Abstract
Burrowing nematodes (Radopholus similis) cause severe harm in many agronomic and horticultural crops and are very difficult to manage. Cathepsin S is one of the most important cysteine proteinases and plays key roles in nematodes and many other parasites. To evaluate the effect of in planta RNAi on the control of this nematode, a specific fragment from the protease gene, cathepsin S (Rs-cps), was cloned into the binary vector pFGC5941 in the forward and reverse orientations to construct recombinant plant RNAi vectors. Transgenic Nicotiana benthamiana plants expressing Rs-cps dsRNA were obtained and studied. The transcript abundance of Rs-cps dsRNA appeared to be diverse in the different transgenic lines. Moreover, the bioassay results revealed that Rs-cps transgenic N. benthamiana plants were resistant to R. similis and the transcription level of Rs-cps in R. similis was drastically decreased. In addition, the reproduction and hatching rate of R. similis isolated from the Rs-cps transgenic plants were also significantly reduced. Our results suggest that Rs-cps is essential for the reproduction and pathogenicity of R. similis. This is the first study to employ in planta RNAi approach to target the Rs-cps gene for the control of plant parasitic nematodes.
Similar content being viewed by others
Introduction
Radopholus similis is a migratory endoparasitic nematode that is known to be a destructive pest of bananas, peppers, coffee, citrus crops and many other agronomic and horticultural crops1,2,3,4. Both juvenile and adult nematodes can enter and leave root tissues and feed on the cytoplasm of cortex cells. Infection by R. similis deprives host plants of essential nutrients, whilst entry wounds make host roots more susceptible to other pathogens present in the soil. This destruction of crops leads to significant growth reduction and severe economic losses5,6. Although R. similis does great damage to agriculture, effective measures to control it are still lacking. The focus of control strategies for plant parasitic nematodes has depended on the application of expensive and environmentally unfriendly chemical nematicides7,8. Therefore, it is particularly important to explore effective approaches for controlling the nematode. One such strategy involves the application of plant-mediated RNA interference (in planta RNAi), which confers resistance to plants engineered to express specific dsRNA to target and silence specific genes involved in the reproduction, development, parasitism and pathogenesis of nematodes.
RNAi is an effective gene-silencing mechanism in eukaryotes, which was first discovered in Caenorhabditis elegans, and has provided a significant new tool to study gene function in many organisms including nematodes, plants, fungi, insects and mammals8,9,10,11,12,13,14,15,16. The dsRNA produced by transgenic plants against key pest genes has been regarded as a safeguard that endows transgenic pest-resistant plants with new innovations17,18. In recent years, in planta RNAi has emerged as an efficient tool to research gene functions and manage crop pathogens17,18,19,20,21,22,23,24. It also has been used to study gene functions in plant parasitic nematodes. The first successful demonstration of in planta RNAi was accomplished by targeting splicing factors and integrase genes of Meloidogyne incognita. Transgenic Nicotiana benthamiana plants constitutively expressing special dsRNA of these genes resulted in a reduction of root knots25. In another report, transgenic Arabidopsis expressing 16D10 dsRNA had a wide resistance against four major root-knot nematode species19. Some of the nematode genes were knocked down using in planta RNAi, causing reduction in the parasitic success of cyst and root-knot nematodes in different plants26. However, there are limited reports about the use of in planta RNAi in research against migratory plant parasitic nematodes27,28,29. Therefore, the further use of the in planta RNAi method to research the functions of parasitic or pathogenesis-related genes will enrich the understanding of the use of RNAi against migratory endoparasites and lay the foundation for the control of these nematodes.
Cysteine proteinases are essential for a wide range of physiological processes in all living organisms30. Cysteine proteinases play key roles in embryogenesis, development, invasion, parasitism and evasion of host immune responses, and most of them are the main digestive enzymes in the intestines of nematodes and many other animal parasites16,31. Therefore, these were identified as the primary targets for the control of parasites. In parasitic helminths, the papain superfamily of cysteine proteinases (i.e., cathepsins) has drawn the most attention32. According to the absence and presence of a distinctive set of amino acids within the polypeptide, there are more than 10 cathepsins within the cysteine proteinase family, including cathepsin B, C, L, S, F, K, and Z33. The most studied are cathepsin B and cathepsin L-like proteases, which have been studied in many parasitic nematodes in recent years. For the functional analysis of cathepsin genes in plant parasitic nematodes, special dsRNA delivery was accomplished by soaking the nematodes in a dsRNA solution (in vitro RNAi). As reported by Li et al., silencing of the cathepsin B gene (Rs-cb-1) using in vitro RNAi not only significantly inhibited the reproduction and development of R. similis but also greatly reduced its pathogenicity29,34. Targeting the cysteine proteinases of Globodera pallida gpcp-I and Heterodera glycines hgcp-I led to a decreased recovery of egg-laying females and altered sexual fate11. When in vitro RNAi was used to research the gene function of Mi-cpl-1 in M. incognita, a reduction in gene transcript abundance was observed and the number of nematodes infecting plants was reduced by almost 60% at 21 days post-infection35. In our previous studies, we demonstrated that the cathepsin S gene of R. similis (Rs-cps) plays important roles in reproduction and pathogenesis using in vitro RNAi16. However, the functions of cathepsin S gene (cps) have rarely been researched, and only the cps genes of H. glycines and H. avenae have been cloned in other plant nematodes30,31.
Despite reports of in vitro RNAi studies, cps genes of plant parasitic nematodes have not yet been targeted using the in planta RNAi approach. At the same time, Rs-cps gene plays important roles in the reproduction and pathogenesis of R. similis 16. Therefore, we selected Rs-cps gene as a promising target for in planta RNAi research to control R. similis. In this study, the plant expression vector pFGC-Rs-cps2 was constructed, which can generate a hairpin RNAi construct. Nicotiana benthamiana transgenic lines producing Rs-cps dsRNA were generated from transformed callus tissues by Agrobacterium-mediated transformation. Putative transgenic N. benthamiana plants were detected by PCR, Southern blot and RT-PCR. In addition, single-copy Rs-cps transgenic plants were chosen for resistance studies. The feeding bioassay clearly showed that the resistance of Rs-cps transgenic N. benthamiana plants to R. similis was significantly improved, and the transcription level of Rs-cps in R. similis was drastically suppressed. The reproduction and hatching of R. similis isolated from Rs-cps transgenic N. benthamiana plants were also significantly inhibited. This is the first study to employ in planta RNAi targeting of the Rs-cps gene to control the plant parasitic nematode.
Results
Construction of plant RNAi expression vectors and production of transgenic plants
A 438-bp partial cDNA fragment of Rs-cps was chosen as the target for RNAi (Fig. 1A). The constructed plant RNAi vector pFGC-Rs-cps2 contained a 438-bp sense and antisense Rs-cps cDNA fragment, a CHSA intron, an OCS terminator, and the cDNA fragments as inverted repeats under the control of CaMV35S promoter to produce the hairpin Rs-cps dsRNA (Fig. 1B). The constructed vectors were introduced into Agrobacterium tumefaciens strain EHA105 via the freeze-thaw method. The aseptic seedling was cut into small pieces of approximately 0.5 cm × 0.5 cm after four leaves grew, and the veins were removed. After pre-culture, the plant RNAi vectors were introduced into N. benthamiana explants by Agrobacterium-mediated transformation. The transgenic seedlings germinated from transformed N. benthamiana calli were selected for kanamycin resistance, and then, the seedlings were transferred into the rooting medium when they grew to approximately 2 cm. After acclimatization, the transgenic plantlets were transplanted into sterilized nutritive soil in a greenhouse for normal growth. We obtained a total of 26 independent kanamycin-resistant transgenic plants, including 15 Rs-cps transgenic plants and 11 egfp transgenic plants. These transgenic plants had wild-type N. benthamiana morphology and growth (result not shown).
Target sequence of the Rs-cps gene for RNA interference and plant RNAi vector for Nicotiana benthamiana genetic transformation. (A) The sequence of Rs-cps was used for RNAi target sequences in this study. Blue font: protective bases; the specific primers are indicated in boxes. (B) Schematic representation of the plant RNAi vector expressing hairpin Rs-cps dsRNA in transgenic N. benthamiana plants.
Molecular analysis of Rs-cps transgenic N. benthamiana plants
The independently generated T0 generation transgenic lines were analysed by PCR. A 467-bp DNA fragment of the target gene was amplified from most of the putative T0 generation Rs-cps transgenic plants, and a 315-bp DNA fragment was amplified from most of the putative egfp transgenic plants, while no specific band was amplified from the wild-type N. benthamiana plants (Fig. 2A,B). These results showed that the target DNA fragment was successfully inserted into the N. benthamiana genomic DNA. Although 13 of the 15 the kanamycin-resistant transgenic plants test positive for the RNAi construct in PCR, some detected plants showed nonspecific bands. Therefore, we only select part of transgenic plants (Nos. 3, 4, 6, 7, 12, 13 and 14) with specific bands for the Southern blot. The results showed that these Rs-cps transgenic plants had one to four insertion loci (Plants No. 4 and 13 had a single insertion locus, plants No. 6 and 14 had two insertion loci, plants No. 7 and 12 had three insertion loci, and plant No. 3 had four insertion loci), but no Rs-cps hybridization bands were observed with genomic DNA from the egfp transgenic plants and wild-type N. benthamiana plants (Fig. 2C). RT-PCR was performed to detect the expression of Rs-cps dsRNA in positive transgenic N. benthamiana lines (Nos. 3, 4, 6, 7, 12, 13 and 14). A 467-bp fragment corresponding to the sequence of Rs-cps was amplified from these positive Rs-cps transgenic plants, while no specific amplification was observed from the wild-type N. benthamiana plants (Fig. 2D). These results indicated that the integrated Rs-cps dsRNA was successfully expressed in transgenic N. benthamiana plants. Genetic stability analysis indicated that the integrated Rs-cps could be inherited steadily in the genomic DNA of the T1 generation transgenic N. benthamiana plants (result not shown).
Molecular analysis of the putative transgenic Nicotiana benthamiana plants. (A) PCR analysis for putative Rs-cps transgenic plants using the primers RNAi-F/RNAi-R (lanes 1-15: different Rs-cps transgenic lines). (B) PCR analysis for putative egfp transgenic plants using the primers eGFP-F/eGFP-R (lanes 1-11: different egfp transgenic lines). (C) Southern blot analysis of EcoRI-digested genomic DNA from leaves of the T0 generation transgenic plants (lanes 3, 4, 6, 7, 12, 13 and 14: DNA from the 3, 4, 6, 7, 12, 13 and 14 Rs-cps transgenic lines; lanes e and W: DNA from the egfp transgenic plant and wild-type N. benthamiana plant as the control). (D) Different transgenic plants were analysed by RT-PCR using the primers RNAi-F/RNAi-R (lanes 3, 4, 6, 7, 12, 13 and 14: RNA from the 3, 4, 6, 7, 12, 13 and 14 Rs-cps transgenic N. benthamiana plants). M, DL2000 DNA marker; P, positive plasmid control; B, blank control without template; W, wild-type N. benthamiana plant (negative control).
Expression analysis of Rs-cps in T2 generation transgenic lines
To detect the expression of the Rs-cps dsRNA transcript and its abundance, qPCR analysis of the selected single copy T2 generation transgenic lines (No. 4 and 13) was carried out. The result showed that Rs-cps dsRNA was expressed in two positive transgenic lines, while the transcript abundance of dsRNA varied between the two lines. The Rs-cps dsRNA expression level in line No. 4 was 3.07 times higher, which was significant (p < 0.05), than that in line No. 13 of the Rs-cps transgenic N. benthamiana plants (Fig. 3). Previous studies have shown that the effectiveness of RNAi is higher in single-copy lines than in other lines36,37. Therefore, the single-copy Rs-cps transgenic line (No. 4) with a relatively high expression level of dsRNA was selected for a nematodes feeding bioassay to analyse the reproduction and pathogenicity of R. similis and the efficiency of RNAi.
Expression analysis of Rs-cps in T2 generation transgenic plants. The mRNA abundance was determined by qPCR, and Actin was amplified as a reference gene. Bars indicate the standard errors of the mean data (n = 3), and different letters indicate significant differences (p < 0.05) between treatments. CPS-N4 and CPS-N13: No. 4 and No. 13 Rs-cps transgenic Nicotiana benthamiana plants; WT, wild-type N. benthamiana plants.
T2 generation transgenic N. benthamiana plants expressing Rs-cps dsRNA show enhanced resistance to R. similis
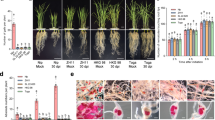
To evaluate whether the resistance to R. similis was improved in T2 generation Rs-cps transgenic plants in comparison with egfp transgenic plants and wild-type plants, nematodes were introduced to the roots of N. benthamiana plants. At 75 d, it was evident that the transgenic plants expressing Rs-cps dsRNA exhibited much higher resistance to R. similis than the control groups (egfp transgenic plants and wild-type N. benthamiana plants). The control N. benthamiana plants were smaller and had fewer tillers than the Rs-cps transgenic plants. There were no obvious infection symptoms aboveground for the Rs-cps transgenic plants compared with the uninoculated wild-type N. benthamiana plants (Fig. 4). The pot experiment results showed that the plant height, fresh above-ground plant weight and fresh root weight of the Rs-cps transgenic plants (No. 4) were significantly higher than those in the control groups (p < 0.05). There was a significant difference in the fresh root weight between uninoculated wild-type and Rs-cps transgenic N. benthamiana plants (p < 0.05); however, no significant differences in the plant height and fresh above-ground plant weight were observed between them (p > 0.05). There was no significant difference in the three growth parameters between the control groups (p > 0.05) (Fig. 5A–C). Additionally, the number of nematodes in the rhizosphere of the Rs-cps transgenic plants was 1158, which was significantly lower than that of the egfp transgenic plants (3954) and wild-type N. benthamiana plants (4182) (p < 0.05) (Fig. 5D). The pot inoculation trials clearly demonstrated resistance to R. similis in the T2 generation Rs-cps transgenic N. benthamiana plants was significantly improved.
Infection symptoms of T2 generation transgenic plants after being inoculated with R. similis for 75 days. The selected Nicotiana benthamiana plantlets were approximately 20 cm in height, and each plantlet was inoculated with 2000 mixed stage nematodes and cultivated in a greenhouse. Seventy-five days after inoculation, Rs-cps transgenic plants with less damage exhibited higher resistance to R. similis than the control N. benthamiana plants. CK, uninoculated wild-type N. benthamiana plants; CPS, No. 4 Rs-cps transgenic plants; GFP, egfp transgenic plants; WT, wild-type N. benthamiana plants.
Resistance to R. similis in T2 transgenic Nicotiana benthamiana plants expressing Rs-cps dsRNA was significantly improved. Plant height (A), fresh above-ground plant weight (B), fresh root weight (C) and the number of nematodes in the rhizosphere (D) of the different N. benthamiana plants after being inoculated with 2000 mixed stage nematodes for 75 days. Bars indicate the standard errors of the mean data (n = 5), and different letters indicate significant differences (p < 0.05) between treatments. CK, uninoculated wild-type N. benthamiana plants; CPS, No. 4 Rs-cps transgenic plants; GFP, egfp transgenic plants; WT, wild-type N. benthamiana plants.
Transcription of Rs-cps in tested R. similis was dramatically suppressed by T2 generation transgenic N. benthamiana plant-derived Rs-cps dsRNA
We assume that the significant reduction in pathogenicity of R. similis resulted from suppression of its Rs-cps mRNA levels by feeding on transgenic plants expressing Rs-cps dsRNA. To confirm this hypothesis, the Rs-cps mRNA levels in R. similis isolated from the transgenic N. benthamiana roots were detected by qPCR. The results showed that (Fig. 6) the transcription level of Rs-cps in R. similis isolated from transgenic N. benthamiana plants expressing Rs-cps dsRNA was significantly lower (p < 0.05) than that from the control groups (egfp transgenic plants and wild-type N. benthamiana plants) and was as much as 74.5% lower than that from wild-type N. benthamiana plants. There was no significant (p > 0.05) difference in the Rs-cps expression between the two control groups. Taken together, we conclude that the suppression of Rs-cps expression in R. similis by feeding on the roots of transgenic plants expressing Rs-cps dsRNA causes weaker pathogenic ability.
Rs-cps expression in R. similis was significantly suppressed by transgenic Nicotiana benthamiana plants-derived Rs-cps dsRNA. The mRNA abundance was determined by qPCR analysis, and Actin was amplified as a reference gene. Bars indicate the standard errors of the mean data (n = 3), and different letters indicate significant differences (p < 0.05) among groups. CPS, No. 4 Rs-cps transgenic plants; GFP, egfp transgenic plants; WT, wild-type N. benthamiana plants.
Transgenic N. benthamiana plants expressing Rs-cps dsRNA inhibited the reproduction and hatching of R. similis
As described previously, the reproductive capability of R. similis treated with Rs-cps dsRNA (in vitro RNAi) was significantly decreased compared with the control groups16, and the number of nematodes in the rhizosphere of the Rs-cps transgenic plants was significantly lower than that of the control groups. To evaluate the effect of transgenic N. benthamiana plant-derived dsRNAs (in planta RNAi) on the reproduction and hatching of R. similis, the nematodes were inoculated onto carrot callus. After being cultured for 50 d, the nematodes isolated from the roots of Rs-cps transgenic N. benthamiana plants exhibited significantly lower reproduction than those from the roots of egfp transgenic and wild-type N. benthamiana plants. The number of nematodes in the Rs-cps transgenic N. benthamiana groups (CPS) was 2136, which was significantly (p < 0.05) lower than those in the egfp transgenic N. benthamiana groups (16296) and wild-type N. benthamiana groups (15536), but there was no significant (p > 0.05) difference observed between the latter two groups (Fig. 7A). In terms of the infection symptoms, the carrot callus presented brown hygrophanous lesions because of the reproduction of a large number of nematodes derived from the roots of egfp transgenic and wild-type N. benthamiana plants. The carrot callus inoculated with nematodes derived from Rs-cps transgenic N. benthamiana roots only presented a slightly brown infection spot, and the brown hygrophanous lesion was not observed in this experiment (Fig. 7C–F). The hatching rate of eggs derived from Rs-cps transgenic N. benthamiana plants was 32%, which was significantly lower (p < 0.05) than that in the control groups (eggs derived from egfp transgenic and wild-type N. benthamiana plants). The hatching rates of eggs derived from the egfp transgenic and wild-type N. benthamiana plants were 88% and 90%, respectively, and there was no significant difference (p > 0.05) between these groups (Fig. 7B). All these results suggested that the Rs-cps transgenic plant-derived dsRNAs not only inhibited reproduction but also suppressed the hatching of R. similis.
The reproduction and hatching of R. similis were decreased significantly by Rs-cps transgenic Nicotiana benthamiana plant-derived dsRNA. (A) The number of nematodes on carrot callus 50 d after the inoculation of 30 females isolated from No. 4 Rs-cps transgenic N. benthamiana roots. (B) The average hatching rate of eggs derived from No. 4 Rs-cps transgenic N. benthamiana plants. Bars indicate the standard errors of the mean data (n = 5), and different letters indicate significant differences among the different treatments (p < 0.05). Infection symptoms of carrot callus 50 d after being inoculated with 30 females isolated from No. 4 Rs-cps (C) and egfp (D) transgenic N. benthamiana plants and wild-type (E) N. benthamiana plants. (F) Blank control without nematodes.
Discussion
Using RNAi in crop pathogens and pest control to suppress target genes using specific dsRNA has been well documented in recent years. Among the early studies, dsRNA was usually delivered by ingestion20, the injection method38 and feeding via bacteria expressing dsRNAs39,40. In spite of the huge costs, the injection method was still widely used for insect pests due to its high efficiency and accuracy. However, using a similar method for plant parasitic nematodes has been a major challenge. The introduction of special dsRNAs into plant parasitic nematodes could not be achieved consistently using microinjection26. The ingestion method (performed by soaking with synthesized dsRNA in vitro) and another approach to deliver special dsRNA into nematodes both showed potential for application in nematode control once special dsRNA was verified to down-regulate target gene expression. However, the efficiency of the latter two methods may not be as satisfactory for the instability of dsRNA when exposed to diverse environments. As a result, the above three methods are not applicable for the control of plant parasitic nematodes in farmland conditions. As an alternative, feeding the nematodes with transgenic plants expressing special dsRNA has been developed as an effective method to deliver dsRNA and control plant parasitic nematodes in agriculture. Transgenic plants possess relatively stable expression level of dsRNAs, providing nematodes with continual stress. Using in planta RNAi against plant parasitic nematodes was first described for root-knot nematodes41. It has also been used to study the control methods of many nematodes, including M. incognita 19,26,42,43, M. javanica 44, H. glycines 45,46, R. similis 28,29 and Pratylenchus vulnus 27. However, using an in planta RNAi method to target the cps gene for the control of plant parasitic nematodes has not yet been reported.
In this study, the cps gene from R. similis was selected as the special dsRNA construct, which was then transferred into N. benthamiana plants. The exact length of the dsRNA fragment needed to trigger RNAi in plants and other eukaryotes is still not entirely clear47. The targeting sequence for gene silencing in plants is approximately 300 to 700 bp in length48. In feeding experiments, most sequences range from 300 to 520 bp49. Previous research has shown that long dsRNA sequences may lead to greater RNAi effect compared to short dsRNAs50. Therefore, in the present study, a 438-bp partial cDNA sequence of Rs-cps was chosen as the target fragment for RNAi. As reported by Mao et al.51, the long dsRNA produced in plants could effectively suppress insect gene expression. Our study also showed that the transcription of Rs-cps in tested R. similis was dramatically suppressed by T2 generation transgenic N. benthamiana plant-derived Rs-cps dsRNA. Therefore, the result of the Rs-cps expression in R. similis suggests that the use of the 438-bp target fragment was successful in generating RNAi in this study.
Cysteine proteinases play key roles in nematodes and many other animal parasites31. Nematode cysteine proteinases mainly include cathepsin B-, L-, S-, K-, and Z-like cysteine proteinases, and cathepsin L and cathepsin B have been extensively studied in recent years29,31,33,34,35. Cathepsin S is one of the most important cysteine proteinases in nematodes and many other parasites. However, the functions of cathepsin S have not been researched using in planta RNAi in plant nematodes. This study was designed to investigate the functions of Rs-cps in R. similis and to find new targets for its control. Therefore, we used the similar method and strategy in this manuscript as described previously. The strategy of in planta RNAi against plant nematodes has been tested in several economically important crops25,28,33,44,45,46. All these works were using in planta RNAi to study gene functions of plant nematodes. However, only one economically important crop has been tested in each study. What’s more, the main purpose of this study is to investigate the functions of Rs-cps and to find new targets for its control. Therefore, we used a single model plants (N. benthamiana) for the RNAi research in this study. Previous studies have shown that the effectiveness of RNAi is higher in single-copy lines than in other lines36,37. Our results showed that No. 4 and No. 13 Rs-cps transgenic N. benthamiana plants had a single copy insertion. The expression level of Rs-cps dsRNA in line No. 4 was significant (p < 0.05) higher than that in line No. 13 of the Rs-cps transgenic plants. Therefore, we only selected the higher expression single-copy No. 4 Rs-cps transgenic line for the nematodes feeding bioassay.
Rs-cps, the target gene chosen in this study, plays important roles in the reproduction and pathogenesis of R. similis. In our previous studies, we verified the feasibility of RNAi for the Rs-cps gene in R. similis by soaking with in vitro synthesized dsRNA16. Therefore, the Rs-cps gene is expected to be a promising target for controlling R. similis. In this study, N. benthamiana plants were transformed to express the special Rs-cps dsRNA and challenged with nematodes to research the Rs-cps function in R. similis using in planta RNAi. Previous studies have shown that delivering dsRNA to plant parasitic nematodes by feeding them with transgenic plants results in the reduction of the target mRNA expression by approximately 65% to 80%28,29,44. In this study, the transcription level of Rs-cps in R. similis fed with Rs-cps transgenic N. benthamiana plants was significantly reduced by 74.5% compared with the control group (p < 0.05). However, this dsRNA-mediated RNAi did not completely eliminate expression of the gene products. That is, the target genes can be knocked down, rather than knocked out, through plant-mediated RNAi. This was consistent with the results seen using in vitro RNAi-mediated gene silencing11,14,15,16. There are several possible reasons for this phenomenon. The first reason for the phenomenon may be due to the lower accumulation of dsRNA in the transgenic plants. Dicer-like enzymes in plants could have decreased the expression levels of the generated dsRNA in these transgenic lines47. The second reason may be that there are energy metabolic pathways that compensate for the RNAi targeting genes.
In this study, we confirmed that the resistance of T2 generation Rs-cps transgenic N. benthamiana plants to R. similis was significantly improved. The result was consistent with the roles of Rs-cps in R. similis, which was verified by in vitro RNAi16. In planta RNAi has been used to research gene functions in cyst nematodes, root-knot nematodes, R. similis and other plant parasitic nematodes. As reported by Klink et al.46, in planta RNAi targeting of four genes of H. glycines caused 81–93% fewer females to develop in transgenic soybean roots. In another report, transgenic Arabidopsis expressing 16D10 dsRNA had a wide resistance against four major root-knot nematode species19. Steeves et al. demonstrated that MSP transgenic soybean plants significantly reduced the reproductive potential of H. glycines 45. All these studies were done using in planta RNAi to research gene functions and demonstrate the efficacy of an RNAi-based strategy for controlling sedentary plant parasitic nematodes. There is limited information available regarding in planta RNAi against migratory endoparasites compared to sedentary endoparasites, even though Li et al. reported that the T2 generation transgenic plants expressing special dsRNA show enhanced resistance to R. similis 28,29. In this study, we first confirmed the feasibility of in planta RNAi in controlling R. similis by targeting the Rs-cps gene. Steeves et al.45 first confirmed the inheritability of gene silencing induced by in planta RNAi in the sedentary plant nematode H. glycines. In another report, burrowing nematodes infecting transgenic N. benthamiana plants expressing specific dsRNA targeting the Rs-cb-1 gene showed a significant reduction in pathogenicity. Interestingly, the progeny nematodes (F1 generation) that hatched from the eggs also showed a significant reduction in pathogenicity and reproduction when allowed to infect the wild-type N. benthamiana plants. Our study showed that the hatching rate of eggs derived from Rs-cps transgenic plants was 32%, which significantly lower than that in control groups. The result was consistent with the inheritability of gene silencing induced by in planta RNAi described above. In conclusion, this study demonstrated that specific dsRNA targeting Rs-cps was effective at suppressing the reproductive capability and hatching of R. similis, and the resistance to R. similis was significantly improved in Rs-cps transgenic N. benthamiana plants. To the best of our knowledge, this is the first study to report the use of an in planta RNAi approach targeting the Rs-cps gene to control R. similis. The current results will enrich the understanding of the use of RNAi against migratory plant parasitic nematodes and provide a scientific basis for the control of other nematodes and pests.
Methods
Plant materials and growth conditions
Nicotiana benthamiana was used as the background line for transformation in this study. For germination, seeds were soaked in sterile water for 1 h at 26 °C, treated with 75% (v/v) absolute ethanol for 1 min and with 0.75% NaOCl for 30 min, washed several times with sterile water, and then sowed on MS medium (pH 5.8) solidified with 0.3% phytagel52,53. The aseptic seeds germinated, and the seedlings were cultured in a growth chamber at 26 ± 1 °C under a photoperiod of 16 h-light /8 h-dark22.
Culturing of nematodes
Carrot discs were prepared as previously described54. The banana burrowing nematode was collected from the roots of Anthurium andraeanum and cultured on carrot discs at 25 °C in a dark incubator55. Approximately 50 d after inoculation, the cultured nematodes were extracted from the carrot discs according to the method of Zhang et al.14.
Cloning and sequencing of the Rs-cps gene
Total RNA was isolated from mixed-stage nematodes using TRIzol reagent (Invitrogen, USA) and digested with DNase (RQ1, Promega) to remove the contaminating DNA molecules. The quality of the extracted RNA was assessed using a spectrophotometer (Nanodrop ND-2000C, Thermo Scientific). The full-length cDNA sequence of Rs-cps was amplified using the specific primers Rs-cps-S1 and Rs-cps-A1 (Table 1), and the purified PCR product was cloned and confirmed by sequencing as previously described16. The recombinant pMD20-Rs-cps plasmid was extracted for later use.
Plant expression vector construction and N. benthamiana transformation
For the Rs-cps plant RNAi expression vector, the partial coding region of Rs-cps was amplified from the pMD20-Rs-cps plasmid containing the full-length Rs-cps cDNA using the specific primers RNAi-F and RNAi-R (Table 1)16. XhoI and XbaI restriction enzyme sites were added to the 5′ end of the forward primer RNAi-F. At the same time, NcoI and BamHI restriction enzyme sites were added to the 3′ end of the reverse primer RNAi-R. The digested Rs-cps fragments were inserted into the XbaI/BamHI and XhoI/NcoI enzyme sites of the binary vector pFGC5941 at inverted repeat sequences to form a plant RNAi expression vector, pFGC-Rs-cps2, which can generate a hairpin RNAi construct. A similar non-endogenous control vector, pFGC-egfp2, was constructed using the primers eGFP-F/eGFP-R (Table 1)29. Then, the constructed pFGC-Rs-cps2 and pFGC-egfp2 were introduced into the competent cells of Agrobacterium tumefaciens (EHA105) by the freeze-thaw method56. The co-cultivation of a N. benthamiana leaf disc was followed using previously described methods57,58. The plant transformation was performed as described previously59, and the transformants were selected for kanamycin resistance on MS medium. After one month of tissue culture, the well-rooted N. benthamiana plants were transferred into sterilized nutritive soil for normal growth and reproduction under the greenhouse conditions described above.
Molecular analysis of putative transformed N. benthamiana plants
Putative transgenic N. benthamiana plant lines were identified by PCR, Southern blot hybridization and RT-PCR. Genomic DNA (gDNA) was extracted from the leaves of T0 generation transgenic and wild-type N. benthamiana plants using the cetyltrimethyl ammonium bromide (CTAB) method60. Rs-cps transgenic N. benthamiana plants were confirmed by PCR with the specific primers RNAi-F and RNAi-R (Table 1). The egfp transgenic plants were similarly checked using the specific primers eGFP-F/eGFP-R as described above (Table 1). To confirm integration of the transformed plants, Southern blot hybridization was performed using the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche). The extracted gDNA (15 μg) of each PCR-positive N. benthamiana line was digested with EcoRI (Thermo Scientific) at 37 °C and then transferred to a Hybond N+ membrane (Amersham)29. The DIG-labelled probe was prepared using the specific primers Southern-F/Southern-R (Table 1)16. Hybridisation and detection were performed as previously described16,29. Equal amounts of gDNA from the T0 generation egfp transgenic and wild-type N. benthamiana plants were used as the controls. For the RT-PCR, total RNAs of PCR and Southern-positive Rs-cps transgenic N. benthamiana leaves were extracted and reverse transcribed using the PrimeScriptTM 1st Strand cDNA Synthesis Kit (Takara). The PCR reactions were carried out using the specific primers RNAi-F/RNAi-R (Table 1) as described above. The positive transgenic N. benthamiana plants were cultured in the greenhouse to obtain seeds for further research.
Detection the transcription level of Rs-cps in transgenic N. benthamiana leaves by qPCR
qPCR was performed to detect the production of Rs-cps dsRNA in different single copy T2 generation transgenic and wild-type N. benthamiana plants. Total RNA of these fresh N. benthamiana leaves was extracted and reverse transcribed as described above. The specific primers qPCR-F and qPCR-R (Table 1) were designed to detect Rs-cps expression levels. The actin gene of N. benthamiana (GenBank accession No: AY179605) was amplified as a reference using the primers Nb-actin-F and Nb-actin-R (Table 1). qPCR tests were performed with a Mastercycler EP Realplex qPCR machine (Eppendorf) in a reaction volume of 20 μl using iTaq Universal SYBR Green Supermix (Bio-Rad). Data were collected at the end of each extension step. The initial data analysis was carried out using the manager software, which calculated Ct values, and the relative levels of Rs-cps were extrapolated from standard curves. Melt curves were obtained routinely, which allowed the possibility of both contamination and primer dimers to be discounted. All expression experiments were performed in triplicate with three biological replicates29.
Resistance studies of T2 generation transgenic N. benthamiana plants expressing Rs-cps dsRNA against R. similis
To investigate the resistance of the transgenic plants, the single-copy Rs-cps transgenic line was chosen for the feeding bioassay. Wild-type N. benthamiana and egfp dsRNA-expressing N. benthamiana lines were used as controls. A total of 2,000 mixed-stage nematodes were inoculated to the roots of each of the selected N. benthamiana plantlets, which had been grown under the same conditions (approximately 20 cm in height). Plants were harvested 70 d post-inoculation, and roots were washed free of soil. The infection symptoms, plant height, fresh above-ground plant weight and fresh root weight of these different inoculated N. benthamiana plants were measured and recorded. In addition, observations were taken on the number of nematodes in the rhizosphere for each N. benthamiana plant under a microscope as described previously14,61. There were 5 replicates for each experiment, and each experiment was conducted twice. Representative data from one round of the experiments were presented.
Expression analysis of Rs-cps in R. similis extracted from T2 generation transgenic N. benthamiana plants
Approximately 100 mixed-stage nematodes feeding on the roots of the wild-type and transgenic N. benthamiana plants were isolated. Extracted nematodes were washed with DEPC water, frozen immediately in liquid N2 and stored at −80 °C. Total RNA was extracted using the RNeasy Micro Kit (Qiagen) and reverse transcribed as described above. Transcript accumulation of Rs-cps in R. similis was analysed by qPCR using the specific primers qPCR-F/qPCR-R and Actin-F/Actin-R (Table 1) as previously described28,29. All the results were obtained from three independent biological replicates. The relative gene expression data were analysed as described above.
Analysis of reproduction and hatching of nematodes extracted from transgenic N. benthamiana plants
The female nematodes isolated from the roots of T2 generation Rs-cps transgenic N. benthamiana plants were sterilized with streptomycin sulfate (3 g/L) for 6 h and then washed with sterile water. After that, the sterilized nematodes were used for the following experiments: (I) A total of 30 females were inoculated onto carrot callus, and the reproduction was evaluated after 50 d at 25 °C. (II) A total of 100 females were cultured on carrot callus for 25 d to obtain nematode eggs. The collected eggs were observed and collected by a dropper under a Ti inverted microscope (Nikon), then placed in Petri dishes (3 cm in diameter, 10/dish) with sterile water and cultivated at 25 °C. After being cultured for 7 d, eggs were observed every 12 h till hatching, and the hatching rates (hatching rate = number of hatched eggs/total number of eggs) were calculated. Nematodes isolated from the roots of egfp transgenic and wild-type N. benthamiana plants were used as the controls. There were 5 replicates for each experiment, and each experiment was conducted twice. Representative data from one round of the experiments were presented.
Statistical analyses
All statistical analyses were conducted using SAS 9.2 (SAS Institute, Cary, NC, USA) in this study. The data collected from the experiments were subjected to one-way analysis of variance (ANOVA) and tested for differences among treatments at the 5% level using Duncan’s Multiple Range Test (DMRT).
References
Gowen, S. R. Some considerations of problems associated with the nematode pests of bananas. Nematropica 9, 79–91 (1979).
Richardson, P. N. & Grewal, P. S. Nematode pests of glasshouse crops and mushrooms. In: Plant parasitic nematodes in temperate agriculture (ed. Evans, K., Trudgill, D. L. & Webster, J. M.) 515–516 (CAB International, 1993).
Luc, M., Sikora, R. A. & Bridge, J. Plant parasitic nematodes in subtropical and tropical agriculture 467–482 (CAB International, 2005).
Jones, J. T. et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol 46, 946–961 (2013).
Sarah, J. L. Banana nematodes and their control in African. Nematropica 19, 199–216 (1989).
Ssango, F., Speijer, P. R., Coyne, D. L. & Waele, D. D. Path analysis: a novel approach to determine the contribution of nematode damage to East African Highland banana (Musa spp., AAA) yield loss under two crop management practices in Uganda. Field Crop Res 90, 177–187 (2004).
Jeger, M. J., Waller, J. M., Johanson, A. & Gowen, S. R. Monitoring in banana pest management. Crop Prot 15, 391–397 (1996).
Tan, J. A., Jones, M. G. & Fosunyarko, J. Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp Parasitol 133, 166–178 (2013).
Fraser, A. G. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 (2000).
Bucher, G., Scholten, J. & Klingler, M. Parental RNAi in Tribolium (Coleoptera). Curr Biol 12, R85–R86 (2002).
Urwin, P. E., Lilley, C. J. & Atkinson, H. J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol Plant Microbe Interact 15, 747–752 (2002).
Cottrell, T. R. & Doering, T. L. Silence of the strands: RNA interference in eukaryotic pathogens. Trends Microbiol 11, 37–43 (2003).
Rosso, M. N., Dubrana, M. P., Cimbolini, N. & Abad, P. Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol Plant Microbe Interact 18, 615–620 (2005).
Zhang, C. et al. Differential expression of Rs-eng-1b in two populations of Radopholus similis (Tylenchida: Pratylenchidae) and its relationship to pathogenicity. Eur J Plant Pathol 133, 899–910 (2012).
Cheng, X. et al. Molecular characterization and functions of fatty acid and retinoid binding protein gene (Ab-far-1) in Aphelenchoides besseyi. PLoS One 8, e66011 (2013).
Wang, K. et al. The cathepsin S cysteine proteinase of the burrowing nematode Radopholus similis is essential for the reproduction and invasion. Cell Biosci 6, 39 (2016).
Mao, Y. B. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25, 1307 (2007).
Tian, G. et al. Transgenic cotton plants expressing double-stranded RNAs target HMG-CoA reductase (HMGR) gene inhibits the growth, development and survival of Cotton Bollworms. Int J Biol Sci 11, 1296–1305 (2015).
Huang, G. Z., Allen, R., Davis, E. L., Baum, T. J. & Hussey, R. S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci USA 103, 14302–14306 (2006).
Baum, J. A. et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25, 1322–1326 (2007).
Ma, J. et al. Production of transgenic rice new germplasm with strong resistance against two isolations of Rice stripe virus by RNA interference. Transgenic Res 20, 1367–1377 (2011).
Zhu, J. Q. et al. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PloS One 7, e38572 (2012).
Xiong, Y., Zeng, H., Zhang, Y., Xu, D. & Qiu, D. Silencing the HaHR3 Gene by Transgenic Plant-mediated RNAi to Disrupt Helicoverpa armigera Development. Int J Biol Sci 9, 370–381 (2013).
Mao, J. J. & Zeng, F. R. Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res 23, 145–152 (2014).
Yadav, B. C., Veluthambi, K. & Subramaniam, K. Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol Biochem Parasit 148, 219–222 (2006).
Dutta, T. K., Banakar, P. & Rao, U. The status of RNAi-based transgenics in plant nematology. Front Microbiol 5, 760 (2015).
Walawage, S. L. et al. Stacking resistance to crown gall and nematodes in walnut rootstocks. BMC Genomics 14, 668 (2013).
Li, Y. et al. A nematode calreticulin, Rs-CRT, is a key effector in reproduction and pathogenicity of Radopholus similis. PLoS One 10, e0129351 (2015).
Li, Y. et al. Cathepsin B cysteine proteinase is essential for the development and pathogenesis of the plant parasitic nematode Radopholus similis. Int J Biol Sci 11, 1073–1087 (2015).
Thakur, P. K., Kumar, M., Kumar, J., Gantasala, N. P. & Rao, U. Structural and functional analysis of cathepsin S of Heterodera spp: a promising candidate for its control. Indian J Exp Biol 52, 223–231 (2014).
Urwin, P. E., Lilley, C. J., McPherson, M. J. & Atkinson, H. J. Characterization of two cDNAs encoding cysteine proteinases from the soybean cyst nematode Heterodera glycines. Parasitology 114, 605–613 (1997).
Tort, J., Brindley, P. J., Knox, D., Wolfe, K. H. & Dalton, J. P. Proteinases and associated genes of parasitic helminths. Adv Parasit 43, 161–266 (1999).
Dutta, T. K. et al. Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front Microbiol 6, 260 (2015).
Li, Y., Xie, H., Xu, C. L., Li, D. L. & Zhang, C. RNAi effect of Cathepsin B gene on reproduction of Radopholus similis. Scientia Agricultura Sinica 43, 1608–1616 (2010).
Shingles, J., Lilley, C. J., Atkinson, H. J. & Urwin, P. E. Meloidogyne incognita: molecular and biochemical characterisation of a cathepsin L cysteine proteinase and the effect on parasitism following RNAi. Exp Parasitol 115, 114–120 (2007).
Kerschen, A., Napoli, C. A., Jorgensen, R. A. & Müller, A. E. Effectiveness of RNA interference in transgenic plants. FEBS Lett 566, 223–228 (2004).
Peng, H. et al. Establishment and functional analysis of high efficiency RNA interference system in rice. Scientia Agricultura Sinica 39, 1729–1735 (2006).
Mutti, N. S., Park, Y., Reese, J. C. & Reeck, G. R. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid. Acyrthosiphon pisum. J insect sci 6, 38 (2006).
Tian, H. et al. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PloS one 4, e6225 (2009).
Timmons, L. & Fire, A. Specific interference by ingested dsRNA. Nature 395, 854 (1998).
Michaeli, S., Kenigsbuch, D., Livneh, O., Levy, D. & Khayat, E. Plants resistant to cytoplasm-feeding parasites. WIPO Patent No. 2005019408 (2005).
Ibrahim, H. M. et al. Post-transcriptional gene silencing of root-knot nematode in transformed soybean roots. Exp Parasitol 127, 90–99 (2011).
Papolu, P. K. et al. Expression of a cystatin transgene in eggplant provides resistance to Root-knot nematode, Meloidogyne incognita. Front Plant Sci 7, 1122 (2016).
Hu, L. et al. Molecular and biochemical characterization of the β-1,4-endoglucanase gene Mj-eng-3 in the root-knot nematode Meloidogyne javanica. Exp Parasitol 135, 15–23 (2013).
Steeves, R. M., Todd, T. C., Essig, J. S. & Trick, H. N. Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct Plant Biol 33, 991–999 (2006).
Klink, V. P. et al. A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of Glycine max. Planta 230, 53–71 (2009).
Liu, F. et al. Silencing the HaAK gene by transgenic plant-mediated RNAi impairs larval growth of Helicoverpa armigera. Int J Biol Sci 11, 67–74 (2015).
Preuss, S. & Pikaard, C. S. Targeted gene silencing in plants using RNA interference. In: RNA Interference (RNAi): nuts & bolts of RNAi technology (ed. Engelke, D.) 23–36 (DNA Press, 2003).
Huvenne, H. & Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56, 227–235 (2010).
Sharp, P. A. RNA interference–2001. Gene Dev 15, 485–490 (2001).
Mao, Y. B. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechol 25, 1307–1313 (2007).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol plantarum 15, 473–497 (1962).
Chaudhury, A. & Qu, R. Somatic embryogenesis and plant regeneration of turf-type bermudagrass: effect of 6-benzyladenine in callus induction medium. Plant Cell Tiss Org Cult 60, 113–120 (2000).
Reise, R. W., Huettel, R. N. & Sayre, R. M. Carrot callus tissue for culture of endoparasitic nematodes. J Nematol 19, 387–389 (1987).
Fallas, G. A. & Sarah, J. L. Effect of storage temperature on the in vitro reproduction of Rahodpholus similis. Nematropica 24, 175–177 (1994).
Chen, H., Nelson, R. S. & Sherwood, J. L. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16, 664–670 (1994).
Gallois, P. & Marinho, P. Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. Methods Mo. Biol 49, 39–48 (1995).
Sunilkumar, G., Vijayachandra, K. & Veluthambi, K. Preincubation of cut tobacco leaf explants promotes Agrobacterium-mediated transformation by increasing vir gene induction. Plant Sci 141, 51–58 (1999).
Horsch, R. B. et al. A simple and general method for transferring genes into plants. Science 227, 1229–1231 (1985).
Murray, M. G. & Thompson, W. F. Rapid isolation of high molecular weight plant DNA. Nucleic acids Res 8, 4321–4326 (1980).
Kaplan, D. T., Vanderspool, M. C. & Opperman, C. H. Sequence tag site and host range assays demonstrate that Radopholus similis and R. citrophilus are not reproductively isolated. J Nematol 29, 421–429 (1997).
Acknowledgements
We are grateful to Prof. GZ Kang (The National Engineering Research Centre for Wheat, Henan Agricultural University, China) for the contribution of binary vector pFGC5941, to Prof. HP Li (South China Agricultural University) for the contribution of Nicotiana benthamiana and Agrobacterium tumefaciens strain EHA105. This work was funded by the National Natural Science Foundation of China (No. 31601619), Special Fund for Agro-Scientific Research in the Public Interest of China (No. 201503112) and China Postdoctoral Science Foundation (No. 2015M582185).
Author information
Authors and Affiliations
Contributions
H.L., B.S. and Y.L. conceived of the study, participated in its design and coordination and drafted the manuscript. Y.L., K.W., Q.L. and J.D. performed the experiments; Y.L., K.W., Z.W. and D.W. analyzed the data; Y.L., K.W., D.W. and H.L. wrote the manuscript. All of the authors read and approved of the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Y., Wang, K., Lu, Q. et al. Transgenic Nicotiana benthamiana plants expressing a hairpin RNAi construct of a nematode Rs-cps gene exhibit enhanced resistance to Radopholus similis . Sci Rep 7, 13126 (2017). https://doi.org/10.1038/s41598-017-13024-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-13024-9
This article is cited by
-
Molecular cloning and functional characterization of AcGST1, an anthocyanin-related glutathione S-transferase gene in kiwifruit (Actinidia chinensis)
Plant Molecular Biology (2019)
-
Transgenic potato lines expressing hairpin RNAi construct of molting-associated EcR gene exhibit enhanced resistance against Colorado potato beetle (Leptinotarsa decemlineata, Say)
Transgenic Research (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.