Abstract
The holotype (MHM-K2) of the Eocene cheloniine Tasbacka danica is arguably one of the best preserved juvenile fossil sea turtles on record. Notwithstanding compactional flattening, the specimen is virtually intact, comprising a fully articulated skeleton exposed in dorsal view. MHM-K2 also preserves, with great fidelity, soft tissue traces visible as a sharply delineated carbon film around the bones and marginal scutes along the edge of the carapace. Here we show that the extraordinary preservation of the type of T. danica goes beyond gross morphology to include ultrastructural details and labile molecular components of the once-living animal. Haemoglobin-derived compounds, eumelanic pigments and proteinaceous materials retaining the immunological characteristics of sauropsid-specific β-keratin and tropomyosin were detected in tissues containing remnant melanosomes and decayed keratin plates. The preserved organics represent condensed remains of the cornified epidermis and, likely also, deeper anatomical features, and provide direct chemical evidence that adaptive melanism – a biological means used by extant sea turtle hatchlings to elevate metabolic and growth rates – had evolved 54 million years ago.
Similar content being viewed by others
Introduction
Marine deposits of the early Eocene Fur Formation (Jutland, Denmark) have yielded a diverse biota of exceptionally preserved plant and animal fossils1. Noteworthy finds include a speciose assemblage of early neornithes2 and a small, but significant, collection of chelonioid turtles3,4. Of the latter group, by far the best preserved specimen is MHM-K2: a diminutive (about 74 mm when measured from the nuchal emargination to the posterior rim of the pygal), ontogenetically young cheloniine referred to as Tasbacka danica by Karl & Madsen5. MHM-K2 was collected in 2008 from within a limestone concretion in the Ejerslev Mo-clay pit on the Isle of Mors, Denmark, by one of us (HM). Initially, the fossil was prepared using a combination of mechanical tools and buffered acetic acid. However, following the discovery of soft tissue remains, only a pneumatic air scribe was employed to expose the dorsal aspect of a virtually intact turtle skeleton showing true bone-to-bone relationships, save for the eighth right costal and suprapygal (Fig. 1a). Although the fossil has suffered some compaction damage affecting parts of the skull, the individual bones are generally in pristine condition and retain their original three-dimensional shape. The carapace has a broadly ovate outline with a concave anterior margin, and it is about as wide as it is long in its current, compressed state (Fig. 1a). Large spaces (fontanelles) occur between the ribs and peripherals, and sculpturing in the form of four neural nodes can be seen along its antero-posterior midline (Fig. 1a—arrowheads). The plastron is mostly obscured by the carapace; however, parts of the lateral processes of both the hyo- and hypoplastron are visible in the fontanelles enclosed by the second and third, and fourth and fifth costal, respectively. Adaptations for an obligate marine existence (e.g. elongate phalanges) are apparent in the paddle-shaped limbs (Fig. 1a,b); notably though, the extremities are proportionally short by sea turtle standards. Vestiges of soft tissues include buff-coloured marginal scutes lining the outside of the peripherals (Fig. 1a,c—arrowheads), and dark matter forming well-defined outlines of the neck and flippers (Fig. 1a,b), as well as covering a number of bones and fontanelles (Fig. 1c—arrows and stars).
Holotype of Tasbacka danica. (a) Photograph of the fossil. Fo, fontanelle (the light colour is a result of sediment infill); Hyo, hyoplastron; Hyp, hypoplastron; Ne, neural; Nu, nuchal; Pe, peripheral; Py, pygal. Arrowheads indicate neural nodes. (b) Detail of the carapace with the sampled area demarcated by a circle. Co, costal; Hu, humerus; Sc, scapula. (c) Higher magnification image showing marginal scutes (arrowheads), pigmentations on bones (arrows), and a brown-black film covering the fontanelles (stars).
Here we examine, at the molecular level, details of the soft tissue anatomy of MHM-K2. Renewed preparations of the fossil in 2013 uncovered soft tissue residues from a ‘fresh’ sub-surface layer located within a sub-triangular area bordered by the dorsal process of the left scapula, proximal end of the left humerus, left costal 1, and left peripherals 1 and 2 (Fig. 1b). Five small samples representing the counterpart of MHM-K2 were collected for ultrastructural and biomolecular analysis; these were untreated with preservatives and form the basis of this study (the fossil has since been embedded in consolidants while making replica casts, thereby precluding the recovery of additional samples for molecular analysis).
To determine the ultrastructural and chemical composition of the preserved soft tissues, both untreated and demineralised (using ethylenediaminetetraacetic acid, EDTA) samples were subjected to a selection of high-resolution analytical techniques, including field emission gun scanning electron microscopy (FEG-SEM), transmission electron microscopy (TEM), in situ immunohistochemistry, time-of-flight secondary ion mass spectrometry (ToF-SIMS), and infrared (IR) microspectroscopy.
Results
Ultrastructural and elemental analyses
Initial macroscopic examination of the preserved soft tissues showed a dark, well-defined film that is distinct from the surrounding sediment in both texture and colour. Subsequent FEG-SEM analysis revealed that the thin layer comprises accumulations of sub-spherical to elongate microbodies with a homogenous interior and botryoidal, irregularly pitted exterior (Fig. 2a–c). These are encapsulated within a matrix consisting of two main types of fabric: a fine-grained material interpreted as an authigenic precipitation (Fig. 2c—black star), and, more commonly, a three-dimensional meshwork with a sheet-like to vesicular (frothy) texture (Fig. 2c—white star and Fig. 2d). Elemental analysis using energy-dispersive X-ray microspectroscopy established that the microbodies and sheet-like substrate are enriched in carbon, indicating the possible retention of organic compounds.
Ultrastructure of MHM-K2 soft tissues. (a) FEG-SEM micrograph of demineralised tissue showing microbodies and adhering matrix. (b) At higher magnification, the microbodies possess a rough surface texture and scattered pits (arrowheads). (c) FEG-SEM micrograph of untreated soft tissue depicting microbodies embedded in a mineral precipitate (black star) and sheet-like matter (white star). (d) Microbodies (arrowheads) in a sheet-like substrate. (e) TEM micrograph of electron-dense microbodies and fibrous matrix (black arrowheads) after demineralisation. White star indicates epoxy resin, whereas black star marks an artificial rupture.
TEM imaging revealed that the film, although only 15 µm in depth, has a three-dimensional configuration. Furthermore, the electron-dense microbodies are neither stacked nor overlapping, but rather separated by a fibrous matrix that defines the interstitial spaces (Fig. 2e—arrowheads).
Immunohistochemical analyses
In living chelonians, the bony carapace is covered by an epidermis comprised mainly of keratinous proteins. Both α- and β-keratins are present; however, the latter is only expressed in sauropsid cornified tissues6,7, and thus cannot arise from human or microbial contamination. Accordingly, we employed the specificity of the vertebrate immune system (Figs 3, 4 and Supplementary Fig. S1) to distinguish between endogenous animal remains and invasive microorganisms that naturally colonise decaying organic matter8,9,10. We used antibodies raised against chemically extracted white feathers (which almost exclusively comprise β-keratin) from chicken, Gallus gallus domesticus 9,11, to test the hypothesis that the structurally distinct brown-black residue associated with the bony shield of MHM-K2 was consistent with β-keratin of the epidermal coating of the carapace12,13,14. Regions of this protein are homologous in turtles and birds7,12,13,15; therefore, if original molecules (or remains thereof) were preserved, we predicted that they would be recognised by these antibodies. Antibody-antigen complexes were identified by green fluorescence in situ on the ancient matter (Fig. 3a,b), and compared to the pattern observed in scute material obtained from an extant green sea turtle, Chelonia mydas, using the same antibodies and experimental parameters (Fig. 3c,d and Supplementary Fig. S1). Although the binding was less intense and more punctate in MHM-K2 relative to its modern counterpart, it was well above background levels, indicating that epitopes (that is, three to five amino acid residues derived from the protein and retaining their original three-dimensional conformation) remained in the fossil tissues. Furthermore, these antibodies bound to the ancient matter with the same pattern as that observed in comparable extant tissues, and were restricted to areas in between the fossil microbodies (Fig. 3a).
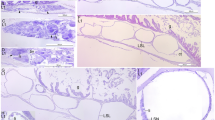
Immunoreactivity of fossil and extant turtle tissues. Immunohistochemical staining results for (a,b,e,f,i,j,m,n,q,r; columns 1 and 2) MHM-K2 and Chelonia mydas (c,d; columns 3 and 4) carapace scute and (g,h,k,l,o,p,s,t; columns 3 and 4) muscle tissue to antibodies raised against (a–d; row 1) Gallus gallus domesticus feathers (anti-Gallus fth), (e–h; row 2) Alligator mississippiensis haemoglobin (anti-Alligator Hb), (i–l; row 3) Struthio camelus haemoglobin (anti-Struthio Hb), (m–p; row 4) G. g. domesticus tropomyosin (anti-Gallus trop), and (q–t; row 5) bacterial peptidoglycan (anti-bac pep). a,c,e,g,i,k,m,o,q,s are overlay images, superimposing fluorescent signal on transmitted light image of sectioned tissue to reveal the localisation of antibody-antigen complexes to tissue. b,d,f,h,j,l,n,p,r,t are imaged using a FITC filter. Antibody-antigen complexes are indicated by green fluorescent signal.
Comparison of immunoreactivity between fossil and extant turtle tissues using antibodies raised against chicken feathers conjugated to 12 nm gold beads. (a–d) Low and (e–h) high resolution localisation of gold beads to fibrous matter, but not microbodies/melanosomes, in (a,b,e,f) fossil tissues and (c,d,g,h) modern Chelonia mydas carapace scute material. Insets in b and d demarcate areas depicted in e,f and g,h, respectively. The data support the specificity of the chicken feather antibodies used in this study, and provide independent validation of the immunofluorescent results.
To obtain higher resolution of the antibody-antigen complexes, binding was also detected using a secondary antibody conjugated to 12 nm gold beads. Specific localisation of beads to the filamentous matrix was evident in both the fossil (Fig. 4a,b,e,f) and modern (Fig. 4c,d,g,h) turtle tissues.
To test the hypothesis that structures deeper than the cornified epidermis might be preserved in MHM-K2, we exposed fossil material to antibodies raised against haemoglobin from both the American alligator, Alligator mississippiensis, and ostrich, Struthio camelus, as well as antibodies raised against chicken tropomyosin (an eukaryotic cytoskeletal protein16). Reactivity to antibodies against haemoglobin and tropomyosin was evident in MHM-K2 (Fig. 3e,f,i,j,m,n) in patterns similar to those in C. mydas muscle tissue (Fig. 3g,h,k,l,o,p), but with reduced intensity.
Finally, to address the possibility that some of the microbodies and/or organic matrix in which they are housed could be microbially derived, we tested the fossil and modern tissues against antibodies raised against peptidoglycan – a common component of bacterial biofilms17. Whereas reactivity was absent in the control samples (Fig. 3s,t), weak antibody response was visualised in MHM-K2 (Fig. 3q,r), suggesting a minor (but not unexpected) microbial contribution.
Time-of-flight secondary ion mass spectrometric analyses
ToF-SIMS data obtained from the fossil soft tissues provided evidence for the presence of heme, eumelanin and proteinaceous matter (Figs 5 and 6), thereby independently validating the immunological results.
Molecular characterisation of MHM-K2 tissues by ToF-SIMS analysis. (a) Positive ion spectrum from a region with strong signal from heme-related ions. (b) Positive ion spectrum of a heme (hemin) standard. (c) Negative ion image showing the signal intensity distribution of ions representing heme (red; 65 + 108 + 134 u), eumelanin (green; 66 + 73 + 74 + 97 + 98 + 121 + 122 u) and silica (blue; 60 + 76 + 77 u). Field of view: 200 × 200 µm2. (d) FEG-SEM micrograph showing a fracture edge. Note abundant microbodies in the crack wall and vesicular texture of the surface. (e) Negative ion spectrum from a region with mixed signal from eumelanin- and heme-related ions. (f) Negative ion spectrum from a region dominated by signal from eumelanin-related ions. (g) Positive ion image showing the signal intensity distribution of ions representing heme (red; 436–488 u), aromatics (blue; 91 + 115 u) and proteinaceous materials (green; 30 + 44 + 70 u). Field of view: 328 × 328 µm2. (h) FEG-SEM micrograph of the demarcated area in g depicting sheet-like matter with high signal from amino acid-related peaks.
ToF-SIMS spectra obtained from MHM-K2 soft tissues together with selected standards. (a) Negative ion spectra of hemin (top) and Sepia eumelanin (bottom) standard compounds. (b) Expanded negative ion spectra of (from top to bottom): MHM-K2, from an area dominated by eumelanin-related ions; MHM-K2, from an area with mixed signal from eumelanin- and heme-related ions; Sepia eumelanin, and; hemin. The depicted masses (65, 108 and 134 u) all show strong signal in the hemin reference spectrum. (c) Positive ion spectra acquired from regions of MHM-K2 with strong signal from amino acid- (top) and heme-related (bottom) ions, respectively. Peaks marked in green represent ions characteristic of proteinaceous materials.
Heme was identified in positive ion mode as a distinct set of undulating peaks in the mass range between 400 and 500 u (Fig. 5a); this fragmentation pattern was reproduced in detail in spectra acquired from a purified heme control sample (Fig. 5b). The peaks derive from ions comprising the entire iron-containing porphyrin unit of heme after the successive loss of various CHx entities18,19. However, the molecular ion (that is, the intact heme molecule in its ionised form) was not detected in the fossil spectra, indicating partial, likely diagenetically mediated degradation of the heme molecular structure (including loss of various -COOH units18). Compounding this identification, heme-derived molecular fragments were observed in negative ion spectra e.g. at 65 (C3N2H−), 108 [Fe(CN)2 −] and 134 u [Fe(CN)3 −]; these peaks occurred at high signal intensities also in the heme standard spectra (Fig. 6a,b).
The identification of eumelanin on the fossil surfaces was based on its detailed spectral agreement in the mass range between 45 and 175 u (negative ions) with a Sepia eumelanin standard sample (Figs 5f and 6a,b), both with regard to the relative signal intensity distribution and exact mass positions10,20,21,22.
In addition to heme and eumelanin, a third compound showing spectral features typical of proteinaceous materials (such as strong signal intensity from specific nitrogen-containing ions representing amino acids23) was also recognised (Fig. 6c). Notably, the majority of the amino acid-related peaks were not prominent in ToF-SIMS spectra of either eumelanin or heme, to suggest a different molecular origin of these ions.
The eumelanin- and heme-related ions showed relatively homogeneous distributions on the fossil surfaces, but with some noteworthy differences. Particularly, areas deficient in heme, but with strong signal from eumelanin-related peaks, could be found at edges of desiccation cracks running across the sample surfaces (Fig. 5c). Subsequent FEG-SEM analysis revealed the occurrence of densely packed microbodies in association with a vesicular film; the latter originally occurring medial to the microbody-rich layer (Fig. 5d). Negative ion spectra taken from regions comprising a mixture of bodies and vesicular matter showed characteristics of both eumelanin and heme (Figs 5e and 6a,b), whereas areas dominated by microbodies produced spectra typical of eumelanins (Figs 5f and 6a,b). Regions with strong signal intensity from amino acid-related peaks were relatively scarce and their distribution was distinct from that of both heme and eumelanin, to suggest the presence of scattered proteinaceous residues on the fossil surfaces (Figs 5g and 6c). FEG-SEM imaging of areas with amino acid signal showed sheet-like material (Fig. 5h), interpreted as partially degraded keratin plates.
IR microspectroscopic analyses
IR microspectroscopic measurements of the fossil tissues produced broad-band absorbance in the 900–1,750 and 2,700–3,700 cm−1 regions, consistent with natural eumelanin (Fig. 7).
IR absorbance spectra of Tasbacka soft tissues and Sepia eumelanin. The Tasbacka spectrum exhibits broad-band absorbance in the 900–1,750 and 2,700–3,700 cm−1 regions, consistent with the natural eumelanin standard. Differences (marked with ‘+’) can be attributed to contributions from the sedimentary matrix (mainly calcite)20,68. These occur also in previously analysed samples from the Fur Formation20. For example, the intense and broad peak at ~1,450 cm−1 likely includes absorbance from the asymmetric stretch mode of calcium carbonate68.
Discussion
The methods employed herein were chosen to obtain maximal information while minimising sample destruction. They are highly specific and extremely sensitive, yet capitalise on different aspects of the molecules they characterise. Accordingly, the results of our integrated ultrastructural and biochemical approach provide compelling independent lines of evidence for the presence of an unprecedented number of endogenous biomolecular residues, including haemoglobin-, β-keratin-, tropomyosin-, and melanin-derived compounds (as well as residual peptidoglycan), in the soft tissue remains recovered from MHM-K2. Moreover, the anatomical localisation, geometry and size of the fossil microbodies and adhering matrix compare favourably with those of remnant melanosomes (Fig. 2c in ref.21) and degraded keratin plates (Supplementary Fig. S2; see also Fig. 10c–e in ref.14), respectively, to suggest that the preserved organic matter comprises vestiges of the pigmented keratinous epidermal coating of the shell. This interpretation is supported not only by the presence of β-keratin epitopes in the fibrous substrate surrounding the microbodies (Figs 3a,b and 4a,b,e,f), but also by their electron-dense interior (Fig. 2e) – a distinguishing feature of mature melanosomes10. Additionally, marginal scutes are visible alongside the peripherals (Fig. 1a,c—arrowheads), and stains on some bones indicate that the soft tissues are located outside of the ribcage (Fig. 1c—arrows). Incomplete decomposition of the carapace due to oxygen deprivation and/or early sealing of the entombing sediment by concretion formation2,24 (perhaps facilitated by microbial mat growth25) presumably reduced the keratin laminate (that is, the stacked layers of cornified cells and β-pleated sheet keratin forming the scutes14,26), thereby concentrating the more decay-resistant (refs27,28, but see also ref.9) melanosomes into a condensed layer, which in turn underwent compactional flattening. Preferential preservation of integumental features may seem unlikely; however, skin-derived structures are second only to biomineralised tissues in the vertebrate fossil record (ref.29) and references therein), and decay experiments on modern carcasses have revealed the persistence of integumental tissues even after long-term exposure to microbial decomposers25,30. In the Fur Formation, carbonised animal soft tissues, with the notable exception of orbital (retinal) pigmentations10,20, chiefly comprise integumental features (often with preserved colour patterns; Figs 38, 39a and 46 in ref.31, Fig. 1 in ref.32).
In extant vertebrates, skin forms a tough physico-chemical barrier that performs multiple functions33. Additionally, turtle scutes constitute a resilient shielding material which provides the shell with a compliant coating that enhances its resistance to mechanical damage26,34. Recalcitrant biopolymers, including keratins and melanins, are key components of integumental appendages; their hydrophobic nature and inherent ability to form robust intra- and intermolecular cross-links likely contribute to the survival of epidermal structures across geological time9,22,29,35. Besides being impermeable, most epidermally-derived features are also avascular33, thus reducing the possible entry of degrading microbes.
Another factor contributing to the selective preservation of skin and its derivatives may be the close spatial juxtaposition between these anatomies and the host rock. Sorption of organics onto clay minerals (a common component of the Fur Formation36) promotes fixation37,38,39; clays also possess antibiotic properties40. Furthermore, association of biological matter with mineral nano- and microfabrics adds protection from oxidative and hydrolytic damage through the creation of stabile organometallic complexes41,42. Chemical evidence for this process is seen in MHM-K2 as a series of molecular fragments involving calcium adducts (Table 1). We hypothesise that calcium ions (and other trace elements) adsorbed onto the surface of the carcass during the microbially mediated24 formation of the calcareous concretion in which MHM-K2 was found. Mild geothermal conditions43 might then have limited further breakdown of the stabilised organics.
Haemoglobin also imparts tissue fixation by iron-catalysed free radical reactions and/or inhibition of bacterial growth (ref.44 and references therein), possibly contributing to preservation of anatomical features deeper than the cornified epidermis. Blood breakdown products released from erythrocytes during hemolysis can seep into surrounding tissues, causing a reddish-brown discolouration45. Impregnation by haemoglobin-derived compounds has been recorded not only in bones46, but also in scales and teeth45. Consequently, it is possible that the outer integument was infiltrated by blood residues diffusing from underlying (and now almost completely degraded) dermal or deeper tissues sometime during the early stages of decomposition of MHM-K2. Detection of haemoglobin- and tropomyosin-derived compounds supports this possibility.
Extant neonate sea turtles are generally predominantly dark-coloured, at least on their dorsal surface (Supplementary Fig. S3). In most species, this dusky patterning changes with age47,48. Thus, its selective advantage is not immediately apparent, especially since the brown-black pigmentation increases the risk of overheating while on land (owing to enhanced absorbance of solar radiation)47, but also because it makes the hatchlings stand out against the beach sand during their dash towards the sea (Supplementary Fig. S3a,b). However, potential selective benefits are illustrated by the green sea turtle, Chelonia mydas. Hatchlings of this species are dark brown to black dorsally, save for white edgings on the carapace and flippers (Supplementary Fig. S3c). The plastron, on the other hand, is lightly coloured (ref.48, but see also ref.49). When emerging from the nest, hatchling C. mydas face the same challenges as do other sea turtles; however, the functional significance of the dark dorsum becomes more readily apparent once in their pelagic habitat. While resting, neonate C. mydas float at the sea surface with large parts of the body above the waterline (Fig. 1 in ref.47). In this posture, the dark upper surface provides ultraviolet (UV) ray protection and camouflage50, but also more efficient absorption of solar radiation, thereby promoting an elevation of the body temperature47. The resulting rise in basal metabolic rates contributes to increased growth rates during this vulnerable early life stage47.
In ectotherms, incident radiation is converted into thermal energy primarily by integumental eumelanins51; there is also a direct relationship between the density of these pigments and skin darkness51,52,53. In addition to melanins, non-endothermic vertebrate skins often contain a number of other biochromes, which together with finely organised nanostructures causing light interference, produce the variety of colour patterns seen in fish, amphibians and reptiles today32,52,53,54. However, contrary to recent claims55, non-melanic pigments and photonic nanosurfaces are not ubiquitous to all reptile skins. For instance, the colour of neonate C. mydas, Caretta caretta (loggerhead sea turtle) and Dermochelys coriacea (leatherback sea turtle) is determined exclusively by melanins: dark portions of the integument are defined by abundant melanosomes and melanophores (Supplementary Fig. S4a,b,d,e,g–n), whereas light portions possess few melanophores or lack pigment cells altogether (Supplementary Fig. S4c,f,o). Similar observations have previously been made by Solomon et al.26, to suggest that contributions from light-reflecting biochromes and materials are negligible in melanised marine reptile skins21. Additionally, countershading is consistent with life in the featureless pelagic zone, where there are few hiding places and thus little need for bright colours or disruptive markings56.
With a carapace length roughly corresponding to that of four- to five-week old C. mydas 49, MHM-K2 represents an ontogenetically very young individual5. Apart from size, skeletal immaturity is manifested by the incomplete development of the costals – resulting in large fontanelles – and presence of neural nodes along the carapace midline (Fig. 1a—arrowheads). These juvenile features, together with the findings of this study, combine to suggest strongly that the type of Tasbacka danica originally had a predominantly dark dorsum with pale edges to the carapace (although concentrated by scute reduction, the sheer number of melanosomes in MHM-K2 indicates that the original density was high, equalling that of modern chelonioids). Thus, it was likely similar in appearance to extant sea turtle hatchlings, particularly C. mydas (Supplementary Fig. S3c), and may even have relied on melanism as an integral part of its thermoregulation. Although we cannot completely exclude the possibility that there may have been biochromes and/or nanostructures that did not survive long enough to enter the fossil record, our interpretation is corroborated by the fact that the colouration of most (if not all) modern neonate chelonioids is produced solely by melanins (Supplementary Fig. S4). We also acknowledge the inherent uncertainty associated with extrapolations of large-scale pigment patterns from minute samples57. However, in this case, a partial colour reconstruction is justifiable because: (1) the fossil preserves a distinct patterning incorporating both light (marginal scutes) and dark (fontanelle coverings) areas; (2) a relationship exists between melanosome density and apparent darkness of preserved soft tissues in Fur Formation vertebrate fossils10,20,21,58,59; (3) there is a close phylogenetic relationship between T. danica and living cheloniines5; and (4) extant neonate sea turtles exhibit relatively uniform, darkly pigmented carapaces48,60. Likewise, we exclude major contributions from dispersed internal melanosomes61 because the fossil pigment organelles in MHM-K2 are embedded within remnant β-keratin, consistent with the epidermal covering of the shield. Additionally, blackish soft tissues occur also in the distal end of the flippers (Fig. 1a,b), which are far from any potential melanin-harbouring internal source.
Our results suggest that a biological means of UV protection and/or melanin-based thermoregulation existed in hatchling cheloniines at least 54 million years ago (notably in the pronounced greenhouse climate following the Palaeocene-Eocene Thermal Maximum). However, because melanism is widespread among extant neonate chelonioids and because they share a common ancestor in the Early Cretaceous (or possibly Late Jurassic)62,63, phylogenetic bracketing64 implies that this adaptation may have appeared already during the latter part of the Mesozoic. If so, then it was secondarily lost in the evolutionary lineage leading to the modern flatback sea turtle, Natator depressus (hatchlings of this species have a light grey dorsum; Fig. 1b in ref.65), perhaps due to marked size differences at birth47 or because neonate N. depressus employ survival strategies different from those of other marine turtles65.
Methods
Fossil material
MHM-K2 is housed in the collections at the Mo-clay Museum, Denmark (the holotype of Tasbacka danica has recently been transferred to the Natural History Museum of Denmark. New accession number: NHMD 141598). The fossil tissue samples were triple-washed in 96% ethanol and Milli-Q water to remove potential contaminants from human handling. They were then dried under a hood, wrapped loosely in fresh aluminium foil and stored in a sealed, sterile container. Fresh aluminium foil was used to cover all work areas, and surgical gloves were used during all handling and treatment.
Modern reference materials
The body of a hatchling green sea turtle, Chelonia mydas (KPC16030901; housed in Kamezaki public collection), that had died from natural causes, was collected on the Amami Island of Japan in August of 2011 and has since been stored in a freezer. Similarly, a second (juvenile) specimen (unnumbered, Department of Biological Sciences, North Carolina State University) of C. mydas (also naturally deceased) was collected from the New River, North Carolina, USA, in the fall of 2010 and has since been kept frozen. A naturally deceased hatchling loggerhead sea turtle, Caretta caretta (KPC16030906), was collected in the Kyoto Prefecture of Japan in August of 2010. Initially, KPC16030906 was fixed in 10% formaldehyde, but it has since been treated with 70% ethanol. A neonate leatherback sea turtle, Dermochelys coriacea (ZMUC R2106), stored in 70% ethanol, was acquired as a gift from ‘Danmarks Akvarium’ (former Danish Aquarium) in 1962, and has since been part of the vertebrate collection at the Zoological Museum, Natural History Museum of Denmark.
Scanning electron microscopy (SEM)
Initial screening was performed using a Hitachi S-3400N SEM on uncoated fossil samples under low vacuum, and the elemental composition was determined via elemental mapping using EDX analysis (1,900 sec scanning time at 15 keV, 62.0 μA and a working distance of 10 mm). Following ToF-SIMS analysis, the samples were sputter-coated with a gold/palladium mixture and re-examined using a Zeiss Supra 40VP FEG-SEM (2 keV, working distance 3–5 mm, Everhart-Thornley secondary electron detector). Modern reference samples were sputter-coated with gold and analysed using a Tescan Mira3 High Resolution Schottky FEG-SEM (30 keV, working distance 3–5 mm, in-beam secondary electron detector).
Transmission electron microscopy (TEM)
Both untreated and demineralised fossil matter was embedded in epoxy resin (AGAR 100 Resin kit, R1031), which was left to polymerise at room temperature for 72 hours, followed by 48 hours at 60 °C. Ultrathin sections (50 nm) of the infiltrated material were produced with a Leica EM UC7 Ultramicrotome using a diamond knife. The sections were then mounted to pioloform-coated copper grids.
Representative samples from the neck, carapace, plastron, and forelimbs of three extant sea turtle hatchlings (Chelonia mydas, Caretta caretta and Dermochelys coriacea) were collected using sterile scissors. The C. mydas and C. caretta samples were fixed overnight at 4 °C in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3). Following this treatment, the tissues were rinsed in 0.25 M sucrose in 0.1 M phosphate buffer (pH 7.3), and then fixed a second time for two hours at room temperature using 0.1 M buffer containing 1% osmium tetroxide. Thereafter, the integumental tissues were rinsed with a buffer solution and dehydrated in stepwise increased concentrations of ethanol. Finally, the tissues (together with D. coriacea skin dehydrated in a graded ethanol series) were embedded in epoxy resin (Quetol 651 and AGAR 100 Resin kit). Semithin sections (1.5 μm) were produced using a glass knife, whereas ultrathin sections (50 nm) were cut with a Leica Ultracut UC7 ultrotome using a diamond knife.
Ultrathin sections of all samples were examined in a JEOL JEM-1400 PLUS transmission electron microscope at 80 and 120 kV without further treatment or staining. Areas of interest were photographed using a JEOL Matataki CMOS camera.
In situ immunohistochemistry
All manipulations of Tasbacka samples were conducted in a laboratory dedicated specifically to analyses of fossil materials, in which no modern tissues were handled or stored. All instruments, buffers and solutions were kept separated from those used in analyses of the modern controls. An aseptic protocol was applied; gloves, gowns and face masks were worn at all times. In a separate laboratory, a sterile scalpel was used to remove small pieces of muscle, claw and carapace scute material from a juvenile Chelonia mydas. The collected tissues were kept frozen at −80 °C in a freezer dedicated exclusively to modern samples, and in which no fossil material has ever been stored, until analysed.
Fossil samples or fixed C. mydas tissues (scute, claw and muscle) were dehydrated in 70% ethanol, then infiltrated with a 2:1 mixture of LR White (EMS Cat # 14383, hard grade) and 70% ethanol for one hour. Samples were then incubated, separately, in two changes of concentrated LR White for one hour, each time to equilibrate. All specimens were then embedded in concentrated LR White and allowed to polymerise at 60 °C for 24 hours. They were then sectioned to 200 nm using a Leica EM UC6 Ultramicrotome. Sections were collected and transferred to six-well, Teflon-coated slides, dried on a warming plate, then dried to completion in 45 °C oven overnight. Sections were etched with Proteinase K (PCR grade, Roche, 25 µg/ml) in ×1 phosphate buffered saline (PBS) buffer at 37 °C to expose epitopes, followed by two incubations in 500 mM EDTA (pH 8.0) and two incubations in 1 mg/ml sodium borohydride for 10 min each for epitope retrieval. All incubations were separated by sequential washes (two times for five min each) in PBS. Sections were then incubated for two hours with 4% normal goat serum (NGS) in PBS to occupy non-specific binding sites and prevent spurious binding. Sections were then incubated overnight at 4 °C in either primary antibody [Polyclonal Rabbit X-Alligator Haemoglobin (Biosynthesis BSYN6941) 1:100; Polyclonal Rabbit X-Ostrich Haemoglobin (GenScript 70594-1) 1:75; Polyclonal Rabbit X-Chicken Feather (Biosynthesis BSYN6734) 1:500; Polyclonal Rabbit X-Tropomyosin (Abcam ab11190) 1:50; or Monoclonal Mouse X-Peptidoglycan (AbD Serotec, 7263–1006) 1:75], diluted to final concentration in primary dilution buffer [(1% Bovine Serum Albumin (BSA) (Fisher, BP1660-100), 0.1% Cold Fish Skin Gelatine (Sigma G7765), 0.05% Sodium Azide (Sigma S-8032), 0.01 M PBS, pH 7.2], or in primary dilution buffer only, without primary antibody added to control for non-specific secondary antibody binding. All sections (test and control, fossil and modern) were washed thoroughly to remove unbound antibody, then incubated with secondary antibody [(Biotinylated Goat Anti-Rabbit IgG (H + L) (Vector BA-1000) diluted 1:500 for rabbit primary antibodies, Biotinylated Goat Anti-Mouse IgG (H + L) (Vector BA-9200), diluted 1:500 for monoclonal mouse anti-peptidoglycan] for two hours at room temperature. Fluorescein Avidin D (FITC, Vector Laboratories A-2001) diluted 1:1,000 was applied to all sections and allowed to bind for one hour at room temperature. All incubations were separated by sequential washes (two times for 10 min each) in PBS w/Tween 20 (ACROS Organics) followed by two 10 min rinses in PBS. Finally, the sections were mounted with Vectashield Anti-Fade mounting medium (Vector H-1000), and coverslips were applied. The sections were examined with a Zeiss Axioskop 2 Plus biological microscope and captured using an AxioCam MRc 5 (Zeiss) with ×10 ocular magnification; data were collected using the Axiovision software package (version 4.7.0.0).
The post-embedding TEM immunogold labelling protocol was applied to fossil soft tissue matter and, separately, to C. mydas scute material. Ultrathin (90 nm) sections were collected on carbon-coated nickel grids (EMS Cat # CFT200-NI). The sections were rinsed for 10 min by placing the grids on large droplets of PBS Tween-20. They were then incubated in 4% normal donkey serum (NDS) for one hour in PBS at room temperature to occupy non-specific binding sites and prevent spurious binding. Grids were incubated in primary antibody (Polyclonal Rabbit X-Chicken Feather; Biosynthesis BSYN6734) diluted to 1:20 in primary dilution buffer (as described above) for three hours at room temperature. The grids were washed on large droplets of PBS Tween-20 for 10 × 2 min, and then incubated with secondary antibody [12 nm Colloidal Gold AffiniPure Donkey Anti-Rabbit IgG (H + L) 1:20 (Jackson Immuno Research Inc Cat # 711-205-152)] for one hour. The grids were rinsed with PBS Tween-20 for 10 × 2 min, then in E-pure water for 3 × 30 sec, and finally dried with filter paper. The sections were observed using the Aberration Corrected STEM-FEI Titan 80–300 electron microscope at the Analytical Instrumentation Facility of North Carolina State University.
Time-of-flight secondary ion mass spectrometry (ToF-SIMS)
ToF-SIMS is a surface analysis technique that provides spatially resolved molecular information from solid materials. During examination, a focussed beam of high-energy (primary) ions bombards the sample and mass spectra of the emitted (secondary) ions are recorded66. Ion images showing the spatial distribution of specific molecular species and mass spectra from selected structures are generated by scanning the primary ion beam across the sample surface.
In this study, both untreated and demineralised fossil samples were fixed for ToF-SIMS analysis on silicon wafers using either double-sided tape or Milli-Q water. Additionally, natural eumelanin from the cephalopod Sepia officinalis (Sigma-Aldrich) and hemin (MP Biomedicals) were fixed on solid substrates using double-sided tape. The fossil data were compared also to a number of other standard compounds, including various pheo- and pyomelanins20,21,22, hopanoids67, peptidoglycans (from Micrococcus luteus and Methanobacterium sp., provided by Sigma-Aldrich), and porphyrins [chlorophyll a20, protoporphyrin IX (Sigma-Aldrich), protoporphyrin X with copper (Sigma-Aldrich), uroporphyrin I dihydrochloride (Sigma-Aldrich), coproporphyrin I dihydrochloride21, and copper II phthalocyanine21].
ToF-SIMS analyses in the static SIMS mode were conducted in a TOFSIMS IV instrument (IONTOF GmbH) using 25 keV Bi3 + primary ions and low energy electron flooding for charge compensation. Positive and negative ion data were acquired with the instrument optimised either for high mass resolution (m/∆m ~5,000, spatial resolution ~3–4 µm) or high image resolution (m/∆m ~300, spatial resolution ~0.2–0.5 µm). The pulsed primary ion current was 0.10 pA for the high mass resolution data and 0.04 pA for the high image resolution data.
Infrared (IR) microspectroscopy
Fossil tissues and sediments were removed from MHM-K2 using a sterile scalpel, suspended in Milli-Q water, and placed on a sterile CaF2 infrared window and left to air dry under a hood at room temperature. IR microspectroscopic measurements were recorded at beamline D7, MAX-IV laboratory, Sweden. The set-up combines a Hyperion 3000 microscope and a Bruker IFS66/v FTIR spectrometer. The infrared microscope was operated in transmission mode using a 170 × 170 µm aperture, a single element MCT, a 100 × 100 µm detector, and a ×15 objective/condenser. This arrangement gives a visible magnification of ×215 for the video camera in the microscope, which was used to locate relevant structures in the sample.
Measurements were taken also at the Department of Biology, Lund University. Here, a Hyperion 3000 microscope combined with a Tensor 27 spectrometer was used together with a single element MCT detector (250 × 250 µm) and a Globar light source. The microscope was operated in transmission mode at 4 cm−1 resolution, and a ×15 objective was employed. 128 scans were averaged to give a good signal to noise ratio.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
References
Bonde, N., Andersen, S., Hald, N. & Jakobsen, S. L. Danekræ – Danmarks Bedste Fossiler (Gyldendal, 2008).
Dyke, G. & Lindow, B. Taphonomy and abundance of birds from the Lower Eocene Fur Formation of Denmark. Geol. J. 44, 365–373 (2009).
Nielsen, E. Eocene turtles from Denmark. Meddelelser fra Dansk Geologisk Forening 14, 96–115 (1959).
Karl, H.-V. & Lindow, B. E. K. Eocene leatherback turtle material of the genus Egyptemys (Testudines: Dermochelyoidea) from Denmark. Stud. Geol. Salmant. 46, 55–63 (2010).
Karl, H.-V. & Madsen, H. Tasbacka danica n. sp., a new Eocene marine turtle of Denmark (Testudines: Chelonioidea). Studia Palaeocheloniologica 4, 193–204 (2012).
Alibardi, L. & Toni, M. Immunolocalization and characterization of beta-keratins in growing epidermis of chelonians. Tissue Cell 38, 53–63 (2006).
Greenwold, M. J. et al. Dynamic evolution of the alpha (α) and beta (β) keratins has accompanied integument diversification and the adaptation of birds into novel lifestyles. BMC Evol. Biol. 14, 249 (2014).
Moyer, A. E. et al. Melanosomes or microbes: testing an alternative hypothesis for the origin of microbodies in fossil feathers. Sci. Rep. 4, 4233 (2014).
Moyer, A. E., Zheng, W. & Schweitzer, M. H. Keratin durability has implications for the fossil record: results from a 10 year feather degradation experiment. PLoS ONE 11, e0157699 (2016).
Lindgren, J. et al. Interpreting melanin-based coloration through deep time: a critical review. Proc. R. Soc. B. 282, 20150614 (2015).
Moyer, A. E., Zheng, W. & Schweitzer, M. H. Microscopic and immunohistochemical analyses of the claw of the nesting dinosaur, Citipati osmolskae. Proc. R. Soc. B. 283, 20161997 (2016).
Dalla Valle, L., Nardi, A., Toni, M., Emera, D. & Alibardi, L. Beta-keratins of turtle shell are glycine-proline-tyrosine rich proteins similar to those of crocodilians and birds. J. Anat. 214, 284–300 (2009).
Li, Y. I., Kong, L., Ponting, C. P. & Haerty, W. Rapid evolution of beta-keratin genes contribute to phenotypic differences that distinguish turtles and birds from other reptiles. Genome Biol. Evol. 5, 923–933 (2013).
Achrai, B. & Wagner, H. D. Micro-structure and mechanical properties of the turtle carapace as a biological composite shield. Acta Biomaterialia 9, 5890–5902 (2013).
Greenwold, M. J. & Sawyer, R. H. Molecular evolution and expression of archosaurian β-keratins: diversification and expansion of archosaurian β-keratins and the origin of feather β-keratins. J. Exp. Zool. Mol. Dev. Evol. 320, 393–405 (2013).
Perry, S. V. Vertebrate tropomyosin: distribution, properties and function. J. Muscle Res. Cell Motility 22, 5–49 (2001).
Vollmer, W., Blanot, D. & de Pedro, M. A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 (2008).
Sou, Z., Avci, R., Schweitzer, M. H. & Deliorman, M. Porphyrin as an ideal biomarker in the search for extraterrestrial life. Astrobiology 7, 605–615 (2007).
Greenwalt, D. E., Goreva, Y. S., Siljeström, S. M., Rose, T. & Harbach, R. E. Hemoglobin-derived porphyrins preserved in a Middle Eocene blood-engorged mosquito. Proc. Natl. Acad. Sci. 110, 18496–18500 (2013).
Lindgren, J. et al. Molecular preservation of the pigment melanin in fossil melanosomes. Nat. Commun. 3, 824 (2012).
Lindgren, J. et al. Skin pigmentation provides evidence of convergent melanism in extinct marine reptiles. Nature 506, 484–488 (2014).
Lindgren, J. et al. Molecular composition and ultrastructure of Jurassic paravian feathers. Sci. Rep. 5, 13520 (2015).
Muramoto, S. et al. ToF-SIMS analysis of adsorbed proteins: principal component analysis of the primary ion species effect on the protein fragmentation patterns. J. Phys. Chem. 115, 24247–24255 (2011).
Pedersen, G. K. & Buchardt, B. The calcareous concretions (cementsten) in the Fur Formation (Paleogene, Denmark): isotopic evidence of early diagenetic growth. Bull. Geol. Soc. Denmark 43, 78–86 (1996).
Iniesto, M. et al. Involvement of microbial mats in early fossilization by decay delay and formation of impressions and replicas of vertebrates and invertebrates. Sci. Rep. 6, 25716 (2016).
Solomon, S. E., Hendrickson, J. R. & Hendrickson, L. P. The structure of the carapace and plastron of juvenile turtles, Chelonia mydas (the green turtle) and Caretta caretta (the loggerhead turtle). J. Anat. 145, 123–131 (1986).
Borovanský, J., Hach, P. & Duchoň, J. Melanosome: an unusually resistant subcellular particle. Cell Biol. Int. Rep. 1, 549–554 (1977).
Borovanský, J. & Elleder, M. Melanosome degradation: fact or fiction. Pigment Cell Res. 16, 280–286 (2003).
Schweitzer, M. H. Soft tissue preservation in terrestrial Mesozoic vertebrates. Annu. Rev. Earth Planet. Sci. 39, 187–216 (2011).
Weigelt, J. Recent Vertebrate Carcasses and their Implications (University of Chicago Press, 1989).
Pedersen, G. K. et al. Molerområdets geologi – sedimenter, fossiler, askelag og glaicaltektonik. Geologisk Tidsskrift 2011, 41–135 (2011).
Lindgren, J. Fossil pigments. Curr. Biol. 26, R451–R452 (2016).
Landmann, L. In Biology of the integument. 150–187 (Springer, 1986).
Achrai, B., Bar-On, B. & Wagner, H. D. Biological armors under impact – effect of keratin coating, and synthetic bio-inspired analogues. Bioinspir. Biomim. 10, 016009 (2015).
Edwards, N. P. et al. Infrared mapping resolves soft tissue preservation in 50 million year-old reptile skin. Proc. R. Soc. B. 278, 3209–3218 (2011).
Pedersen, G. K., Pedersen, S. A. S., Steffensen, J. & Pedersen, C. S. Clay content of a clayey diatomite, the Early Eocene Fur Formation, Denmark. Bull. Geol. Soc. Denmark 51, 159–177 (2004).
Drouin, S., Boussafir, M., Robert, J.-L., Alberic, P. & Durand, A. Carboxylic acid sorption on synthetic clays in sea water: in vitro experiments and implications for organo-clay behaviour under marine conditions. Org. Geochem. 41, 192–199 (2010).
Forchielli, A., Steiner, M., Kasbohm, J., Hu, S. & Keupp, H. Taphonomic traits of clay-hosted early Cambrian Burgess Shale-type fossil Lagerstätten in South China. Palaeogeogr., Palaeoclim., Palaeoecol. 398, 59–85 (2014).
Wilson, L. A. & Butterfield, N. J. Sediment effects on the preservation of Burgess Shale-type compression fossils. Palaios 29, 145–153 (2014).
McMahon, S., Anderson, R. P., Saupe, E. E. & Briggs, D. E. G. Experimental evidence that clay inhibits bacterial decomposers: implications for preservation of organic fossils. Geology 44, 867–870 (2016).
DeNiro, M. J. & Weiner, S. Organic matter within crystalline aggregates of hydroxyapatite: a new substrate for stable isotopic and possibly other biogeochemical analyses of bone. Geochim. Cosmochim. Acta 52, 2415–2423 (1988).
Yu, B. et al. The role of clay minerals in the preservation of organic matter in sediments of Qinghai Lake, NW China. Clays Clay Min. 57, 213–226 (2009).
McNamara, M. E., Briggs, D. E. G., Orr, P. J., Field, D. J. & Wang, Z. Experimental maturation of feathers: implications for reconstructions of fossil feather colour. Biol. Lett. 9, 20130184 (2013).
Schweitzer, M. H. et al. A role for iron and oxygen chemistry in preserving soft tissues, cells and molecules from deep time. Proc. R. Soc. B. 281, 20132741 (2014).
Wiemann, J. et al. From white to black: Maillard reaction products and endogenous porphyrins stain fossil hard tissues. J. Vert. Paleontol. Program Abstr. 2016, 250 (2016).
Schweitzer, M. H. et al. Heme compounds in dinosaur trabecular bone. Proc. Natl. Acad. Sci. 94, 6291–6296 (1999).
Bustard, H. R. The adaptive significance of coloration in hatchling green sea turtles. Herpetologica 26, 224–227 (1970).
Wyneken, J. The Anatomy of Sea Turtles. U.S. Department of Commerce NOAA Technical Memorandum NMFS-SEFSC-470 (2001).
Balazs, G. H. Ontogenetic changes in the plastron pigmentation of hatchling Hawaiian green turtles. J. Herpetol. 20, 280–282 (1986).
Penacchio, O., Lovell, P. G., Cuthill, I. C., Ruxton, G. D. & Harris, J. M. Three-dimensional camouflage: exploiting photons to conceal form. Am. Nat. 186, 553–563 (2015).
Clusella Trullas, S., van Wyk, J. H. & Spotila, J. R. Thermal melanism in ectotherms. J. Therm. Biol. 32, 235–245 (2007).
Kuriyama, T., Morimoto, G., Miyaji, K. & Hasegawa, M. Cellular basis of anti-predator adaptation in a lizard with autotomizable blue tail against specific predators with different colour vision. J. Zool. 300, 89–98 (2016).
Kuriyama, T., Okamoto, T., Miyaji, K. & Hasegawa, M. Iridophore- and xanthophore-deficient melanistic color variant of the lizard Plestiodon latiscutatus. Herpetologica 72, 189–195 (2016).
McNamara, M. E. et al. Reconstructing carotenoid-based and structural coloration in fossil skin. Curr. Biol. 26, 1075–1082 (2016).
Vinther, J. A guide to the field of palaeo colour: melanin and other pigments can fossilise: reconstructing colour patterns from ancient organisms can give new insights to ecology and behaviour. BioEssays 37, 643–656 (2015).
Marshall, J. & Johnsen, S. In Animal camouflage, mechanisms and functions. 186–211 (Cambridge University Press, 2011).
Edwards, N. P., Manning, P. L. & Wogelius, R. A. Pigments through time. Pigment Cell Melanoma Res. 27, 684–685 (2014).
Vinther, J., Briggs, D. E. G., Prum, R. O. & Saranathan, V. The colour of fossil feathers. Biol. Lett. 4, 522–525 (2008).
Field, D. J. et al. Melanin concentration gradients in modern and fossil feathers. PloS ONE 8, e59451 (2013).
Spotila, J. R. Sea Turtles – a Complete Guide to their Biology, Behavior, and Conservation (Johns Hopkins University Press, 2004).
McNamara, M. E., Kaye, J. S., Benton, M. J. & Orr, P. J. Non-integumentary melanosomes can bias reconstructions of the colours of fossil vertebrate skin. Paper presented at 4th International Palaeontological Congress. The history of life: a view from the Southern Hemisphere, Mendoza, Argentina. Abstract volume: Ianigla, CCT-Conicet (September 28–October 3) (2014).
Bardet, N. et al. A giant chelonioid turtle from the Late Cretaceous of Morocco with a suction feeding apparatus unique among tetrapods. PLoS ONE 8, e63586 (2013).
Cadena, E. A. & Parham, J. F. Oldest known sea turtle? A new protostegid from the Lower Cretaceous of Colombia. PaleoBios 32, 1–42 (2015).
Witmer, L. M. In Functional morphology in vertebrate paleontology. 19–33 (Cambridge University Press, 1995).
Salmon, M., Hamann, M., Wyneken, J. & Schauble, C. Early swimming activity of hatchling flatback sea turtles Natator depressus: a test of the ‘predation risk’ hypothesis. Endang. Species Res. 9, 41–47 (2009).
Thiel, V. & Sjövall, P. In Principles and practice of analytical techniques in geosciences. 122–170 (Royal Society of Chemistry, 2014).
Leefmann, T. et al. Spectral characterization of ten cyclic lipids using time-of-flight secondary ion mass spectrometry. Rap. Commun. Mass Spectrom. 27, 565–581 (2013).
Reig, F. B., Adelantado, J. V. G. & Moreno, M. C. M. M. FTIR quantitative analysis of calcium carbonate (calcite) and silica (quartz) mixtures using the constant ratio method. Application to geological samples. Talanta 58, 811–821 (2002).
Acknowledgements
Ola Gustafsson assisted during the TEM analyses. James Parham provided information on sea turtle anatomy, whereas Edwin Cadena provided samples of an extant analogue for controls. This research was supported through a Grant for Distinguished Young Researchers (award number 642-2014-3773; Swedish Research Council) to J.L. Part of this work was performed at the Analytical Instrumentation Facility at North Carolina State University, which is supported by the State of North Carolina and National Science Foundation (award number ECCS-1542015). Further support was provided through a National Science Foundation INSPIRE grant (award number EAR-1344198) to M.H.S. and W.Z., and the National Science Foundation Graduate Research Fellowship Program (award number DGE-1252376) to A.E.M.
Author information
Authors and Affiliations
Contributions
J.L. designed the research. J.L., M.H.S. and P.S. wrote the manuscript with input from all other authors. H.M. collected and prepared MHM-K2. T.K., J.L., P.S., and J.A.G. carried out the FEG-SEM and TEM analyses, M.H.S. and W.Z. performed the immunohistochemical investigation, P.S. and J.L. conducted the ToF-SIMS experiments, and P.U., A.E. and J.L. recorded the IR microspectroscopic measurements. A.E.M. developed the chicken feather antibodies used in this study, whereas N.K. and S.U. provided reference materials.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lindgren, J., Kuriyama, T., Madsen, H. et al. Biochemistry and adaptive colouration of an exceptionally preserved juvenile fossil sea turtle. Sci Rep 7, 13324 (2017). https://doi.org/10.1038/s41598-017-13187-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-13187-5
This article is cited by
-
Taphonomic experiments reveal authentic molecular signals for fossil melanins and verify preservation of phaeomelanin in fossils
Nature Communications (2023)
-
An ancestral hard-shelled sea turtle with a mosaic of soft skin and scutes
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.