inform strain surveillance and research studies on relapse and reinfection where specimens are limited. Collectively, these findings may reduce the need for culture.
Although our data suggest that the MTBDRsl will work on CE from cartridges with an Xpert semiquantitation category of at least “low”, we suggest that, in laboratories where CT cannot be readily calculated, a category of at least “medium” is used to guide use of this strategy unless the laboratory is comfortable with some semiquantitation low specimens not having a valid MTBDRsl result. Alternatively, if smear microscopy is available, smear-positivity may be used to guide use of CE, however, some smear-negative specimens in whom this approach would work (103-104 CFU/ml) would be unnecessarily excluded.
When considering the CE approach, it is important to identify a safe and sterile environment to avoid contamination. Although Xpert sample reagent as well as the sonication lysis step within the cartridge helps ensure M. tuberculosis is no longer culturable (and therefore poses minimal infectious risk40), steps to minimise the risk of rpoB amplicon cross-contamination should be implemented. These can include working in a dedicated cabinet or room and sterilising the work area with UV and disinfectant after CE is collected. Importantly, however, cross-contamination of other Xpert cartridges with rpoB amplicons appears unlikely. Although Xpert’s automated pre-amplification wash step does not remove large pieces of debris-associated genomic DNA, it does efficiently remove high concentrations of contaminating rpoB amplicons from assays like MTBDRplus 41,42. NAATs without such a wash step may be more vulnerable to CE rpoB amplicon cross-contamination.
Our study differed from a previous study which showed that sequencing, MTBDRplus, spoligotyping and MIRU-VNTR typing are feasible on the sputum mixed with Xpert sample reagent43. However, this sample reagent method has a number of disadvantages: 1) often no volume remains, 2) prolonged exposure to sample reagent degrades DNA and potentially introduces mutations9,40, and 3) it still requires DNA extraction prior to PCR. Furthermore, DNA extraction adds cost and is not always feasible in laboratories in high burden countries; whereas the CE method yields directly usable material and does not need additional extraction or purification steps. An advantage, however, of using the sputum mixed with Xpert reagent buffer, is that it likely avoids high MTBDRplus error rates (TUB-band negative, indeterminate, false-positive) seen with CE. This could be due to the large amount of rpoB amplicons in Xpert TB-positive CE, which share binding sites with MTBDRplus probes and confound the assay resulting in non-valid results. Furthermore, the rpoB PCR that occurs as part of MTBDRplus may sequester reagents away from the multiplex inhA and katG amplification reactions. Testing for mutations conferring INH resistance using CE might hence be possible with the Genoscholar INH II line probe assay (which does not contain rpoB probes)44. Sequencing from CE thus primarily appears to be driven by rpoB amplicon interference (although a PCR clean-up was done prior to sequencing, this would have co-purified rpoB amplicons). Further investigation with primers optimised for minimal-input DNA may be warranted, however, it appears that, for sequencing, the best approach to avoid contaminating amplicons might be to PCR from the specimen-Xpert sample reagent mixture43. Given the rates of non-valid CE results below the defined threshold, we suggest that specimen-Xpert sample reagent mix be kept in the event that CT falls >24.
The results presented here should be interpreted in context of their limitations. For the clinical specimens tested from the NHLS, matched culture isolates were not available for Xpert RIF-susceptible specimens, as per the national algorithm. However, the dilution series experiments showed very high concordance between MTBDRsl on CE vs. the isolates. The utility of CE depends on the downstream test used and MTBDRsl susceptible or non-valid results should be interpreted from CE the same as when they are done on patient specimens (i.e., further investigation, including culture, is recommended)45. Realistically, cartridges may need to be transported from remote locations and so the effect of storing cartridges for prolonged duration (>5 days) and at ambient temperature requires further systematic testing. Using bacilli in buffer can have limitations, which is why we also used patient clinical specimens, which are a better material to test than bacilli added to sputum (the former has bacilli within a sputum matrix, whereas in the latter bacilli are typically freely floating in bubbles).
In conclusion, CE contains template DNA for second-line DST using MTBDRsl, resulting in accurate results highly concordant with those from isolates, provided bacillary load in the specimen corresponds to at least a “medium” Xpert semiquantitation category of CT ≤ 24. This potentially facilitates XDR-TB detection within days from a single specimen. Spoligotyping is also feasible on CE and works consistently at this threshold. Our method provides an opportunity to potentially reduce the burden associated with addition specimen collection, such as patient treatment delay, pre-treatment loss-to-follow-up, and increased patient and provider costs. Furthermore, it shows that material that would otherwise be discarded still holds diagnostic utility.
References
World Health Organization. Global tuberculosis report (2016).
Pooran, A., Pieterson, E., Davids, M., Theron, G. & Dheda, K. What is the cost of diagnosis and management of drug resistant tuberculosis in South Africa? PloS one 8, e54587 (2013).
Cox, H., Ramma, L., Wilkinson, L., Azevedo, V. & Sinanovic, E. Cost per patient of treatment for rifampicin-resistant tuberculosis in a community-based programme in Khayelitsha, South Africa. Tropical medicine & international health: TM & IH 20, 1337–1345, https://doi.org/10.1111/tmi.12544 (2015).
Dheda, K. et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. The lancet Respiratory medicine 5, 291–360 (2017).
Cox, H. et al. Community-based treatment of drug-resistant tuberculosis in Khayelitsha, South Africa. The International Journal of Tuberculosis and Lung Disease 18, 441–448 (2014).
Moyo, S. et al. Loss from treatment for drug resistant tuberculosis: risk factors and patient outcomes in a community-based program in Khayelitsha, South Africa. PloS one 10, e0118919 (2015).
US Food Drug Administration. New data shows test can help physicians remove patients with suspected TB from isolation earlier (2015).
Boehme, C. C. et al. Rapid molecular detection of tuberculosis and rifampin resistance. New England Journal of Medicine 363, 1005–1015 (2010).
World Health Organization. Xpert MTB/RIF implementation manual: technical and perational ‘how-to’: practical considerations [Internet]. Geneva: World Health Organization (2014).
World Health Organization. Xpert Rollout Status (2016). accessed at: http://www.who.int/tb/areas-of-work/laboratory/mtb-rif-rollout/en/, 19 June 2017.
Health Department of the Republic of South Africa. National Tuberculosis Management Guidelines (2014).
Sachdeva, K. S. Management of Tuberculosis: Indian Guidelines. API Medicine Update, Section 15, 479e483 (2013).
Jacobson, K. R. et al. Implementation of GenoType MTBDR plus Reduces Time to Multidrug-Resistant Tuberculosis Therapy Initiation in South Africa. Clinical infectious diseases 56, 503–508 (2012).
MacPherson, P., Houben, R. M., Glynn, J. R., Corbett, E. L. & Kranzer, K. Pre-treatment loss to follow-up in tuberculosis patients in low-and lower-middle-income countries and high-burden countries: a systematic review and meta-analysis. Bulletin of the World Health Organization 92, 126–138 (2014).
Peter, J. G. et al. Comparison of two methods for acquisition of sputum samples for diagnosis of suspected tuberculosis in smear-negative or sputum-scarce people: a randomised controlled trial. The Lancet Respiratory Medicine 1, 471–478 (2013).
Sharath, B. & Shastri, S. India’s new TB diagnostic algorithm-far from reality? Public health action 6, 206–206 (2016).
Peter, J. G., Theron, G., Singh, N., Singh, A. & Dheda, K. Sputum induction to aid diagnosis of smear-negative or sputum-scarce tuberculosis in adults in HIV-endemic settings. European Respiratory Journal 43, 185–194 (2014).
Tomasicchio, M. et al. The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates. Scientific reports 6 (2016).
Theron, G. et al. The diagnostic accuracy of the GenoType (®) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database of Systematic Reviews 10 (2014).
Nathavitharana, R. R. et al. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis. European Respiratory Journal 49, 1601075 (2017).
Global Laboratory Initiative. Practicial Guide to TB Laboratory Strenghtening (2017).
Metcalfe, J. Z. et al. Mycobacterium tuberculosis subculture results in loss of potentially clinically relevant heteroresistance. Antimicrobial agents and chemotherapy, AAC. 00888–00817 (2017).
Dheda, K. et al. TB drug resistance in high-incidence countries. Tuberculosis 58, 95–110 (2012).
Dlamini-Mvelase, N. R., Werner, L., Phili, R., Cele, L. P. & Mlisana, K. P. Effects of introducing Xpert MTB/RIF test on multi-drug resistant tuberculosis diagnosis in KwaZulu-Natal South Africa. BMC infectious diseases 14, 442 (2014).
Cox, H. et al. Delays and loss to follow-up before treatment of drug-resistant tuberculosis following implementation of Xpert MTB/RIF in South Africa: A retrospective cohort study. PLoS medicine 14, e1002238 (2017).
Engel, N. et al. Compounding diagnostic delays: a qualitative study of point-of-care testing in South Africa. Tropical medicine & international health: TM & IH 20, 493–500, https://doi.org/10.1111/tmi.12450 (2015).
Naidoo, P. et al. Estimation of losses in the tuberculosis care cascade in South Africa and methodological challenges. Under revision.
Truant, J., Brett, W. & Thomas, W. Jr Fluorescence microscopy of tubercle bacilli stained with auramine and rhodamine. Henry Ford Hospital Medical Bulletin 10, 287–296 (1962).
Hain Lifescience. GenoType MTBDRplus VER 2.0 Instructions for Use (2014).
Hain Lifescience. GenoType MTBDRsl VER 2.0 Instructions for Use (2015).
Streicher, E. et al. Spoligotype signatures in the Mycobacterium tuberculosis complex. Journal of clinical microbiology 45, 237–240 (2007).
Kamerbeek, J. et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. Journal of clinical microbiology 35, 907–914 (1997).
Demay, C. et al. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infection, Genetics and Evolution 12, 755–766 (2012).
Lawn, S. D. & Nicol, M. P. Xpert® MTB/RIF assay: development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance. Future microbiology 6, 1067–1082 (2011).
Metcalfe, J. Z. et al. Xpert® MTB/RIF detection of rifampin resistance and time to treatment initiation in Harare, Zimbabwe. The International Journal of Tuberculosis and Lung Disease 20, 882–889 (2016).
Iruedo, J., O’Mahony, D., Mabunda, S., Wright, G. & Cawe, B. The effect of the Xpert MTB/RIF test on the time to MDR-TB treatment initiation in a rural setting: a cohort study in South Africa’s Eastern Cape Province. BMC infectious diseases 17, 91 (2017).
Cox, H. S. et al. Impact of Xpert MTB/RIF for TB diagnosis in a primary care clinic with high TB and HIV prevalence in South Africa: a pragmatic randomised trial. PLoS medicine 11, e1001760 (2014).
van Kampen, S. C. et al. Effects of introducing Xpert MTB/RIF on diagnosis and treatment of drug-resistant tuberculosis patients in Indonesia: a pre-post intervention study. PloS one 10, e0123536 (2015).
Theron, G. et al. GenoType® MTBDRsl assay for resistance to second‐line anti‐tuberculosis drugs. The Cochrane Library (2016).
Banada, P. P. et al. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. Journal of clinical microbiology 48, 3551–3557 (2010).
Theron, G. et al. Xpert MTB/RIF results in patients with previous tuberculosis: can we distinguish true from false positive results? Clinical Infectious Diseases 62, 995–1001 (2016).
Blakemore, R. et al. Evaluation of the analytical performance of the Xpert MTB/RIF assay. Journal of clinical microbiology 48, 2495–2501 (2010).
Alame-Emane, A. K. et al. The use of GeneXpert remnants for drug resistance profiling and molecular epidemiology of tuberculosis in Libreville, Gabon. Journal of clinical microbiology, JCM. 02257–02216 (2017).
Mitarai, S. et al. Comprehensive multicenter evaluation of a new line probe assay kit for identification of Mycobacterium species and detection of drug-resistant Mycobacterium tuberculosis. Journal of clinical microbiology 50, 884–890 (2012).
World Health Organization. The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs: policy guidance (2016).
Acknowledgements
The authors would like to thank Dr Michael Whitfield who provided the strains for the in vitro study. The financial assistance of the National Research Foundation (NRF) towards this research is hereby acknowledged. Opinions expresses and conclusions arrived at, are those of the author(s) and are not necessarily to be attributed to the NRF. Research reported in this publication was supported by the South African Medical Research Council. The content is solely the responsibility of the authors and does not necessarily represent the official views of the South African Medical Research Council. Research reported in this publication was also supported by the Faculty of Medicine and Health Sciences, Stellenbosch University and the European & Developing Countries Clinical Trials Partnership 2.
Author information
Authors and Affiliations
Contributions
G.T., R.W. and M.D.V. conceived the experiments. R.V., B.D., S.P., N.K. and A.R. conducted the experiments. J.S. and T.D. provided specimens and data from the NHLS. R.V. and B.D. analysed the data. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Venter, R., Derendinger, B., de Vos, M. et al. Mycobacterial genomic DNA from used Xpert MTB/RIF cartridges can be utilised for accurate second-line genotypic drug susceptibility testing and spoligotyping. Sci Rep 7, 14854 (2017). https://doi.org/10.1038/s41598-017-14385-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-14385-x
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.