Abstract
Recent efforts to combat increasing greenhouse gas emissions include their capture into advanced biofuels, such as butanol. Traditionally, biobutanol research has been centered solely on its generation from sugars. Our results show partial re-assimilation of CO2 and H2 by n-butanol-producer C. beijerinckii. This was detected as synchronous CO2/H2 oscillations by direct (real-time) monitoring of their fermentation gasses. Additional functional analysis demonstrated increased total carbon recovery above heterotrophic values associated to mixotrophic assimilation of synthesis gas (H2, CO2 and CO). This was further confirmed using 13C-Tracer experiments feeding 13CO2 and measuring the resulting labeled products. Genome- and transcriptome-wide analysis revealed transcription of key C-1 capture and additional energy conservation genes, including partial Wood-Ljungdahl and complete reversed pyruvate ferredoxin oxidoreductase / pyruvate-formate-lyase-dependent (rPFOR/Pfl) pathways. Therefore, this report provides direct genetic and physiological evidences of mixotrophic inorganic carbon-capture by C. beijerinckii.
Similar content being viewed by others
Introduction
Current societal efforts require solutions to address increasing greenhouse gas emissions1. Accordingly, carbon-capture and its biotransformation into useful value-added commodities, including biofuels, has become a growing area of research. Thanks to a better understanding of pathways such as the Wood-Ljungdahl (WL), and the recently described reversed-pyruvate ferredoxin oxidoreductase (rPFOR)/pyruvate-formate-lyase-dependent (Pfl) carbon assimilation2, more microorganisms can potentially be screened for presence of these pathways using physiological signals. These pathways allow the incorporation of one-carbon (C-1) molecules into Acetyl-CoA. This is a key molecule that can be subsequently bio-transformed into value-added molecules such as acetate or ethanol3,4,5,6,7. Currently, butanol is considered one of the ideal advanced renewable fuel due to a number of favorable properties and applications8,9,10. For example, it can be used unblended in unmodified car engines and is compatible with current oil infrastructure11. However, only recently the assimilation of synthesis gas (containing H2, CO and CO2) into butanol, by natural or genetically modified microbes, has been assessed, and remains in the early stages of development5,6,12,13. As a result, seeking to achieve cost-competitive butanol production, most research has focused on assessing the heterotrophic biotransformation of renewable feedstock by traditional solventogenic Clostridia. However, heterotrophic fermentations have the inherent limitation that 1/3 of carbon is lost in the form of CO2. Interestingly, the reported data shows significant variability in apparent final product yields, pointing towards overlooked metabolic capabilities9,14,15,16,17. With this in mind, we performed a deeper examination of the evolving fermentation gases as physiological signals, while assessing the assimilation of synthesis gas by the natural n-butanol producer C. beijerinckii.
Results
Real-time (in-line) fermentation gas monitoring reveals CO2 and H2 oscillations
We performed a series of fed-batch fermentations of C. beijerinckii while monitoring, in real-time (in-line), the evolving endogenous gasses. Interestingly, we observed in-phase, synchronous H2 and CO2 oscillations coinciding with late log-phase and the onset of solventogenesis (when H2 and CO2 reached ≈3% [v/v]) (Fig. 1). These types of fluctuations are normally observed in feedback-loop controls as a response to metabolic pathway changes18,19. Indeed, diauxic growth was evident from the decrease in the specific growth rate as H2 and CO2 resumed their accumulation (Fig. S1). Although we tested three feed compositions, (i.e. all while containing additional sucrose: (i) fresh complete medium; (ii) 2X trace components20; (iii) sucrose only) these had no effect on the gas oscillations. This suggested to us that neither organic carbon nor another medium component modulated this phenotype. Furthermore, the cells failed to utilize all the provided sugar, and its utilization varied significantly among experiments (Fig. S2, D, D1). In contrast, the kinetic and yield parameters for products and biomass did not vary significantly (Fig. S2–A1, B1, and C1; and Table S1). We used nitrogen (F = 12.48 L/h) as carrier/stripping for gas measurements. In batch (i.e. closed-system) without stripping CO2 and H2 accumulated in the head-space and reached 48 and 23%, respectively (not shown).
Direct monitoring of hydrogen and carbon dioxide evolution in the gas-phase during fed-batch fermentations by C. beijerinckii. H2 and CO2 evolution from three independent experiments performed in a Biostat B+ reactor using defined medium20 containing 6% (w/v) sucrose as limiting carbon and energy source with an initial and final volumes of 1000 mL and 1400 mL, respectively. Feeds (400 mL) contained 80 g sucrose fed at 0.08 mL/h, to reach a final concentration of 100 g/L (w/v) along with: Red line: only sugar was added; Black line: fresh whole medium and; Blue line: 2X trace components. Yellow boxes show detail of the H2 and CO2 oscillation. Fermentations were controlled at 250 rpm, 37 °C and pH 6.5 and constantly sparged (12.48 L/h) with nitrogen gas was achieved using mass flow controllers. Output gas-phase composition was continuously monitored and recorded using two analyzers: An EasyLine continuous analyzer, model EL3020 (ABB, Germany) and a Pfeiffer OmniStar quadrupole mass spectrometer.
It was previously shown that recirculating endogenous H2 and CO2 during butanol fermentation (for maintaining anoxic conditions) allows for more sugar consumption and acid generation14. Additionally, increased product biosynthesis has been shown when electrochemical bioreactors were cultured with C. acetobutylicum in complex medium along with CO2 21. Interestingly, the C-1 assimilation in bacteria is also a mechanism for redox balance, helping to sustain substrate uptake22. Nevertheless, C-1 assimilation has not been previously described in Clostridium beijerinckii 6,7.
Genomic and indirect transcriptomic analysis indicates that C. beijerinckii has the genetic potential for C-1 assimilation
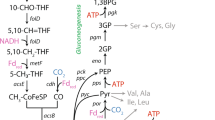
To explain the gas oscillations and their potential assimilation, we explored the C. beijerinckii genome searching for genes related to C-1 assimilation, such as those associated to the WL or rPFOR/Pfl pathways2,5,7,23. We found open reading frames that putatively code for CO dehydrogenase (CODH) (Cbei_5054 and Cbei_3020), formate dehydrogenase and accessory genes (Cbei_3798 to Cbei_3801), formyl-THF ligase (Cbei_0101), methylene-THF dehydrogenase/cyclohydrolase (Cbei_1702) and methylene-THF reductase (Cbei_1828). The putative proteins encoded by these genes have high sequence identity to those of Clostridium ljungdahlii, the species most often utilized for ethanol generation from synthesis gas5,24 (Fig. 2A–B). However, as opposed to this species, C. beijerinckii does not contain annotated a gene coding specifically for an acetyl-CoA synthase, which is essential within the WL pathway. Furthermore, in C. ljungdahlii, most of the WL pathway genes are clustered, except a gene coding for a Ni-Fe-S containing CODH (CLJU_c17910), and a formate dehydrogenase (CLJU_c08930). The former is the main enzyme within the carbonyl branch of the WL pathway, and the final step of the methyl branch (or initial step, if CO is supplied). The latter initiates the methyl branch, allowing CO2 capture into formate. Together, they lead to the generation of acetyl-CoA via an acetyl-CoA synthase. Conversely, C. beijerinckii contains the homologous genes scattered through its chromosome (Fig. 2A). Interestingly, the annotated CODH and the formate dehydrogenase from C. beijerinckii have 77.62 and 72.23% sequence identity, respectively, to the corresponding genes of C. ljungdahlii localized outside its WL cluster. In addition to these genes, C. beijerinckii contains Fe-only and NiFe-hydrogenases (Cbei_1773, Cbei_3796, Cbei_4110 and Cbei_3013) with similarities to those of C. ljungdahlii. In this species, along with H2 generation, these enzymes have hydrogen uptake capabilities, providing extra reducing equivalents to its C-1-fixation pathway5.
Partial Wood-Ljungdahl (WL) and reverse pyruvate: ferredoxin oxidoreductase/pyruvate-formate lyase (rPFOR/Pfl) pathways scheme in C. beijerinckii and chromosome localization of corresponding genes. (A) C-1 assimilation genes found in C. beijerinckii and C. ljungdahlii (WL pathway), mapped in their respective chromosomes. (B) Presumed partial WL and rPFOR/Pfl scheme pathways in C. beijerinckii. Red arrows indicate reactions predicted to be catalyzed by the CO dehydrogenase/Acetyl-CoA synthase complex, (acetyl-CoA is not coded in C. beijerinckii genome). Gray and blue arrows indicate reactions belonging to the methyl branch of the WL, and rPFOR/Pfl pathways, respectively. Black and yellow arrows indicate CO2 and H2 assimilation (and evolving) reactions. (C) Genes and their names associated to C-1 assimilation in C. beijerinckii.
C. beijerinckii also contains two putative Pfl-coding genes (Cbs_1009 and Cbs_1011), (both annotated as formate acetyltransferase, as is the case in Clostridium thermocellum [pflB, clo1313_1717)])2, and a putative pyruvate formate-lyase activating enzyme gene (Cbs_1010). The proteins coded by Cbs_1009 and Cbss_1011, and Cbs_1010, have ~63.5 and 44.4% sequence identity to those of C. thermocellum, respectively. This bacterium, while relying on a partial WL pathway (i.e. the methyl branch without the formate dehydrogenase), contains a reverse pyruvate ferredoxin oxidoreductase (clo1313_0673 and others) that combines acetyl-CoA and CO2 to generate pyruvate, which is then transformed into formate and acetyl-CoA by Pfl2. Interestingly, the C. beijerinckii pyruvate ferredoxin oxidoreductase (PFOR) (Cbs_4318) has 64.1% sequence identity to C. thermocellum rPFOR. The reverse reaction of PFOR has also been observed in other acetogenic and methanogenic bacteria, where this enzyme links the WL pathway and glycolysis25. Additional genes related to the rPFOR/Pfl pathway are a serine hydroxymethyltransferase and a methionine synthase, both of which are also encoded in C. beijerinckii chromosome (Cbs_1868, and Cbs_3100, Cbs_2329 and Cbs_1401, respectively).
With these C-1 assimilation genes in focus, we performed an analysis of publicly available transcriptomic data from batch cultures of C. beijerinckii 26. An RNA-seq time-course experiment was previously reported by Wang et al.27 using cells growing in P2 medium sparged with pure nitrogen. After quality trimming and normalization for gene length and number of assembled reads, we found the putative genes required for C1-assimilation being expressed, either constitutively (Cbei_5054, Cbei_1828 and Cbei_4318) or differentially over time (Cbei_1702, Cbei_0101, Cbei_3801, Cbei_3794, Cbei_3798, Cbei_3799, Cbei_3800, Cbei_3020, Cbei_1010 and Cbei_1011) (Figs 3 and S3). After mapping this transcriptomic response to our gas oscillation data, we identified expression changes that coincided with this phenotype, indirectly pointing towards the re-assimilation of these gases (Figs 1 and 3). Among these genes, formate dehydrogenase and its accessory genes showed the lowest expression in the evaluated experimental condition (i.e. low biomass and CO2/H2 concentration, and nitrogen atmosphere); however, increasing towards mid-log-phase, concurring with the time-point where CO2 and H2 accumulation is maximal (Fig. 3). These genes belong to the methyl-branch of the WL, and rPFOR/Pfl C1-assimilation pathways. Among the four annotated hydrogenase genes, two displayed the highest expression levels: the correlation between expression and hydrogen oscillation/evolution suggested that Cbei_3013 and Cbei_1773 are primarily used for H2 uptake and H2 evolution, respectively. Additionally, the genes encoding for putative carbonic anhydrases also showed expression (Cbei_4425/Cbei_1031). This enzyme allows for even more intracellular CO2 availability28. This in-silico analysis provided an indirect overview of the gas oscillations at the transcriptomic level, while suggesting that these gases may regulate the C1-assimilatory phenotype in C. beijerinckii. Similar gas-dependent behavior has been previously observed in cultures of acetogens Clostridium thermoautotrophicum (reclassified as Moorella thermoautotrophica)29 and C. ljungdahlii, which contain a complete WL pathway30,31.
Time-course transcription profile of C-1 assimilation and energy conservation genes in C. beijerinckii. Time-course expression profiles of partial Wood-Ljungdahl and pyruvate: ferredoxin oxidoreductase/pyruvate-formate lyase (rPFOR/Pfl) predicted genes, along with genes related to energy conservation in C. beijerinckii. The FPKM (fragments per kilobase per million) were calculated from publicly available RNA-seq data27. Lines represent CO2 and H2 evolution in the gas-phase of C. beijerinckii growing in defined medium20, 37 °C, 250 rpm, and constantly sparged with pure nitrogen (12.48 L/h).
Functional evaluation shows inorganic carbon capture by C. beijerinckii
Interested in direct evidence of C-1 assimilation, we performed mixotrophic (sucrose 3% and fructose 1.5% [w/v]) chemostat fermentations (D = 0.135 h−1) while steadily sparging CO2 and H2 at high and low concentrations, balanced with nitrogen. We observed steady-state consumption of CO2 and H2 along with proportional increases of apparent product-yield values above theoretical levels (Fig. S4 and Table S2). If sucrose and fructose were the only carbon and energy sources, apparent yields should have remained at or below the theoretical maximum (i.e. 0.66% C-mol, as a result of one decarboxylation from C-3 pyruvate to the C-2 acetyl group of acetyl-CoA). The higher-than-maximum apparent yields indicated additional carbon assimilation that was only possible by inorganic carbon capture.
Considering current efforts to transform surplus synthesis gas into biofuels6,13, we also sparged this gas at increasing step-wise concentrations (Table S3). Specifically, we sparged synthesis gas mixtures from low (9%), to medium (32%), to high (60%) concentrations, balanced with nitrogen (100% synthesis gas contained 20% CO, 20% CO2, 10% H2 and 50% N2). At low concentration, C. beijerinckii oxidized CO, releasing H2 and CO2 as shown by the steady-state values (Fig. 4A, Table S4). Accordingly, there was no significant difference in apparent C-mol yields and carbon-energy recovery balance compared to the control. Additionally, with increasing electron sink availability, the steady-state sugar utilization improved, resulting in higher product titers (Fig. 4B). Similar behavior has been observed by acetogens Clostridium thermoaceticum (Now Moorella thermoacetica)32, C. autoethanogenum, Rhodopseudomonas gelatinosa and also Carboxydothermus hydrogenoformans, according to the following reaction: CO + H2O → CO2 + H2, which is mainly used for redox balance when grown mixotrophically12,22,33. Therefore, in agreement with our transcriptomic analysis (i.e. overexpression of C-1 capture genes when CO2/H2 were maximal), this physiological behavior suggests these gases may be growth-limiting factors during C-1 assimilation by C. beijerinckii. Accordingly, when cultures were exposed to higher synthesis gas concentrations, higher-than-theoretical apparent C-mol yields and carbon/energy mass balances were detected with the concomitant increased consumption of H2, CO, and CO2, (Fig. 4A,C,D). Specifically, at medium and high synthesis gas concentrations, 11 and 17% more carbon, and 19 and 27% more carbon and electrons, respectively, were recovered. Interestingly, butanol and butyric acid increased by 5.5- and 1.85-fold, respectively, while biomass did not change significantly, which is typical for C1-assimilation pathways such as WL, or rPFOR/Pfl. To confirm gas assimilation into products, we collected steady-state cells (D = 0.135 h−1) growing under high synthesis gas concentration and cultured them in batch conditions in the presence of pure 13CO2 in the headspace as tracer. After 48 h of incubation and NMR analysis, peaks at ~ 178 and 180 ppm revealed the presence of 13C-labeled acetate and butyrate, respectively (Fig. 5). These peaks were likely from the enriched 13C quaternary carbon of these compounds. We confirmed this by adding acetate and butyrate standards (12.8 and 15 g/L, respectively), and observed an increase of peaks at 178 and 180 ppm (quaternary Cs of acetate and butyrate), at 22 (primary C of acetate), and 14 and 40 ppm (primary, secondary (also at 14 ppm), and tertiary Cs of butyrate, respectively). This is a typical spectrum of unlabeled compounds, where quaternary Cs are difficult to detect, as seen in the control in panel A (Fig. 5A). Overall, this provides direct evidence of C-1 assimilation by C. beijerinckii.
Kinetic, yield parameters and carbon and energy balances of steady-state cells (D = 0.135 h−1) cultured mixotrophically on presence of synthesis gas (CO, CO2 and H2). (A) Each tested gas mixture was constantly sparged at 12.48 L/h. Steady-state values of gas generation (H2 and CO2) and synthesis gas utilization were obtained after subtracting the net values of H2 and CO2 generated under sparged nitrogen from the values of output gas phase for each condition. Zero value indicates input = output. Negative values indicate more gases being produced than input. Positive values indicate the steady-state amounts continuously assimilated. (B) Sucrose and fructose consumed in steady states under the different tested gas phase condition. (C) Apparent yield (C-mol product/C-mol carbon source utilized) and (D) carbon and energy balance, calculated as \(\sum (\frac{Yp.\gamma p}{\gamma s})+\,\frac{Yx.\gamma x}{\gamma s}=1\), where Yp and Yx represent C-mol ratios on figure (C); γ represents electrons available for each fermentation product58. Synthesis gas mixture was balanced with nitrogen (100% synthesis gas contains 20% CO, 20% CO2, 10% H2 and 50% N2). The results presented here were obtained from three biological replicates and the represented means are values at steady-state from at least three samples extracted at different retention time intervals. Significance at 0.05 refers to comparisons between whole columns.
NMR spectra of C-13 labeled products. Cultures of C. beijerinckii SA-1 growing mixotrophically and exposed to high synthesis gas concentration (60% synthesis gas 40% N2, where 100% synthesis gas contains 20% CO, 20% CO2, 10% H2 and 50% N2) were collected at steady state (D = 0.135 h−1) and grown in batch during 48 h in presence or absence of 13C labeled CO2. NMR 13C spectra of culture media from: (A) Control, C. beijerinckii grown in the presence of 100% [v/v] CO2 in the headspace; (B) C. beijerinckii grown in the presence of 100% [v/v] 13C labeled CO2 tracer in the headspace; (C) Culture media from control cultures supplemented with standards butyric (9 µg) and acetic acid (7.68 µg). All samples contained as internal standard 0.081 µg of C-13 methanol.
Although butanol is the main target in ABE fermentation and the proportion of total carbon in the form of n-butanol increased by 92%, butyric acid is also a value-added product and can be re-assimilated into n-butanol through multi-stage fermentations20,34. The generation of C-4 compounds, such as butyric acid and butanol, require more NADH than C-2 compounds (such as ethanol)16, underscoring the cells emphasis in recycling electrons.
The rate of gas assimilation, larger than the saturation values in each condition, also indicated biological activity (Figs 4A and S5). We also tested higher synthesis gas concentrations (up to 100%, containing 20% CO, 20% CO2, 10% H2 and 50% N2), showing carbon capture but generating lower yields, possibly due to carbon monoxide poisoning35 (Fig. S6). This indicates that the fermentation working-window for mixotrophic capture of synthesis gas by C. beijerinckii is between 30 and 60% (balanced with N2).
We have also performed batch fermentations of C. beijerinckii under a continuous flow of high synthesis gas concentration as sole carbon and energy source. We observed transient cell proliferation and CO assimilation (not shown). However, cell growth and gas assimilation stopped in early exponential growth phase, as the cells initiated sporulation. As a result, no products were detected.
Transcriptomic analysis of the partial WL and rPFOR/Pfl pathways in C. beijerinckii
To complement the time-course transcriptomic data previously described, we performed a RNA-seq experiment using chemostat cultures (D = 0.135 h−1) of C. beijerinckii SA-1, continuously sparged either with nitrogen (control), low or high synthesis gas. Figure 6 shows constitutive expression of each gene under N2 conditions (normalized to transcripts per million [TPM]), but differentially expressed under both synthesis gas conditions. Under low concentration of synthesis gas, there was a significant overexpression of a putative formate-THF ligase (Cbs_0101), which belongs to the WL pathway, a PFOR (Cbs_4318), a carbonic anhydrase (Cbs_4425), and a hydrogenase (Cbs_1773). Conversely, there was a repression of a putative CODH (Cbs_5054), a gene that belongs to the formate dehydrogenase complex (Cbs_3799), a flavodoxin (Cbs_3109), and several genes that putatively code for the Rnf-complex (Cbs_2449/54). Under high concentration of synthesis gas, there was a significant overexpression of the same genes under low synthesis gas, and a Pfl-activating enzyme gene (Cbs_1010), which belongs to the rPFOR/Pf pathway. However, under that condition, the formate dehydrogenase complex, a CODH (Cbs_3020), a hydrogenase (Cbs_3796), and a cytochrome c biogenesis coding protein (Cbs_2976) were completely shut down. Overall, this transcriptomic data suggests that C. beijerinckii constitutively expresses its putative genes associated to C-1 capture, by preferentially activating those belonging to the rPFOR/Pfl pathway. Specifically, when cultured under synthesis gas, the repression of the formate dehydrogenase, and the overexpression of PFOR and Pfl suggest that CO2 is assimilated via rPFOR/Pfl pathway (and the carbonic anhydrase), as observed with C. thermocellum 2.
Transcription analysis of C. beijerinckii C-1 assimilation and energy conservation pathways. RNA was extracted from cultures growing in chemostat (D = 0.135 h−1) sparged (12.48 L/h) either with N2, low (9%), or high (60%) concentrations of synthesis gas. Cells were cultivated, in defined media20, 250 rpm, 37 °C and pH 6.5. (100% synthesis gas contains 20% CO, 20% CO2, 10% H2 and 50% N2).
Nitrite as an electron sink for energy conservation
Under mixotrophic growth, the C-1 assimilation pathways operate mainly for electron recycling2,22,36. Consequently, an alternative way to demonstrate an active pathway is to inhibit CO assimilation by providing an alternative and preferred electron acceptor. Both nitrate and nitrite are known to have this effect on CO assimilation by acetogenic bacteria37,38. To test this hypothesis in C. beijerinckii, we performed chemostat pulse experiments under high synthesis gas concentration (i.e. 60% [v/v]). Interestingly, nitrate showed no effect on C. beijerinckii. However, less-reduced nitrite partially inhibited CO assimilation (1 mol per mol of NO2), while increasing hydrogen consumption (2.5 mol of H2 per mol of NO2). Additionally, biomass increased proportionately, shifting the pathway from catabolism to anabolism (Fig. 7). Both electron acceptors have also been shown to increase biomass in acetogens C. thermoautotrophicum and Moorella thermoacetica 37,38. The H2-dependent CO2, or CO assimilations are thermodynamically unfavorable as they do not generate a gain in ATP36. Thus, nitrite reduction is preferred as a less expensive way to recycle electrons. As such, the nitrite reductase reaction requires only electrons, in the form of hydrogen and reduced ferredoxin (i.e. not ATP). C. beijerinckii contains a putative ferredoxin-nitrite reductase (Cbei_0832), likely responsible for the observed phenotype, which also unveils this species as a facultative nitrite dissimilator.
Transient responses to nitrite pulses by C. beijerinckii growing in chemostat (D = 0.135 h−1). Experiments were performed on defined medium20 containing 3% sucrose and 1.5% fructose (w/v) and sparged with 60% (v/v) synthesis gas balanced with nitrogen (100% synthesis gas contains 20% CO, 20% CO2, 10% H2 and 50% N2) at 37 °C. The additions of sodium nitrite are indicated with vertical dashed lines to reach final concentrations as follow; 3.1 mM (A), 6.2 mM (B) and 12.4 mM (C). The CO and H2 data shown were obtained by monitoring, in real time, with an EasyLine continuous gas analyzer, model EL3020 (ABB, Germany). Nitrite concentrations higher than 24 mM proved toxic and led to washout. Steady-state values were re-established prior to testing each nitrite concentration. Correlations of NO2 added with (D) H2 consumed, (E) amount of CO consumption displaced, and (F) biomass increase, were calculated from the slopes after fitting the data to linear regressions.
Transcription of alternative energy-conservation genes
Considering that there is no net ATP generation through C-1 assimilation pathways, autotrophic bacteria rely either on substrate-level phosphorylation or on chemiosmosis for energy conservation7,22,36. Examples of the latter include cytochromes, Na+ pumps, or the Rnf-complex, whereby acetogens generate an ion gradient for energy generation through ATP-synthases. It has been suggested that B-type cytochromes are responsible for H+-dependent ATP generation, and can be coupled to a membrane-bound methylene-THF reductase. Its location suggests the role of this enzyme in energy conservation39. C. ljungdahlii contains a Rnf-complex but not cytochromes5,40,41. Interestingly, the C. beijerinckii genome encodes cytochromes (also involved in nitrite reduction42) b-type (Cbei_2439), c550 (Cbei_2762), c551 (Cbei_4151), c biogenesis protein (Cbei_2976), cytochrome-bound flavoproteins (Cbei_3109), and also genes coding for the Rnf-complex (Cbei_2449-2454). Additionally, the methylene-THF reductase of C. beijerinckii is predicted43 to contain transmembrane domains. The transcriptomic analysis of the publicly available RNA-seq data27 showed high expression of all these energy-conserving genes, especially the Rnf-complex (Fig. 3). In line with this observation, our transcriptomic analysis of chemostat cultures of C. beijerinckii shows constitutive expression of these genes under nitrogen exposure, and a modest repression under low and high concentrations of synthesis gas (Fig. 6). Since sporulation prevents C. beijerinckii to growth autotrophically, these chemiosmotic mechanisms are potentially useful only during mixotrophic growth. Similarly, C. ljungdahlii requires the Rnf-complex when cultured mixotrophically41.
Discussion
The variability on apparent product yields reported in the literature and the empirical records of microbial solvent production, demonstrated the need for a deeper study of the evolving gas-phase as signals for overlooked pathways. We have shown that C. beijerinckii captures inorganic carbon and hydrogen under mixotrophic conditions, increasing apparent product yields above theoretical heterotrophic values. Among the putative WL pathway genes, C. beijerinckii does not contain annotated an acetyl-CoA synthase, but its CODHs have Fe-S and Ni-Fe-S metal centers, which are typical of CODH subunits of bifunctional CODH/acetyl-CoA synthases44,45,46. However, it is likely that this enzyme in C. beijerinckii does not lead to acetyl-CoA synthesis, and thus autotrophic growth. Indeed, as has been recently shown, a mutant strain of C. ljungdahlii with a SNP (single nucleotide polymorphism) in its CODH gene located in its WL cluster (i.e. the one with lower sequence identity to that of C. beijerinckii, and associated to a acetyl-CoA synthase) loses its autotrophic phenotype47, even when its CODH with similarity to C. beijerinckii was intact. A similar phenotype has been observed with a CODH-mutant strain of C. autoethanogenum 48. Nevertheless, C. beijerinckii contains the genetic potential for an active rPFOR/Pfl-based C-1 capture, including an additional formate dehydrogenase, not present in C. thermocellum 2.
Based on our physiologic data, we propose a logic model to explain the carbon-electron flow during mixotrophic growth of C. beijerinckii cultures (Fig. 8). In the presence of CO and CO2, there are three possible paths for carbon capture: 1) CO2 to carbonate through putative carbonic anhydrase, or 2) CO oxidation to generate CO2 + H2, if the CO2 in the gas-phase is <5% (v/v); or finally, 3) C-1 assimilation into acetyl-CoA, if CO2 > 5%. Simultaneously, supplied sugars proceed to glycolysis. In the absence of an electron bottleneck, ABE-fermentation utilizes all the sugar-derived carbon and electrons. Otherwise, and if no external electron sink is provided, fermentation stops, limiting sugar utilization. In the presence of an external electron sink (such as CO/CO2), #2 or #3 takes place. If #3 takes place, the extra acetyl-CoA generated, along with the still-running ABE-pathway, leads to 17 and 27% more carbon and carbon-energy recovered, respectively. Moreover, this physiological capability improves product titers by increasing sugar utilization. However, it is important to note that under our experimental conditions, we recovered an apparent ~86 and 100% total carbon and carbon/electron, respectively.
Logic model of carbon-electron flow in C. beijerinckii grown mixotrophically. The data suggest that in the presence of CO and CO2, there are three possible paths for carbon capture: 1) CO2 to carbonate through putative carbonic anhydrase, or 2) CO oxidation to generate CO2 + H, if the CO2 in the gas-phase is < 5% (v/v); or finally, 3) the C-1 assimilation, if CO2 > 5%. Simultaneously, supplied sugars proceed to glycolysis. In the absence of an electron bottleneck, ABE-fermentation utilizes all the sugar-derived carbon and electrons. Otherwise, and if no external electron sink is provided, fermentation stops. In the presence of an external electron sink (such as CO/CO2), #2 or #3 take place. If #3 takes place, the extra acetyl-CoA generated, along with the still-running ABE-pathway, leads to up to 17 and 27% more carbon and carbon-energy recovered, respectively.
Mixotrophic C-1 assimilation was previously shown by cultures of acetogen C. ljungdahlii, whereby exogenous CO2 gas increases carbon recovery49. The discovery of the same phenotype by cultures of C. beijerinckii has important implications in our understanding of the biology of this industrial butanol-producer, and adds a new alternative for greenhouse gas-capture. Indeed, C. beijerinckii stands out among traditional solventogenic species because: (i) it contains genetic elements for cytochromes and the Rnf-system; (ii) it contains genes that code for catalytic enzymes that belong to the WL (except acetyl-CoA synthase) and rPFOR/Pfl pathways; and (iii) the synchronous H2/CO2 oscillation is an example of a natural integrated oscillator, that can potentially be used for feedback controls in biosensors50,51. Future work involving knockout/complementation and biochemical studies will expand our understanding of these processes. Our approach for in-line endogenous gas monitoring shows that it can readably be utilized to uncover new pathways, or potentially even survey a culture (or consortia) for volatile metabolic signatures, in real-time.
Materials and Methods
Organisms
Clostridium beijerinckii SA-1 (ATCC 35702)26 was obtained from the American Type Culture Collection (ATCC). Its identity was verified by PCR amplification and sequencing of the 16S rRNA gene using the prokaryotic 16S rDNA universal primers 515F (5-GCGGATCCTCTAGACTGCAGTGCCA-3) and 1492R (5-GGTTACCTTGTTACGACTT-3).
Bacterial medium and inoculum preparation
C. beijerinckii stocks were activated as previously described17 and were grown in a previously designed medium20. The base components were autoclaved and the sugar (6% w/v sucrose) and trace components were added aseptically to the medium reservoir by filtration (0.22 µm). The inocula were prepared as consistently performed by our lab52. Exact fermentation conditions are detailed in the Main Text section.
Bacterial culture conditions
Growth experiments were performed in fed-batch or chemostat modes of operation in a 2-Liter Biostat® B plus fermenter equipped with controllers for pH, temperature, agitation, and gas mass-flow (Sartorius BBI Systems, Germany). The temperature was set at 37 °C, agitation speed at 250 rpm, and pH 6.5 by the automated addition of 0.5 N KOH or 25% (v/v) H3PO4, into a working final volume of 1,400 mL of culture for fed-batch or 700 mL for chemostat. The fed-batch fermentations were started with 6% (w/v) sucrose, and 400 mL containing 80 g of the same sugar were added at constant feed rate (0.08 mL/h) to reach a final concentration of 100 g/L (w/v). The initial volume was 1 L and final 1.4 L. Exact feed components and times of feed start are detailed in the Main Text section. For the chemostat experiments the conditions were identical as described for fed-batch except the carbon and energy source were 3% (w/v) sucrose and 1.5% fructose. Once the cells reached exponential phase under sparged pure nitrogen (OD600 nm ~ 1), the feed and harvest flow were initiated and adjusted to a dilution rate D = 0.135 h−1. Exact sparged gas compositions are detailed in the Main Text section, steady-state conditions were verified for each condition and at least three retention times were allowed before sampling was initiated. Three samples at each steady-state condition were obtained from at least one retention time intervals. The discrete ratios of continuous gas streams were always sparged at 12.48 L/h. Different gas-phase conditions, from pure nitrogen gas to increased synthesis gas concentrations, were achieved by modifying the mix ratios between synthesis gas and nitrogen using two mass flow controllers; the exact concentrations tested are detailed in the Results section (and Table Supplemental 2). Inlet and exhaust gases in the gas-phase (O2, N2, CO, CO2, H2, and Ar) were monitored and recorded in real-time using in-line O2/CO2 and H2/CO EasyLine continuous gas analyzers, model EL3020 (ABB, Germany), and a Pfeiffer OmniStar quadrupole mass spectrometer. Biomass proliferation in the fermentation tank was monitored and recorded using an in-line biomass sensor (Fundalux, Sartorius, BBI Systems, and Germany) and also by (SmartSpec Plus, BioRad, USA). Dry weight concentration was obtained by filtering a portion of sample using vacuum suction through a 0.2-µm-pore-size filter of known mass (mixed cellulose esters; EMD Millipore, Germany); the filter was then dried at 60 to 70 °C for 7 days and reweighed until constant weight. Pre-mixed synthesis gas was obtained from National Welders in Goldsboro, NC, USA.
Sample analysis
Sucrose, fructose, acetic acid and butyric acid were quantified with a high-performance liquid chromatograph (HPLC) under isocratic conditions at 65 °C, and a mobile phase of water at a 0.5 mL/min flow rate using a Supelcogel TM Ca column (300 mm × 7.8 mm, Supelco TM Analytical, Bellefonte, PA, USA) coupled to a refractive-index detector. Solvents (acetone, butanol and ethanol) were separated in a gas-chromatograph (GC) SS Porapak Q 80/100 column (OV, Marrietta, OH, USA) in a GC (GC-8A) fitted with a flame ionization detector (FID) (Shimadzu Corporation, Kyoto, Japan), using 200 kPa of nitrogen as the mobile phase with an injection temperature of 220 °C and a column temperature of 140 °C.
C-13 labeled-CO2 experiments were performed using 50 mL of culture was collected from the steady state (D = 0.135 h−1) under high synthesis gas concentration (11.17% CO, 11.01% CO2 and 4.40% H2 [v/v]), depicted in Fig. 4. Cultures were incubated for 48 h in sealed 250 mL serum bottles containing 100% C-13-labeled CO2 in the head space, at 37 °C, 200 rpm. For analysis, samples were sent to the NMR-Center of the Chemistry Department at NC State University. 13-C NMR spectra were carried out using a Bruker DRX-500 spectrometer equipped with a 5 mm ITD probe, maintaining the temperature constant at 298 K and the acquired process data were measured with the same parameter. The “zgig” pulse sequence was used. The data were acquired with 2600 FIDs. The internal standards used were C-13 methanol, butyric and acetic acid (0.081, 9.051 and 7.68 µg, respectively), into 600 µL of sample, diluted into 10% D2O).
Proteins sequence identity analysis
Protein sequence identities were performed as previously described53.
RNA-seq analysis from Wang et al
The sequence reads from the transcriptional profiling experiment of Wang et al.27 were downloaded from the NCBI Sequence Read Archive (SRA045799) and imported into the Cyverse Collaborative Discovery Environment54. The sequence reads were quality filtered with the trimmomatic program55 using the trimmers, “LEADING:5 TRAILING:5 SLIDINGWINDOW:4:15 MINLEN:36”. Two independent platforms were subsequently used to analyze these normalized data, Cyverse Discovery Environment and Geneious v9 (Biomatters Ltd., New Zealand), while aligning the sequences to the C. beijerinckii NCIMB8052 genome (GenBank accession CP000721.1). In Cyverse, the sequences were aligned with tophat256 using the default parameters, while differences in transcript abundance were determined using the Cuffdiff program, which is part of the Cufflinks software package57. The analyses in Geneious were performed using default parameters.
Transcription expression analysis
To complement the time-course transcript expression analysis of RNA-seq experiments previously described, we performed chemostat cultures (D = 0.135 h−1) of C. beijerinckii SA-1 growing in presence of sparged N2, low, or high synthesis gas (Fig. S4). For each condition cultures growing in steady-state were harvested (10 mL) and immediately flash frozen and sent for RNA extraction and sequencing at the Microbiome Core Facility at the University of North Carolina Chapel Hill, NC.
RNA isolation
RNA from bioreactor-derived bacteria was isolated using PowerMicroBiome RNA isolation kit from MO Bio Laboratories (San Diego, CA). Briefly, the bacteria pellets were combined with lysis buffer and glass beads. Subsequently they were lysed for 5 minutes in Qiagen TissueLyser II (Valencia, CA) at 30 Hz. Further, the process included inhibitor removal step and standard on-column purification was carried out according to manufacturer’s instructions. RNA purification included on-column DNAse treatment for 15 minutes at room temperature. Subsequently RNA concentration and quality were determined by RNA electrophoresis on Agilent bioanalyzer (Santa Clara, CA).
rRNA removal and library preparation
rRNA was removed using Illumina Ribo-Zero Gold Bacteria Kit (San Diego, CA), according to manufacturer’s instructions. Briefly, the rRNA-specific magnetic beads were washed with storage buffer and were mixed with 500 ng of total sample RNA. Subsequently, rRNA removal solution was added and samples were incubated for 10 minutes at 65 °C. Finally, samples were placed on a magnetic stand for 15 minutes in room temperature and coding RNA was aspirated after which it was immediately preceded to mRNA library preparation protocol. We used Illumina TruSeq Stranded mRNA Library Prep Kit (San Diego, CA), according to manufacturer’s instructions. Briefly, RNA was mixed with Fragment-Prime mix and incubated at 94 °C for 8 minutes and then it was immediately subject to first strand and second strand cDNA synthesis reactions, respectively, followed by 3′ end repair, adenylation and adapter ligation. After adapter ligation, the libraries were enriched by polymerase chain reaction using the following thermal cycling conditions: 98 °C for 30 s, followed by 15 cycles of 98 °C for 10 s, 60 °C for 30 s and 72 °C for 30 s. Final extension step of 70 °C for 5 minutes was also carried out. After enrichment by PCR, libraries were purified with Beckman Coulter magnetic beads (Brea, CA) followed by an 80% ethanol wash, validated on Agilent bioanalyzer and DNA concentration was determined using Quant-iT PicoGreen dsDNA Reagent from Thermo Fisher Scientific (Eugene, OR).
RNA-seq analysis
Reads from two separate sequencing runs were concatenated to maximize sequencing depth and coverage. RNA-Seq data were analyzed using CLC Genomics Workbench v9.5 (QIAGEN Bioinformatics, Redwood City, CA). Paired-end reads were combined and reads were trimmed of any remaining adapter sequences using CLC’s Illumina read import feature using default parameters. The Clostridum beijerinckii SA-1 genome (GenBank accession number CP006777) was downloaded from NCBI using the CLC GenBank browser. The SA1 nucleotide sequence was then converted into a genome track and the associated annotations were used to create a track for gene evidence. All reads were then mapped to the reference genome using the CLC RNA-Seq analysis feature with default parameters. Expression level data were reported as transcripts per million (TPM). Finally, differential expression analyses were performed using CLC’s Advanced RNA-Seq plugin. The data generated in these analyses allow for the generation of volcano plots in OriginPro 2015 graphing software (OriginLab Corporation, Northhampton, MA). Bam files were deposited in NCBI (PRJNA390299).
Gas calculations
For gas solubility, we used Henry’s law: C = k × p where: C is concentration, k is Henry’s constant at 37 °C and p is partial pressure.
The k values used were (in g/L): 0.0225 for CO; 1 for CO2; 0.033 for O2 and 0.0014 for H2.
To calculate gas consumption C = (O − I + E) × (−1), where O is output, I is input, and E is the amount the cells endogenously generated under nitrogen. Positive values indicate consumption. Negative values indicate generation.
Stoichiometry
For this calculation we used the methods previously reported58.
Nitrite pulse experiments
We performed continuous culture pulse experiments with different concentrations of nitrite in the form of sodium nitrite (Sigma-Aldrich Inc., Saint Louis, MO, USA). Exact conditions are detailed in the Main Text section.
References
Jang, Y. et al. Butanol production from renewable biomass: rediscovery of metabolic pathways and metabolic engineering. Biotechnol. J. 7, 186–198 (2012).
Xiong, W. et al. CO2-fixing one-carbon metabolism in a cellulose-degrading bacterium Clostridium thermocellum. PNAS 113, 13180–13185 (2016).
Wood, H. G. & Ljungdahl, L. G. In Variations in Autotrophic Life (eds Shively, J. M. & Barton, L. L.) 201–250 (Academic Press, 1991).
Ljungdahl, L. G. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu. Rev. Microbiol. 40, 415–450 (1986).
Köpke, M. et al. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. 107, 15305–15305 (2010).
Dürre, P. Butanol formation from gaseous substrates. FEMS Microbiol Lett 363, 1–7 (2016).
Drake, H. L., Gößner, A. S. & Daniel, S. L. Old acetogens, new light. Ann. N. Y. Acad. Sci. 1125, 100–128 (2008).
Qureshi, N., Ezeji, T. C., Ebener, J., Dien, B. S., Cotta, M. A. & B, H. Butanol production by Clostridium beijerinckii Part I: use of acid and enzyme hydrolyzed corn fiber. Bioresour. Technol. 99, 5915–5922 (2008).
Qureshi, N., Saha, B. C. & Cotta, M. A. Butanol production from wheat straw by simultaneous saccharification and fermentation using Clostridium beijerinckii: Part II—Fed-batch fermentation. Biomass Bioenerg 32, 176–183 (2008).
Kumar, M. & Gayen, K. Developments in biobutanol production: New insights. Appl Energy 88, 1999–2012 (2011).
Branduardi, P., Branduardi, P. & Porro, D. n-butanol: challenges and solutions for shifting natural metabolic pathways into a viable microbial production. FEMS Microbiol. Lett. 263, 1–22 (2016).
Abrini, J., Naveau, H. & Nyns, E.-J. Clostridium autoethanogenum, sp. nov., an anaerobic bacterium that produces ethanol from carbon monoxide. Arch. Microbiol. 161, 345–351 (1994).
Fernández-Naveira, Á., Abubackar, H. N., Veiga, M. C. M. C. & Kennes, C. Efficient butanol-ethanol (B-E) production from carbon monoxide fermentation by Clostridium carboxidivorans. Appl. Microbiol. Biotechnol. 100, 3361–3370 (2016).
Ezeji, T. C., Qureshi, N., B. H., Ezeji, T. C., Qureshi, N. & Blaschek, H. P. Acetone butanol ethanol (ABE) production from concentrated substrate: reduction in substrate inhibition by fed-batch technique and product inhibition by gas stripping. Appl. Microbiol. Biotechnol. 63, 653–658 (2004).
Formanek, J., Mackie, R. & Blaschek, H. P. Enhanced butanol production by Clostridium beijerinckii BA101 grown in semidefined P2 medium containing 6 percent maltodextrin or glucose. Appl. Environ. Microbiol. 63, 2306–2310 (1997).
Papoutsakis, E. T. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol. Bioeng. 26, 174–87 (1984).
Sandoval-Espinola, W. J. et al. Comparative phenotypic analysis and genome sequence of Clostridium beijerinckii SA-1, an offspring of NCIMB 8052. Microbiology 62, 2558–2570 (2013).
Monod, J. Recherches sur la croissance des cultures bacteriennes. Hermann Cie 80 (1942).
Chen, Y., Kim, J. K., Hirning, A. J., Josić, K. & Bennett, M. R. Emergent genetic oscillations in a synthetic microbial consortium. Science (80-.). 349, 986–989 (2015).
Heluane, H., Evans, M. R., Dagher, S. F. & Bruno-Bárcena, J. M. Meta-Analysis and functional validation of nutritional requirements of solventogenic Clostridia growing under butanol stress conditions and coutilization of D-glucose and D-xylose. Appl. Environ. Microbiol. 77, 4473–4485 (2011).
Jeon, B. Y., Jung, I. L. & Park, D. H. Conversion of carbon dioxide to metabolites by Clostridium acetobutylicum KCTC1037 cultivated with electrochemical reducing power. Adv. Microbiol. 2, 332–339 (2012).
Müller, V. Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 69, 6345–6353 (2003).
Savage, M. D., Wu, Z., Daniel, S. L., Lundie, L. L. & Drake, H. L. Carbon monoxide dependent chemolithotrophic growth of Clostridiurn thermoautotrophicum. Appl. Environ. Microbiol. 53, 1902–1906 (1987).
Younesi, H., Najafpour, G. & Mohamed, A. R. Ethanol and acetate production from synthesis gas via fermentation processes using anaerobic bacterium. Clostridium ljungdahlii. Biochem Eng J 27, 110–9 (2005).
Furdui, C. & Ragsdale, S. W. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J. Biol. Chem. 275, 28494–28499 (2000).
Lin, Y. & Blaschek, H. P. Butanol production by a butanol-tolerant strain of Clostridium acetobutylicum in extruded corn broth. Appl. Environ. Microbiol. 45, 966–973 (1983).
Wang, Y., Li, X., Mao, Y. & Blaschek, H. P. Single-nucleotide resolution analysis of the transcriptome structure of Clostridium beijerinckii NCIMB 8052 using RNA-Seq. BMC Genomics 12, 479 (2011).
Smith, K. S. & Ferry, J. G. Prokaryotic carbonic anhydrases. FEMS Microbiol Rev 24, 335–366 (2000).
Collins, M. D. et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44, 812–826 (1994).
Savage, M. D., Wu, Z., Daniel, S. L., Lundie, L. L. & Drake, H. L. Carbon monoxide-dependent chemolithotrophic growth of Clostridiurn thermoautotrophicum. Appl. Environ. Microbiol. 53, 1902–1906 (1987).
Tan, Y., Liu, J., Chen, X., Zheng, H. & Li, F. RNA-seq-based comparative transcriptome analysis of the syngas-utilizing bacterium Clostridium ljungdahlii DSM 13528 grown autotrophically and heterotrophically. Mol. Biosyst. 9, 2775–2784 (2013).
Pierce, E. et al. The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10, 2550–2573 (2008).
Diekert, G. B. & Thauer, R. K. Carbon monoxide oxidation by Clostridium thermoaceticum and Clostridium formicoaceticum. J. Bacteriol. 136, 597–606 (1978).
Papoutsakis, E. T. & Meyer, C. L. Equations and calculations of product yields and preferred pathways for butanediol and mixed-acid fermentations. Biotechnol. Bioeng. 27, 50–66 (1985).
Kim, B. H., Bellows, P., Datta, R. & Zeikus, J. G. Control of carbon and electron flow in Clostridium acetobutylicum fermentations: utilization of carbon monoxide to inhibit hydrogen production and to enhance butanol yields. Appl. Environ. Microbiol. 48, 764–770 (1984).
Drake, H., Kusel, K. & Matthies, C. In The Prokaryotes (eds Eugene, R., Edward, F. D., Stephen, L., Erko, S. & Fabiano, T.) 545–577 https://doi.org/10.1007/978-3-642-30194-0 (Springer Berlin Heidelberg, 2013).
Fröstl, J. M., Seifritz, C. & Drake, H. L. Effect of nitrate on the autotrophic metabolism of the acetogens Clostridium thermoautotrophicum and Clostridium thermoaceticum. J. Bacteriol. 178, 4597–4603 (1996).
Seifritz, C., Drake, H. L. & Daniel, S. L. Nitrite as an energy-conserving electron sink for the acetogenic bacterium Moorella thermoacetica. Curr. Microbiol. 46, 329–33 (2003).
Wohlfarth, G. & Diekert, G. Thermodynamics of methylenetetrahydrofolate reduction to methyltetrahydrofolate and its implications for the energy metabolism of homoacetogenic bacteria. Arch Microbiol 7, 378–381 (1991).
Schuchmann, K. & Müller, V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821 (2014).
Nataf, Y. et al. The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. MBio 4, 1–8 (2013).
Stouthamer, A. H., Boogerd, F. C. & van Verseveld, H. W. The bioenergetics of denitrification. Antonie Van Leeuwenhoek 48, 545–553 (1983).
Cserzo, M., Wallin, E., Simon, I., von Heijne. & Elofsson, A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Prot. Eng. 10, 673–76 (1997).
Ragsdale, S. W. & Kumar, M. Nickel-containing carbon monoxide dehydrogenase/Acetyl-CoA synthase. Chem. Rev. 96, 2515–2540 (1996).
Lindahl, P. A. & Chang, B. The evolution of Acetyl-CoA synthase. Orig. Life Evol. Biosph. 31, 403–434 (2001).
Lindahl, Pa Acetyl-coenzyme A synthase: the case for a Ni(p)(0)-based mechanism of catalysis. J. Biol. Inorg. Chem. 9, 516–524 (2004).
Whitham, J. M. et al. Characterization of Clostridium ljungdahlii OTA1: a non-autotrophic hyper ethanol-producing strain. Appl. Microbiol. Biotechnol. 100, https://doi.org/10.1007/s00253-016-7978-6 (2016).
Liew, F. et al. Insights into CO2 fixation pathway of Clostridium autoethanogenum by targeted mutagenesis. MBio 7, e00427–16 (2016).
Jones, S. W. et al. CO2 fixation by anaerobic non-photosynthetic mixotrophy for improved carbon conversion. Nat. Com. 7, 12800 (2016).
Kobayashi, H. et al. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl. Acad. Sci. 101, 8414–9 (2004).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Sandoval-Espinola, W. J., Chinn, M. S. & Bruno-Barcena, J. M. Inoculum optimization of Clostridium beijerinckii for reproducible growth. FEMS Microbiol. Lett. 362, fnv164 (2015).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Merchant, N. et al. The iPlant Collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 14, e1002342 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics btu170, https://doi.org/10.1093/bioinformatics/btu170 (2014).
K, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Erickson, L. E., Selga, S. E. & Viesturs, U. E. Application of mass and energy balance regularities to product formation. Biotech. Bioeng. 20, 1623–1638 (1978).
Acknowledgements
This work was partially supported by the College of Agriculture and Life Sciences at NC State University, and was partially funded by the USDA Biomass Research and Development Initiative under the award number 2011-10006-30363. MRT was supported by a mobility grant from the Ministerio de Educación, Cultura y Deporte, Spain (PR2015-00328). We thank Dr. Jesse Noar for editing this manuscript, the NMR-Center of the Chemistry Department at NC State University and the Microbiome Core Facility at the University of North Carolina.
Author information
Authors and Affiliations
Contributions
W.J.S.E. performed the experiments and wrote the manuscript; M.R.T. performed the RNA-seq analysis and wrote the manuscript; M.C. conceived and contributed to the synthesis gas experiments and J.M.B.B. conceived and designed the research studies and wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
A patent application describing this new alternative for greenhouse gas-capture has been submitted by NC State University. The authors declare no other competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sandoval-Espinola, W., Chinn, M., Thon, M. et al. Evidence of mixotrophic carbon-capture by n-butanol-producer Clostridium beijerinckii . Sci Rep 7, 12759 (2017). https://doi.org/10.1038/s41598-017-12962-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-12962-8
This article is cited by
-
Application of raw industrial sweetpotato hydrolysates for butanol production by Clostridium beijerinckii NCIMB 8052
Biomass Conversion and Biorefinery (2024)
-
Acidogenesis, solventogenesis, metabolic stress response and life cycle changes in Clostridium beijerinckii NRRL B-598 at the transcriptomic level
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.