Abstract
Seeing physiological processes at the nanoscale in living organisms without labeling is an ultimate goal in life sciences. Using X-ray ptychography, we explored in situ the dynamics of unstained, living fission yeast Schizosaccharomyces pombe cells in natural, aqueous environment at the nanoscale. In contrast to previous X-ray imaging studies on biological matter, in this work the eukaryotic cells were alive even after several ptychographic X-ray scans, which allowed us to visualize the chromatin motion as well as the autophagic cell death induced by the ionizing radiation. The accumulated radiation of the sequential scans allowed for the determination of a characteristic dose of autophagic vacuole formation and the lethal dose for fission yeast. The presented results demonstrate a practical method that opens another way of looking at living biological specimens and processes in a time-resolved label-free setting.
Similar content being viewed by others
Introduction
Studies of nanoscale structures and dynamics of biological matter greatly benefit from observing samples in living state using label-free methods1. X-ray ptychography enables quantitative visualization of whole biological cells with nanoscale resolution based on the natural electron density contrast of the cell content2,3,4,5. An ideal eukaryotic model organism for cellular dynamic studies is fission yeast at the horsetail stage owing to the oscillations of meiotic chromosomes in the time scale of minutes to hours6,7,8. Moreover, intracellular structure changes caused by X-ray radiation are of interest for a direct analysis in situ 9. The major challenge of X-ray imaging of living cellular specimens is the very low lethal radiation dose, and owing to the intense radiation damage and a low electron density contrast, sequential X-ray imaging of live eukaryotic cells was not possible so far10,11,12,13. To record sufficient information, the samples have to be exposed to a certain amount of radiation, which has to be greater than the minimum of the required dose for imaging and less than the maximum tolerable dose for the specimen10. The limiting factor of the resolution is therefore set by the X-ray radiation dose10,11. Natural, aqueous environments of biological specimens significantly decrease the electron density contrast, thus higher X-ray flux is required, which consequently increases the radiation dose needed for a given resolution10. Radiation induced degradation can be reduced by chemical or cryo-fixation. Using cryo-fixation, X-ray ptychography2,4,14 and diffraction microscopy15,16 of frozen hydrated cells were accomplished. More advanced X-ray studies were realized on living cells17,18 and appear to be a promising approach to analyze cellular processes in situ. Imaging of initially alive cells simplifies sample preparation, owing to the fact that fixation steps are not required. Imaged living cells died during the first X-ray scan18 or after a free electron laser (FEL) pulse13, due to the lethal radiation dose. However, first electron density maps measured with X-rays of initially alive bacteria, which were obtained with less than a lethal dose, were recently presented12,19. X-ray ptychography is a coherent diffractive imaging (CDI) technique that combines scanning microscopy with advanced phase retrieval algorithms20,21,22. It relies on scanning the extended sample by the X-ray beam, collecting 2D diffraction patterns from a number of overlapping regions of the specimen and a subsequent iterative reconstruction of a single projection image, which is consistent with all recorded diffraction patterns21.
Results
X-ray ptychography
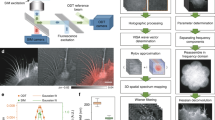
Ptychography is an X-ray imaging technique with spatial resolution limited in principle by the spatial wavelength of the incident beam and the maximum angle at which diffracted signal can be measured with sufficient signal-to-noise ratio, although in practice the resolution can be also limited by scanning precision or radiation induced damage on the specimen under study (Fig. 1). In living samples, intracellular motions are happening during a single scan which can blur the images. A ptychography setup with a pinhole-defined illumination was chosen in order to achieve good contrast and high resolution with a reduced dosage of radiation (Fig. 1A) due to the broad spatial spectrum23. We obtained reconstructed images with pixel sizes of 45 × 45 nm2 and an estimated resolution in the range of 100–200 nm of live cells in aqueous environment and reduced the radiation doses down to about 103–104 Gy per scan. These doses are two to three orders of magnitude less than 1.8 × 106 Gy recorded for recent X-ray images of (initially) alive eukaryotic cells18 and close to the dose of 8.9 × 103 Gy used for holographic imaging of living bacteria19.
Schematic representation of the experimental ptychography setup for imaging living cells. (a) For the X-ray ptychography experiments, a monochromatic (λ = 0.2 nm) beam was used to coherently illuminate a pinhole. The cell sample was scanned to collect a series of diffraction patterns from partially overlapping illuminated regions, which allow for a robust image reconstruction. The high dynamic range and count rate of the detector allows us to record the full dynamic range of the 2D diffraction patterns at the detector and avoid a loss of low spatial-frequency information that would occur if a beamstop was used. (b) A visible light bright-field optical micrograph shows three fission Schizosaccharomyces pombe yeast cells under nitrogen starvation conditions, where two of them were banana-shaped zygotes. (c) Corresponding fluorescence microscopy images of the same cells as in (b) in a time interval of 5 min are shown. In order to distinguish zygotes with moving chromosomes, ‘nuclear oscillations’, among cells with ‘non-oscillating’ ones, the rec25 gene was labeled with green fluorescent protein (GFP) and used as an indirect marker of DNA double strand breaks24. Here, only one of two zygotes was at the horsetail stage.
Here, we explore the dynamics of living fission yeast Schizosaccharomyces pombe cells during meiosis in natural, aqueous environment in situ. Fission yeast cells are ideal eukaryotic model organisms, because many of basic cellular principles and cell regulators are conserved from yeast to humans8. Meiosis in fission yeast is induced by depleting nitrogen sources from the culture medium and haploid cells of the opposite mating type conjugate and form a diploid “banana-shaped” zygote6 (Fig. 1b,c). At the horsetail stage of meiosis strong oscillations of chromosomes can be observed with extended periods of chromosomal back and forth motions along the cell axis25. The period of an individual oscillation is about 10–15 min6. After several hours, at the end of the horsetail stage, the oscillations slow down and finally stop.
X-ray induced autophagy in fission yeast cells
X-ray ptychography micrographs of a successive image sequence of a fission yeast zygote and an analysis of the impact of ionizing radiation on the cell are shown in Fig. 2. During the first four scans, no structural changes - almost homogenous density within the entire cell - of a zygote were observed. A further exposure of X-rays in the successive scans led to the appearance of clear, light and rounded structures in the zygote. These observed cellular structures may be a signature of a radiation induced formation of vacuoles26 and autophagic bodies27, which were described for fission yeast cells and might be a visual indication of autophagy28,29,30. These structural changes coincided with an overall positive shift in the phase shift histograms of zygote images showing autophagic vacuoles in comparison to zygote images without vacuoles (Fig. 2b). Increasing the radiation dose further, a bursting of the membrane and shrinkage of the cell was observed (Fig. 2a -ix,x), which demonstrates that after accumulating a certain amount of radiation the zygote perished. To characterize the dynamics of the autophagic vacuole formation and cell lysis, changes of the projected zygote area and of the projected area of individual vacuoles were analyzed (Fig. 2c). In the first four ptychography scans, a slight increase of the projected area of the zygote was observed. At an accumulated dose of about 2.2 × 104 Gy (sixth scan) for the particular scan shown in Fig. 2c, the area decreased back to its initial size and the autophagic vacuole formation set in, a first critical dose for live fission yeast cells can be defined. Applying more X-ray radiation, the initial number of vacuoles did not change, whereas their volume (projected area) increased until the cell burst. At a dose of about 9.2 × 104 Gy for this particular cell, the bursting of the cell membrane was concurred by a strong decrease of the projected zygote area. The observed formation of vacuoles can be attributed to ionizing effects of the X-ray radiation, which induce radiolysis of water and cause hydrogen peroxide (H2O2) and hydroxyl radical (OH.) formation27. These very reactive oxygen species (ROS) activate protein kinases in yeast9 and can cause autophagy31. Apart from the fact that the zygote was exposed to X-rays, yeast cells grew under nitrogen depletion, which might be a stimulus to facilitate autophagy if cells starve for several hours32. However, the autophagic vacuole formation was observed only after several ptychography scans, and was not observed in comparable experiments using optical microscopy.
X-ray ptychography images of a live fission yeast zygote. (a) Successive image sequence of a live fission yeast zygote obtained by ptychographic CDI scans. (b) Phase shift histograms (plot of the number of pixels with a specific phase shift) of a zygote in the image without autophagic vacuoles (a, iii) and with autophagic vacuoles (a, vi). A positive shift corresponds to a lighter color. (c) Projected cell area and projected vacuole area versus the radiation dose calculated from the ptychographic zygote images. Each set of grey circles connected by a line corresponds to a different vacuole and the filled circles correspond to the mean area of the vacuoles. A characteristic dose of the onset of vacuole formation and a lethal dose can be identified (red dashed lines).
In total, nine individual live fission yeast zygotes were analyzed. The average radiation dose at which autophagic vacuole formation occurred was about (3.30 ± 0.74) × 104 Gy. When the radiation dosages accumulated to (9.6 ± 3.2) × 104 Gy, zygotes were lysed, which can be defined as the lethal radiation dose for fission yeast. Interestingly, non-meiotic cells were more resistant to X-ray radiation, where vacuole formation occurred at higher doses of (8.7 ± 3.8) × 104 Gy and cell death at about (1.20 ± 0.19) × 105 Gy, indicating that the lethal radiation dose of non-meiotic cells is similar to zygotes, whereas the vacuole formation in non-meiotic cells occurs at much higher radiation doses.
X-ray imaging of the chromosome motion
To further demonstrate the potential of ptychography for studying cellular dynamics, we imaged meiotic yeast zygote at the horsetail stage. During this stage, an extended movement of the whole chromatin happens, and at the same time, the motion matches the time scales of the X-ray imaging technique. Freshly prepared samples were firstly analyzed with a fluorescence microscope to find a zygote at the horsetail stage in the sample and then mounted on the X-ray ptychography setup. A temporal sequence of six X-ray ptychography images of a live fission yeast zygote in the horsetail stage is presented in Fig. 3a. Interestingly, a darker (a more negative phase shift) and denser structure in the top part of the zygote was observed. This structure was moving during one scan to another and furthermore changed its shape. We identified this densified structure as moving chromatin, which has the same appearance as the chromosomes in the fluorescence micrograph taken before the X-ray ptychography scans (Fig. 3b). Image processed contours of the chromosomes overlaid on the original ptychography images are shown in Fig. 3c. The motion of the chromosomes was analyzed by calculating their center of mass. Starting from the initial position of the chromosomes the subsequent center of mass positions showed a movement of several hundreds of nanometer away from the upper cell end in the direction of the lower part of the cell (Fig. 3d). For further analysis, we compared the shape changes of the chromosomes over time by calculating the radius of gyration \({R}_{{\rm{G}}}=\sqrt{\frac{1}{N}\sum _{i=1}^{N}{({\overrightarrow{r}}_{i}-{\overrightarrow{r}}_{{\rm{C}}{\rm{M}}})}^{2}}\), where \(N\) is the number of pixels of the chromatin, \(\vec{{r}_{i}}\) are the position vectors and \({\overrightarrow{r}}_{{\rm{C}}{\rm{M}}}\) is the center of mass of the chromosomes. The radius of gyration, which characterizes the packing and shape of the chromosomes, versus time is plotted in Fig. 3f. Firstly, a looser packing of the chromosomes, bigger R G g, was observed, reaching a maximum of R G = (1.32 ± 0.07) µm after 10 min during their motion to the lower cell end. Afterwards, R G steadily decreased to a minimum of (0.85 ± 0.04) µm, which characterizes a strong compaction of the chromosomes. This chromosome compaction was found in conjunction with the formation of autophagic vacuoles, which could be observed after about 20 min (Fig. 3a). The motion of the chromosomes was slowed down in comparison to the observed period of the oscillation of about 10–15 min6. This observed deceleration of the chromosome motion is an indication of an impact of X-ray radiation, which most probably damages the cell at the molecular level; especially the appearance of autophagic vacuoles in concurrence with the chromosomes compaction had a strong impact on the oscillating chromosomes. Moreover, due to a time-consuming sample preparation and mounting procedure, the X-ray ptychography images were presumably taken at the end of the horsetail period, when the oscillations slowed down. The appearance of autophagic vacuoles was observed, when the radiation dose accumulated to about 1.2 × 104 Gy. This dose of the vacuole formation is less than half of the dose, which was found for normal yeast zygotes, which might be an indication that zygotes in the horsetail stage are more sensitive to X-ray radiation.
X-ray ptychography images of chromosomes motion in a live fission yeast zygote at the horsetail stage. (a) Temporal image sequence of a live zygote in the horsetail stage obtained by X-ray ptychography. (b) Fluorescence micrograph of the zygote taken before the X-ray ptychography scans. (c) Overlay of image processed contours of the chromosomes (dark blue) on the original ptychography images. (d) Center of mass motion of chromosomes between the sequential ptychographic images. The x and y-position of the center of mass of the first scan at 0 min was set to 0,0. (e) Calculated radius of gyration of the chromosomes versus time. (f) Schematic representation of autophagic vacuoles formation and cell lysis during meiotic chromosome oscillations.
Conclusions
In these experiments, we optimized the sample preparation and experimental setup to successfully apply ptychography for sequential imaging – producing X-ray movies – of live meiotic yeast cells in aqueous environment and established a method to investigate intracellular nanostructures. Based on the natural electron density contrast, this label-free imaging method allowed us to visualize cellular structures in situ. We discovered autophagic cell death or type II programmed cell death28 and cell lysis, induced by the pathological environment due to the ionizing X-ray radiation. Autophagy in the yeast and mammalian cells is similar30 and it is considered to play an important part in the response of radiation therapy33. Radiation-induced damage on the molecular level most probably occurred already during the first X-ray scan, but it did not cause visible cell changes and the cell stayed alive. Thus an average radiation dose at which visible signs of autophagy occurs by formation of autophagic vacuoles32 and a characteristic dose of membrane bursting of the eukaryotic cells can be obtained. Moreover, the dynamics of denser structures, which are most likely chromatin structures at the end of their oscillatory motion during the horsetail stage of yeast zygotes, can be imaged and analyzed. However, zygotes are most likely already damaged at the molecular level by radiation. This imaging approach also simplifies sample preparation and avoids artifact formation caused by fixation, sectioning or labeling.
We believe that improved sample environments, e.g. microfluidics setup34 to flush fresh medium and remove free radicals35, modified scanning protocols and adjusted ranges of interest will further advance, as here demonstrated, the way of seeing physiological processes of individual eukaryotic cells as well as tissues with subcellular, nanoscale resolution.
Methods
In order to avoid a strong background signal and to create a cytocompatible environment, we used biocompatible and X-ray resistant 200 nm thick silicon nitride (Si3N4) membrane windows (frame: 5 × 5 mm2 × 200 µm, membrane: 1.5 × 1.5 mm2 × 200 nm; Silson Ltd, Blisworth, England)17,18,34. The Si3N4 membrane windows were coated with lectin (Sigma-Aldrich, St. Louis, MO, USA) to increase cell adhesion to the membrane surface, which is crucial for the spatial stability of the cells and thus for the reproducibility of X-ray ptychography scans. Fission yeast Schizosaccharomyces pombe cells were kept in phosphate-buffered saline (PBS) medium at room temperature. For fluorescence optical imaging, we used the genotype of the fission yeast strain h90 rec25::GFP-KanMX6. The strain was a kind gift from C. Martín-Castellanos (CM62, IBFG, Salamanca, Spain). To induce meiosis, fission yeast cells were transferred to an Eppendorf tube with 100 µl of nutrition deficient Edinburgh minimal medium (EMM-N)7 and kept for 30 min at room temperature. Afterwards, a small droplet of medium with cells were put on a lectin-coated Si3N4 membrane window and the device was covered by an uncoated Si3N4 membrane window and accurately glued with UHU plus epoxy quick set adhesive at the edges of the membranes. The described procedure enables the preparation of hydrated living cell samples for X-ray experiments with an intercalated aqueous film of 5 to 10 µm in thickness. The gap was determined based on bright field optical imaging where the distance between the membranes was comparable to the size of the cell as observed by changing the focus. Since the aqueous environment drastically decreases the electron density contrast of the sample, a small sample thickness is crucial to reduce the background signal caused by the medium in the device.
X-ray ptychography experiments were performed at the coherent small-angle X-ray scattering (cSAXS) beamline of the Swiss Light Source, Paul Scherrer Institut, PSI, Villigen, Switzerland. The schematic representation of the setup is shown in Fig. 1a. An X-ray beam of 6.2 keV photon energy, λ = 0.2 nm, was selected using a double crystal Si (111) monochromator. The incident beam was defined by a pinhole with a transverse diameter of about 2.5 µm in order to obtain a coherent spatially confined illumination at the sample, which was placed 3 mm downstream of the pinhole and had at the sample position approximately the same diameter. The sample of hydrated live cells was placed on a piezoelectric scanning stage to allow for nanometer precision scanning. The coherent X-ray beam diffracted by the sample propagates through a helium flushed flight tube to a photon-counting Pilatus 2D detector36, which is located at distance of 7.412 m from the sample. The broad angular spectrum of a pinhole-defined illumination is well suited for minimizing the radiation dose while acquiring images with good contrast and a moderately high resolution23. Before the X-ray ptychography experiments, the cell samples were imaged by fluorescent microscopy in order to identify oscillating zygotes. The membranes were then mounted on the setup and the identified cells were positioned using an on-stage bright field microscope.
In order to avoid the raster grid pathology37 all scans were performed following a Fermat spiral scanning pattern38. In order to find optimal scanning parameters, different step sizes and exposure times were applied. For the measurements in Fig. 2a a scanning field of view of 18 × 14 µm2 and an average step size of 0.7 µm were used with an exposure time of 0.1 s per scanning point. For these parameters the resolution was about 200 nm with an average flux of about 7.4 × 105 photons/µm2. To calculate the average flux we first normalized the reconstructed illumination intensity using the total number of counts arriving at the detector after compensating for absorbing and scattering elements in the path of the beam, then we used the scanning pattern to generate a grid of the distribution of photons incident on the sample for the whole scan. The flux in photons/µm2 is finally calculated by integrating over an area significantly larger than the illumination and dividing by the area, in this manner we included in the calculation the total dose incident on the sample including the overlapping regions of the scan39. The corresponding dose of 3.9 × 103 Gy was estimated as described in ref.16: The dose, D, is calculated based on the surface dose equation \({D}=\mu {N}_{0}h\nu /\rho \) 10, where the attenuation length, 1/μ, was obtained from tabulated values40 assuming an average composition of H128C30N9O49S1 and average density of ρ = 1 g cm−3.
In order to observe the cell behavior (death or ability to recover) between the X-ray scans, the time interval among the images was different: 5 min, 25 min, 20 min, 20 min, 50 min, 30 min, 40 min, 2 h, 3 h. This allows us to assume that the cell death is initiated by the X-ray radiation and does not occur/continue when the X-rays are switched off. The radiation doses used for single ptychography scans in Fig. 2a were different, the 1st scan 1.96 × 103 Gy, 2nd scan 4.01 × 103 Gy, 3rd scan 3.87 × 103 Gy, 4th scan 3.86 × 103 Gy, 5th scan 4.09 × 103 Gy, 6th scan 3.87 × 103 Gy, 7th scan 4.14 × 103 Gy, and 10th scan 8.20 × 103 Gy. The quality of the images in Fig. 2a(viii, ix) and resolution down to about 100 nm was improved using smaller step sizes of 0.5 µm and longer exposure times of 0.4 s, which increased the radiation dose to about 3.3 × 104 Gy.
The images in Fig. 3A were obtained with an exposure time of 0.1 s per scanning point and an average step size of 1 µm. The flux at the sample position was 4.2 × 105 photons/µm2 for the individual scans, corresponding to an estimated resolution of about 100 nm. The radiation doses per scan were 1.93 × 103 Gy (1st), 1.83 × 103 Gy (2nd), 2.71 × 103 Gy (3rd), 2.70 × 103 Gy (4th), 2.76 × 103 Gy (5th), and 2.74 × 103 Gy (6th).
Reconstructions were carried out using the maximum likelihood method through non-linear optimization20,22. In order to reduce the noise in the reconstructions, gradient preconditioning and regularization, as described by Thibault and Guizar-Sicairos22, were used. A good estimate of the incident illumination is important to facilitate the reconstruction of weak contrast specimens41, such as the hydrated live cells presented here. For this purpose, we characterized the incident illumination via ptychography before the experiments using a 2D test patterns similar to those used in42. The illumination phase and amplitude profile were stable for the duration of a single scan. For this case, the resolution of the reconstruction could not be assessed via Fourier shell correlation (FSC)43,44, because two identical datasets were not available due to changes or movement of the live specimens. To assess the resolution of each image we used instead a method based on the angular-averaged power spectral density (PSD) method as described in14.
X-ray ptychography images were analyzed using ImageJ (version 1.47k, Wayne Rasband, National Institute of Health, USA) and MATLAB (version R2012b, The MathWorks, Natick, USA) by applying custom developed scripts. The images were first denoised by conditional mean filtering resulting in an edge preserving smoothing. Further applying edge detection algorithms yield the contour of the cells, which acts as the range of interest in order to find the chromosomes by local thresholding.
References
Landecker, H. Seeing things: from microcinematography to live cell imaging. Nat. Methods 6, 707–709 (2009).
Deng, J. et al. Simultaneous cryo X-ray ptychographic and fluorescence microscopy of green algae. PNAS 112, 2314–2319 (2015).
Miao, J., Ishikawa, T., Robinson, I. K. & Murnane, M. M. Beyond crystallography: Diffractive imaging using coherent x-ray light sources. Science 348, 530–535 (2015).
Diaz, A. et al. Three-dimensional mass density mapping of cellular ultrastructure by ptychographic X-ray nanotomography. J. Struct. Biol. 192, 461–469 (2015).
Heḿonnot, C. J. et al. X‐rays Reveal the Internal Structure of Keratin Bundles in Whole Cells. ACS Nano 10, 3553–3561 (2016).
Chikashige, Y. et al. Telomere-led premeiotic chromosome movement in fission yeast. Science 264, 270–273 (1994).
Forsburg, S. L. & Rhind, N. Basic methods for fission yeast. Yeast 23, 173–183 (2006).
Patterson, J. O., Swaffer, M. & Filby, A. An Imaging Flow Cytometry-based approach to analyse the fission yeast cell cycle in fixed cells. Methods 82, 74–84 (2015).
Spitz, D. R., Azzam, E. I., Li, J. J. & Gius, D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: A unifying concept in stress response biology. Cancer Metastasis Rev. 23, 311–322 (2004).
Howells, M. R. et al. An assessment of the resolution limitation due to radiation-damage in X-ray diffraction microscopy. J. Electron Spectros. Relat. Phenomena 170, 4–12 (2009).
Schneider, G., Guttmann, P., Rehbein, S., Werner, S. & Follath, R. Cryo X-ray microscope with flat sample geometry for correlative fluorescence and nanoscale tomographic imaging. J. Struct. Biol. 177, 212–223 (2012).
Kimura, T. et al. Imaging live cell in micro-liquid enclosure by X-ray laser diffraction. Nat. Commun. 5, 3052 (2014).
van der Schot, G. et al. Imaging single cells in a beam of live cyanobacteria with an X-ray laser. Nat. Commun. 6, 5704 (2015).
Giewekemeyer, K. et al. Quantitative biological imaging by ptychographic x-ray diffraction microscopy. PNAS 107, 529–534 (2010).
Weinhausen, B. et al. X-ray nano-diffraction on cytoskeletal networks. New J. Phys. 14, 085013 (2012).
Lima, E. et al. Cryo-scanning x-ray diffraction microscopy of frozen-hydrated yeast. J. Microsc. 249, 1–7 (2013).
Sedlmair, J. et al. Imaging of Vascular Smooth Muscle Cells with Soft X-Ray Spectromicroscopy. Microsc. Microanal. 17, 991–1001 (2011).
Weinhausen, B. et al. Scanning X-ray nanodiffraction on living eukaryotic cells in microfluidic environments. Phys. Rev. Lett. 112, 88102 (2014).
Bartels, M., Krenkel, M., Haber, J., Wilke, R. N. & Salditt, T. X-Ray Holographic Imaging of Hydrated Biological Cells in Solution. Phys. Rev. Lett. 114, 48103 (2015).
Guizar-Sicairos, M. & Fienup, J. R. Phase retrieval with transverse translation diversity: a nonlinear optimization approach. Opt. Express 16, 7264–7278 (2008).
Thibault, P. et al. High-Resolution Scanning X-ray Diffraction Microscopy. Science 321, 379–383 (2008).
Thibault, P. & Guizar-Sicairos, M. Maximum-likelihood refinement for coherent diffractive imaging. New J. Phys. 14, 63004 (2012).
Guizar-Sicairos, M. et al. Role of the illumination spatial-frequency spectrum for ptychography. Phys. Rev. B 86, 100103 (2012).
Fowler, K. R., Gutiérrez-Velasco, S., Martín-Castellanos, C. & Smith, G. R. Protein Determinants of Meiotic DNA Break Hotspots. Mol. Cell 49, 983–996 (2014).
Vogel, S. K., Pavin, N., Maghelli, N., Jülicher, F. & Tolić-Nørrelykke, I. M. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 7, e1000087 (2009).
Weisman, L. S. Organelles on the move: insights from. Nat. Rev. Mol. Cell. Biol. 7, 243–252 (2006).
Li, L., Ishdorj, G. & Gibson, S. B. Reactive oxygen species regulation of autophagy in cancer: Implications for cancer treatment. Free Radic. Biol. Med. 53, 1399–1410 (2012).
Pyo, J. O. et al. Essential roles of Atg5 and FADD in autophagic cell death: Dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 280, 20722–20729 (2005).
Suzuki, K. & Ohsumi, Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581, 2156–2161 (2007).
Tsukada, M. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 (1993).
Mahmoud, B. S. M. The effects of X-ray radiation on Escherichia coli O157: H7, Listeria monocytogenes, Salmonella enterica and Shigella fl exneri inoculated on whole Roma tomatoes. Food Microbiol. 27, 1057–1063 (2010).
Mizushima, N. Autophagy: process and function. Genes Dev. 21, 2861–2873 (2007).
Palumbo, S. & Comincini, S. Autophagy and ionizing radiation in tumors: The ‘survive or not survive’ dilemma. J. Cell. Physiol. 228, 1–8 (2013).
Köster, S. & Pfohl, T. X-Ray Studies of Biological Matter in Microfluidic Environments. Mod. Phys. Lett. B 26, 1230018 (2012).
Weinhausen, B. & Köster, S. Microfluidic devices for X-ray studies on hydrated cells. Lab Chip 13, 212–215 (2012).
Henrich, B. et al. PILATUS: A single photon counting pixel detector for X-ray applications. Nucl. Instr. Meth. Phys. Res. 607, 247–249 (2009).
Thibault, P., Dierolf, M., Bunk, O., Menzel, A. & Pfeiffer, F. Probe retrieval in ptychographic coherent diffractive imaging. Ultramicroscopy 109, 338–343 (2009).
Huang, X. et al. Optimization of overlap uniformness for ptychography. Opt. Express 22, 12634–12644 (2014).
Wilke, R. N. et al. Hard X-ray imaging of bacterial cells: nano-diffraction and ptychographic reconstruction. Opt. Express 20, 4795–4797 (2012).
Gullikson, E. http://henke.lbl.gov/optical_constants/.
Dierolf, M. et al. Ptychographic coherent diffractive imaging of weakly scattering specimens. New J. Phys. 12, 035017 (2010).
Holler, M. et al. X-ray ptychographic computed tomography at 16 nm isotropic 3D resolution. Scientific Reports 4, 3857 (2014).
Van Heel, M. & Schatz, M. Fourier shell correlation threshold criteria. J. Struct. Biol. 151, 250–262 (2005).
Vila-Comamala, J. et al. Characterization of high-resolution diffractive X-ray optics by ptychographic coherent diffractive imaging. Opt. Express 19, 175–184 (2011).
Acknowledgements
X-ray measurements were carried out at the cSAXS beamline of the Swiss Light Source, Paul Scherrer Institut (PSI), Villigen, Switzerland. We thank Prof. Kurt Ballmer for the use of his fluorescence microscope at PSI during our X-ray ptychography measurements, C. Martin-Castellanos for the kind gift of the Rec25-GFP strain, Zoe Swank and Raphael Urbani for their support during synchrotron experiments. Financial support by the Swiss National Science Foundation (SNF_200020_141270) and Swiss Nanoscience Institute is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
N.S., M.G.-S., A.D., V.Z., and T.P. conceived the research work and designed the experiments. N.S., N.S. (II), M.G.-S., A.D., P.D., V.Z., and T.P. performed the experiments. P.D., M.C. and I.M.T. contributed and modified samples and protocols. N.S., M.G.-S., M.G., V.Z., and T.P. analyzed the data. N.S., M.G.-S., V.Z., and T.P. wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Strelnikova, N., Sauter, N., Guizar-Sicairos, M. et al. Live cell X-ray imaging of autophagic vacuoles formation and chromatin dynamics in fission yeast. Sci Rep 7, 13775 (2017). https://doi.org/10.1038/s41598-017-13175-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-13175-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.