Abstract
The predominant prevalent Mycobacterium tuberculosis (M. tb) lineage was the Beijing genotype family in Beijing. There has been no systematic study on the association between drug resistance and Beijing genotype. Here we collected 268 M. tb strains, analyzed the background information and the bacteriological characteristics. The mean age of the cases was 40.12 years; male patients were almost three times than female patients. After genotyping analyzation, 81.7% (219/268) strains were categorized as Beijing genotype; no significant difference was observed between Beijing and non-Beijing genotype in gender, age and treatment history. Drug susceptibility testing (DST) analyzation demonstrated that 172 (64.2%) strains were fully sensitive to all drugs (Isoniazid, Rifampin, Streptomycin, and Ethambutol), while 96 (35.8%) strains were resistant to at least one of the drugs. Beijing genotype strains exhibited a significantly higher clustering rate. However, no significant association relationship was observed between drug resistance and Beijing genotype family. The study provided insights into the genotype diversity and revealed that the frequencies of drug-resistance of Beijing genotype strains.It would be helpful for the establishment of the efficient tuberculosis (TB) prevention and control strategy in Beijing.
Similar content being viewed by others
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (M. tb) complex, is one of the major threats to the public health around the world and China1. Prevention and control of TB are also confronting with more pressure in Beijing due to the rapid urbanization and economic transition2,3.
The molecular epidemiology methods has been proven playing an important role in TB control and study4,5. Among these, spoligotyping has been considered as a powerful tool to differentiate M. tb complex into various genotypes, especially for identifying Beijing genotype6. Similarly, variable number of tandem repeat (VNTR) genotyping has also been considered as a much better method, as it faster and easier to perform, as well as possesses higher discriminatory power. Both of them can be used friendly and easily compared through generating numerical data. Moreover, the combined application of spoligotyping and VNTR is increasingly common in M. tb molecular epidemiology research7,8.
The molecular typing of M. tb has greatly improved knowledge of TB epidemiology and enabled molecular guided control of the disease. Based on the molecular epidemiology technologies, some studies shown that drug resistance of M. tb was associated with genotype diversity6,9,10,11. However, other studies have not indicated the association relationship between them12,13. It is possible that the results were changed according to the project setting or drug types14. Previous reports15,16,17 and our study18 have demonstrated that Beijing genotype strains were highly prevalent in Beijing, occupied around 82%. However, there has been no systematic study on the association between drug resistance and Beijing genotype. It is quite necessary to clarify the situation in Beijing considering the high tuberculosis prevalence and non-consistent results acquired from different regions and researchers.
In order to clarify the association relationship, a retrospective study was carried out on the genotype diversity and the frequencies of drug-resistance of 268 M. tb strains. We genotyped strains collected from Beijing by spoligotyping and VNTR technology. Additionally, the background information and DST profiles of these cases were also analyzed. The study was conducted to gain insights into the genotype characteristics of M. tb strains and the association with drug resistance. It would be helpful for developing better TB prevention and control strategies in Beijing.
Results
Study population
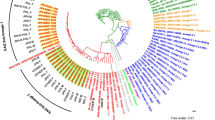
To investigate the characteristics of the study population, we analyzed the basic information of 268 individual strains collected from Beijing in 2008(Fig. 1). Classification of all isolates investigated in this study according to patients’ age and gender were described in Table 1.The enrolled patients included 206 males (76.9%) and 62 females (23.1%), male to female ratio was 3.32; the mean age was 40.12 years old (range from 11 to 89, med 38). Of them, 147 patients were Beijing residents and 121 were migrant population (non-residents) (Table 1). It indicated that the resident population and non-resident population contributed the same to TB prevalence. Additional, we did not find a significant difference between Beijing and non-Beijing genotype strains when considering risk factors such as gender, age, registered residence and treatment history.
The spoligotyping and VNTR genotyping structure of M. tb strains
To clarify the genotype diversity of prevalent strains in Beijing, we analyzed 268 M. tb strains using spoligotyping and VNTR method. Among these strains, 219 (81.7%) were belonged to Beijing genotype families and 49 (18.3%) were belonged to non-Beijing genotype families (see Supplementary Table S1). These results indicated that Beijing genotype strains were the most prevalent strains in Beijing. Of all Beijing-family strains, typical Beijing families (SIT1) accounted for 198 (74.2%), while atypical Beijing families accounted for 21 (7.9%). Among non-Beijing family strains, mainly included T1 (SIT53) family, T2 (SIT52) family, MANU2 (SIT54) family, and H3 (SIT50) family et al. furthermore 16 (5.29%) strains constituted a newly found genotype cluster.
To analyze the above results from clustering angle and demonstrate the genetic linkage map, we made a minimum spanning tree (MST) based on spoligotyping data by the BioNumerics software. MST demonstrated that these strains clustered by a graph. Meanwhile, we calculated the genotypes of the strains using locus variant counts and distances. As shown in see Supplementary Fig. S1, the largest cluster containing 198 strains (SIT1) belonged to the typical Beijing family and was surrounded by small clusters of the atypical Beijing family. Beijing family and non-Beijing family were divided into two main groups. The three larger clusters on the left were the T1, MANU2 family and 16 newfound genotypes surrounded by other genotypes.
The allelic diversity for the 268 strains of each VNTR locus was estimated using the Hunter-Gaston discriminatory index (HGDI) (Table 2). The VNTR 12-locus (BJ) method differentiated the 268 strains into 196 genotypes. A total of 156 strains had unique patterns and the remaining 112 formed 8 clusters (2 to 20 strains per cluster). The 12 VNTR loci exhibited different HGDI scores, ranging from 0.1423 to 0.7255. The cumulative discriminatory power (HGDI) of the total loci set reached 0.998 for Beijing family, HGDI reached 0.999 when applied to all strains. The spoligotyping method differentiated the same strains into 33 genotypes. A total of 28 strains had unique patterns and the remaining 240 formed 40 clusters (2 to 188 strains per cluster).VNTR can separate the Beijing family into different small branches and more less clusters, indicating that it possessed a higher discriminatory power than spoligotyping. The Beijing genotype family strains exhibited a significantly higher clustering rate than the non-Beijing genotype family strains by both spoligotyping and VNTR typing analyzation (Table 3), suggesting transmission of Beijing genotype should be higher than the non-Beijing genotype.
Drug resistance patterns difference and the association with Beijing genotype family
To investigate the profile of drug-resistance of these prevalent strains, drug susceptibility testing (DST) was carried on among the 268 isolated strains. The results showed that 172 isolates (64.2%) were sensitive to the four first-line anti-TB drugs tested and 96 isolates (35.8%) were resistant to at least one of these drugs, including the 44 (16.4%) MDR strains (Table 4).
The total drug resistance rate of the new cases were significantly less than that of the retreatment patients, and the same results were shown in INH, RIF,SM and EMB mono-resistance rate and MDR rate between the two groups (Table 4). In this study, we analyzed the genotypes and the association relationship between drug resistance and Beijing genotype family strains. However, the Beijing genotype and non-Beijing genotype family didn’t show any statistical associations in any drug resistance frequency (Table 4).
Statistic results also showed no significant difference between different genotype when compared in pan-susceptible TB and any drug resistant TB groups (Table 5). These results indicated that there was no statistical association relationship between Beijing genotype strains and the drug resistance of the 4 kinds of first-line anti-tuberculosis drugs.
Discussion
In Beijing, a super megacity, the capital of the People’s Republic of China, TB is a reemerging infectious disease and a substantial public health problem17,19. Beijing genotype family is the most successful lineage in the present tuberculosis epidemic of Beijing. However, there has been no systematic study on the association between drug resistance and Beijing genotype. So, it is very necessary to study the molecular epidemiology and the association with drug-resistance.
Similar with the results of different study in China16, our data demonstrated that Beijing genotype was the most prevalent genotype in Beijing. Based on the Spoligotyping and VNTR data, an interesting finding of this study was the higher clustering rate of Beijing genotype compared with non-Beijing genotype (Table 3). These clustered strains usually suggested the possibility of the recent TB transmission. The result suggested the M. tb isolates within Beijing genotype family demonstrated a relatively high level of transmission in Beijing. Based on the fact, in order to control TB much better in Beijing, these patients carried clustered strains should be given more attention and investigate their social network in the following study.
In this study, we didn’t found any different risk factors between Beijing and non-Beijing genotype strains, even considered clustered and non-clustered patients (see Supplementary Tables S2 and S3). The spoligotyping and VNTR data give us the same result. So, more study and data were needed to precisely identify the nature of transmission between different genotype strains.
Because some new unknown strains were found out in our study, it is important to check out whether they belong to Beijing genotype or non-Beijing genotype. So we analyzed these data using BioNumerics, we got the MST map. According to the MST map, the newfound strains were belonged to the subordinate of non-Beijing genotype. These strains were firstly found and reported in the world, and deeply study should be carried on in the future.
In previous study, genotyping studies were implemented mainly focused on the distribution of M. tb genotypes in Beijing11. Studies on the association relationship between strains genotype and drug resistance were still insufficient in Beijing. After analyzed the frequency of the Beijing genotype among these strains showing different drug resistance, we found the drug-resistant rate of Beijing genotype strains were not higher than that of non-Beijing genotype strains. The result was not consistent with several publications in other regions of the world. Those studies considered that Beijing genotype was associated with drug resistance and those strains showed higher drug resistance level20,21. Meanwhile, other studies also showed no clearly association relationship between drug resistance and strains clade13,22.
Different results obtained from different studies and different regions possibly because of the amount of Beijing genotype sublineages and the percentage of modern or ancient Beijing genotype, which affected the DST results22. In 2006 Mokrousov et al. hypothesized the role of Beijing sublineages that affected DST result which better explained the variability of situations in different countries and districts23. Mathuria JP et al. also concluded that Beijing genotype strains were not so common in north India and these strains were not fully associated with MDR24. Many recent researches also demonstrated the complex DST profiles of Beijing genotype25,26. Secondly, the difference among these patterns might be related to the variation in treatment regimens, compliance to treatment protocols, and varying quality of drugs27. The association relationship between genetic diversity and DST of M. tb is deserved to study intensively in the future.
This study suffered from several limitations. The primary limitation was that the amount of isolated strains was not abundant, which may affect the real association. In order to match the results of genotyping and the results of DST each other, we had to move some samples out. We would furtherly validate the result with larger sample size isolated strains in the future. Secondly, the DST experiments only included the first line anti-TB drugs. The first line anti-TB drugs still were the most common used for initial treatment TB. When this study was carried out, we mainly wanted to clarify the association between them and genotype prevalent. We will investigate all the anti-tuberculosis drugs used in clinical at present in the future. In addition, although 12-locus VNTR typing method was used in this study, but our previous results indicated that this 12-locus VNTR method gives appropriate results and HGDI for genotyping in Beijing, which had almost equivalent discriminatory power to that of the 15-locus or 24-locus VNTR for TB genotyping18, especially when applied to Beijing family strains. So in this study, 12-locus VNTR method was used again.
In summary, these results suggested there was no statistical association relationship between Beijing genotype family strains and the drug resistance of the four first-line anti-tuberculosis drug. This study also offered a correlation picture of drug resistance and strains genotype in Beijing. We will continue to pursue the study of the association relationship between genotype diversity and drug resistance. These work will be helpful to gain a better understanding of the epidemiology of M. tb strains and to improve the TB prevention and control in Beijing.
Methods
Study setting and design
M. tb isolate strains obtained from pulmonary TB patients, whose sputum were culture-positive for M. tb, were delivered to the Beijing Tuberculosis and Thoracic Tumor Research Institute and then were frozen. For any patient with multiple isolates from the culture, only the first isolate was used for performing the bacterial characterization. Six hundred and seventy-nine strains were collected randomly from Beijing from June to December in 2008, which was believed to possess representative and scientific meanings. The strains were recovered from −80 °C stock and were subcultured on solid Lowenstein–Jensen (LJ) medium at 37 °C for three to four weeks. The clinical information of the enrolled patients were retrospectively reviewed and collected, including sex, age, household residence and history of treatment, et al. Informed consent was obtained from all subjects included in the study. The protocols and procedures for the protection of human subjects were approved by the Ethics Committee of Beijing Chest Hospital. Furthermore, all the methods were carried out in accordance with the approved guidelines.
Spacer oligonucleotide typing (spoligotyping)
Spoligotyping was used to identify TB strains genotype according to DR locus as described previously28. Typical Beijing genotype strains were defined with the pattern that hybridized to all of the last nine spacer oligonucleotides (spacers 35 to 43), and Beijing-like genotype strains were the ones that hybridized to only some of the last nine spacers. Both typical Beijing and Beijing-like genotype are Beijing genotype. Genomic DNA was extracted by boiling the freshly cultured bacteria as previously described29, Spoligotyping was carried out based on the standard protocol30. A commercially available kit (Isogen Bioscience BV, Maarssen, and the Netherlands) was used as described by the manufacturer. The results were compared with the SITVIT_WEB database (an international spoligotype database at the Institute Pasteur de Guadeloupe. http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/)31
VNTR typing
12-locus VNTR combination was used in this study, which same to the previous report18. The PCR products were analyzed by electrophoresis in 2% agarose gels, the number of repeats at each locus was interpreted based on the electrophoretic mobility of the products compared with the standard molecular marker (50 bp DNA ladder, TaKaRa-Bio Inc., Dalian, China). H37Rv and double-distilled H2O were used as positive and negative controls, respectively. The copy number of repeats was calculated using the following formula: (length of the PCR product minus length of the flanking regions)/length of one repeat copy unit. Alleles were assigned numerical values according to the number of repeats present at a specific genomic locus. Strains were classified as different genotypes according to VNTR loci set while strains having identical VNTR profile type were defined as a cluster. VNTR dendrograms were constructed by using unweighted pair group method with arithmetic averages (UPGMA).
Drug susceptibility testing (DST)
Mycobacterium tuberculosis drug susceptibility testing (DST) to four first-line anti-TB drugs (Isonazid, Rifampin, Ethambutol, and Streptomycin) was performed using proportion method, as recommended by WHO/IUATLD32. Strains which showed resistance to Isonazid and Rifampin were defined as MDR-TB. Strains declared resistant to drugs when growth rate exceeded 1% compared to the control strain. All drugs were purchased from Sigma-Aldrich (St. Louis, MO).
Data management
Genotyping results showed with binary and octal formats in Microsoft Excel spreadsheets. All spoligotyping data were submitted to the SITVIT_WEB database (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/). VNTR data expressed in decimal format were analyzed. Resulting data was analyzed by BioNumerics (Version 5.0, Applied Maths, Sint-Martens-Latem, Belgium) software. Cluster analysis was performed and a dendrogram was generated in Bionumerics using the Dice similarity coefficient and UPGMA coefficient.
The discriminatory power (the Hunter-Gaston discriminatory index (HGDI)) of each typing method was calculated according to a previously published method33:
where N is the total number of isolates in the typing method, s is the number of distinct patterns discriminated by VNTR, and n j is the number of isolates belonging to the jth pattern. The percentage clustering was calculated with the following formula: (n c − C)/N, where N is the total number of isolates, C is the number of clusters, and n c is the total number of clustered isolates34.
Statistical analysis
Statistical analysis of genotype and phenotype were performed in SPSS 21.0 software. Chi-square test or Fisher’s exact probability test was used to compare the proportions of different groups. A p value less than 0.05 was considered statistically significant. Odd ratios (ORs) and 95% confidence intervals (CI) were calculated to measure the association relationship between genotype and drug susceptibility results.
Availability of data and materials
The online version of this article contains supplementary material, which is available to authorized users.
References
WHO. Global tuberculosis report 2016 (Geneva,Switzerland:WHO, 2016).
Li, L., Xi, Y. & Ren, F. Spatio-Temporal Distribution Characteristics and Trajectory Similarity Analysis of Tuberculosis in Beijing, China. International journal of environmental research and public health 13, doi:https://doi.org/10.3390/ijerph13030291 (2016).
Wang, S. F., Zhou, Y., Pang, Y., Zheng, H. W. & Zhao, Y. L. Prevalence and Risk Factors of Primary Drug-Resistant Tuberculosis in China. Biomedical and environmental sciences: BES 29, 91–98, https://doi.org/10.3967/bes2016.010 (2016).
Barnes, P. F. & Cave, M. D. Molecular epidemiology of tuberculosis. The New England journal of medicine 349, 1149–1156, https://doi.org/10.1056/NEJMra021964 (2003).
Singhal, P. et al. A study on pre-XDR & XDR tuberculosis & their prevalent genotypes in clinical isolates of Mycobacterium tuberculosis in north India. The Indian journal of medical research 143, 341–347, https://doi.org/10.4103/0971-5916.182625 (2016).
Singh, J. et al. Genetic diversity and drug susceptibility profile of Mycobacterium tuberculosis isolated from different regions of India. The Journal of infection 71, 207–219, https://doi.org/10.1016/j.jinf.2015.04.028 (2015).
Egbe, N. F. et al. Molecular epidemiology of Mycobacterium bovis in Cameroon. Scientific reports 7, 4652, https://doi.org/10.1038/s41598-017-04230-6 (2017).
Chen, S. T. et al. The Clinical Features and Bacteriological Characterizations of Bone and Joint Tuberculosis in China. Scientific reports 5, 11084, https://doi.org/10.1038/srep11084 (2015).
European Concerted Action on New Generation Genetic, M., Techniques for the, E. & Control of, T.. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerging infectious diseases 12, 736–743, https://doi.org/10.3201/eid1205.050400 (2006).
Pang, Y. et al. Risk factors and clinical phenotypes of Beijing genotype strains in tuberculosis patients in China. BMC infectious diseases 12, 354, https://doi.org/10.1186/1471-2334-12-354 (2012).
Pang, Y. et al. Spoligotyping and drug resistance analysis of Mycobacterium tuberculosis strains from national survey in China. PloS one 7, e32976, https://doi.org/10.1371/journal.pone.0032976 (2012).
Yang, C. et al. Mycobacterium tuberculosis Beijing strains favor transmission but not drug resistance in China. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 55, 1179–1187, https://doi.org/10.1093/cid/cis670 (2012).
Yuan, L. et al. There is no correlation between sublineages and drug resistance of Mycobacterium tuberculosis Beijing/W lineage clinical isolates in Xinjiang, China. Epidemiology and infection 143, 141–149, https://doi.org/10.1017/S0950268814000582 (2015).
Guo, Y. L. et al. Genotyping and drug resistance patterns of Mycobacterium tuberculosis strains in five provinces of China. The international journal of tuberculosis and lung disease: the official journal of the International Union against Tuberculosis and Lung Disease 15, 789–794, https://doi.org/10.5588/ijtld.10.0403 (2011).
Jia, Z. W. et al. Spatial analysis of tuberculosis cases in migrants and permanent residents, Beijing, 2000-2006. Emerging infectious diseases 14, 1413–1419, https://doi.org/10.3201/eid1409.071543 (2008).
Wan, K. et al. Investigation on Mycobacterium tuberculosis diversity in China and the origin of the Beijing clade. PloS one 6, e29190, https://doi.org/10.1371/journal.pone.0029190 (2011).
Yin, Q. Q. et al. Prevalence and molecular characteristics of drug-resistant Mycobacterium tuberculosis in Beijing, China: 2006 versus 2012. BMC microbiology 16, 85, https://doi.org/10.1186/s12866-016-0699-2 (2016).
Liu, Y. et al. Genotypic diversity analysis of Mycobacterium tuberculosis strains collected from Beijing in 2009, using spoligotyping and VNTR typing. PloS one 9, e106787, https://doi.org/10.1371/journal.pone.0106787 (2014).
Ma, Y. et al. Clinical outcomes for multi- and extensively drug resistant tuberculosis patients with adjunctive resectional lung surgery in Beijing, China. Journal of thoracic disease 9, 841–845, https://doi.org/10.21037/jtd.2017.02.54 (2017).
Sun, H. et al. Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis isolates in Sichuan, China and the association between Beijing-lineage and dual-mutation in gidB. Tuberculosis 96, 102–106, https://doi.org/10.1016/j.tube.2015.09.004 (2016).
Nhu, N. T. et al. Association of streptomycin resistance mutations with level of drug resistance and Mycobacterium tuberculosis genotypes. The international journal of tuberculosis and lung disease: the official journal of the International Union against Tuberculosis and Lung Disease 16, 527–531, https://doi.org/10.5588/ijtld.11.0202 (2012).
Li, Y. et al. Characterization of Mycobacterium tuberculosis isolates from Hebei, China: genotypes and drug susceptibility phenotypes. BMC infectious diseases 16, 107, https://doi.org/10.1186/s12879-016-144—1-2 (2016).
Mokrousov, I. et al. Evolution of drug resistance in different sublineages of Mycobacterium tuberculosis Beijing genotype. Antimicrobial agents and chemotherapy 50, 2820–2823, https://doi.org/10.1128/AAC.00324-06 (2006).
Mathuria, J. P. et al. Prevalence of Mycobacterium tuberculosis Beijing genotype and its association with drug resistance in North India. Journal of infection and public health, doi:https://doi.org/10.1016/j.jiph.2016.06.007 (2016).
Liu, H. C. et al. Molecular Typing Characteristic and Drug Susceptibility Analysis of Mycobacterium tuberculosis Isolates from Zigong, China. BioMed research international 2016, 6790985, https://doi.org/10.1155/2016/6790985 (2016).
Gupta, A., Kulkarni, S., Rastogi, N. & Anupurba, S. A study of Mycobacterium tuberculosis genotypic diversity & drug resistance mutations in Varanasi, north India. The Indian journal of medical research 139, 892–902 (2014).
Parwati, I., van Crevel, R. & van Soolingen, D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. The Lancet. Infectious diseases 10, 103–111, https://doi.org/10.1016/S1473-3099(09)70330-5 (2010).
Desikan, P. et al. Genetic diversity of Mycobacterium tuberculosis isolates from central India. The Indian journal of medical research 143, 481–486, https://doi.org/10.4103/0971-5916.184287 (2016).
Aldous, W. K., Pounder, J. I., Cloud, J. L. & Woods, G. L. Comparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. Journal of clinical microbiology 43, 2471–2473, https://doi.org/10.1128/JCM.43.5.2471-2473.2005 (2005).
Kamerbeek, J. et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. Journal of clinical microbiology 35, 907–914 (1997).
Demay, C. et al. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 12, 755–766, https://doi.org/10.1016/j.meegid.2012.02.004 (2012).
Organization, W. H. Anti-tuberculosis drug resistance in the world, www.who.int/tb/publications/2008/drs_report4_26feb08.pdf (2008).
Hunter, P. R. & Gaston, M. A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. Journal of clinical microbiology 26, 2465–2466 (1988).
Small, P. M. et al. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. The New England journal of medicine 330, 1703–1709, https://doi.org/10.1056/NEJM199406163302402 (1994).
Acknowledgements
We gratefully acknowledge Dr. Tao Luo for performing the BioNumerics analysis, and Dr. Chongguang Yang for helping with setting up the software. This work was supported by the Capital Medical Development Foundation, grant #2014-4-2163; and Beijing Municipal Administration of Hospitals’ Youth Programme, grant# QML20161601.The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: Yi Liu, Chuanyou Li. Performed the experiments: Yi Liu, Xiaoying Jiang, Wensheng Li, Xuxia Zhang, and Wei Wang. Analyzed the data: Yi Liu, Xiaoying Jiang. Contributed reagents/materials: Yi Liu, Chuanyou Li. Wrote the paper: Yi Liu, Xiaoying Jiang. All authors read and approved the final manuscript before submission.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, Y., Jiang, X., Li, W. et al. The study on the association between Beijing genotype family and drug susceptibility phenotypes of Mycobacterium tuberculosis in Beijing. Sci Rep 7, 15076 (2017). https://doi.org/10.1038/s41598-017-14119-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-14119-z
This article is cited by
-
Characterization of Mycobacterium tuberculosis strains in Beijing, China: drug susceptibility phenotypes and Beijing genotype family transmission
BMC Infectious Diseases (2018)
-
Genetic diversity and distribution dynamics of multidrug-resistant Mycobacterium tuberculosis isolates in Nepal
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.