Abstract
Mutations in the RPGR-interacting protein 1 (RPGRIP1) gene cause recessive Leber congenital amaurosis (LCA), juvenile retinitis pigmentosa (RP) and cone-rod dystrophy. RPGRIP1 interacts with other retinal disease-causing proteins and has been proposed to have a role in ciliary protein transport; however, its function remains elusive. Here, we describe a new zebrafish model carrying a nonsense mutation in the rpgrip1 gene. Rpgrip1homozygous mutants do not form rod outer segments and display mislocalization of rhodopsin, suggesting a role for RPGRIP1 in rhodopsin-bearing vesicle trafficking. Furthermore, Rab8, the key regulator of rhodopsin ciliary trafficking, was mislocalized in photoreceptor cells of rpgrip1 mutants. The degeneration of rod cells is early onset, followed by the death of cone cells. These phenotypes are similar to that observed in LCA and juvenile RP patients. Our data indicate RPGRIP1 is necessary for rod outer segment development through regulating ciliary protein trafficking. The rpgrip1 mutant zebrafish may provide a platform for developing therapeutic treatments for RP patients.
Similar content being viewed by others
Introduction
Inherited photoreceptor dystrophies (IPD) are a group of genetically and phenotypically heterogeneous retinal diseases, characterized by the progressive death of photoreceptor cells1. Leber congenital amaurosis (LCA) is the most severe form of childhood IPD, causing early severe visual defects after birth2. LCA is also genetically heterogeneous with at least 17 genes implicated (2, the Retinal Information Network (RetNet) database). The RPGRIP1 gene, encoding retinitis pigmentosa GTPase regulator interacting protein 1, was first identified as an RPGR interacting partner by three independent research groups3,4,5. Defects in the RPGR gene result in severe X-linked retinitis pigmentosa (XLRP), accounting for over 70% of XLRP cases6. RPGRIP1 is a recessive LCA-causing gene that is mutated in 5% of all LCA cases7,8. Mutations in RPGRIP1 also caused juvenile retinitis pigmentosa and cone-rod dystrophy in humans9,10. Similarly naturally occurring recessive mutations in RPGRIP1 cause cone-rod dystrophy in dogs11.
RPGRIP1 localizes predominantly to the connecting cilium of mouse photoreceptor cells and to outer segments of human photoreceptors12,13. RPGRIP1 also localizes at the centrosome of non-ciliated cells and at the basal body of ciliated cells14,15. RPGRIP1 is a multi-domain protein, containing an N-terminal coiled-coil domain, two protein kinase C conserved domains (C2) and a highly-conserved C-terminal RPGR-interacting domain (RID)16,17. RPGRIP1 is reported to interact with different proteins. The N-terminal coiled-coil domain of RPGRIP1 interacts with SPATA7, which is mutated in LCA3 and juvenile RP18,19, the central C2 domain interacts with NPHP4 and notably disease-associated mutations in RPGRIP1 or NPHP4 disrupt this interaction20; the RID domain interacts with RPGR3,4,5 and Nek4 serine/threonine kinase15. RPGRIP1 is an anchor for RPGR localization to the connecting cilium21, but its targeting to the connecting cilium depends on SPATA718. RPGRIP1 is also required for ciliary targeting of NPHP4 and SDCCAG8, which are associated with renal-retinal ciliopathy22. These data suggest RPGRIP1 plays a critical role in ciliary protein trafficking and ciliopathies presenting with the photoreceptor cell death.
An RPGRIP1 knock-out (KO) mouse model exhibited early retinal degeneration with almost complete loss of photoreceptor cells by three months of age. The photoreceptors of these KO mice initially developed with a normal structure of the connecting cilium, but the outer segments were disorganized with oversized outer segment disks21. An N-Ethyl-N-Nitrosourea (ENU)-induced RPGRIP1 null mouse model showed a more severe photoreceptor degeneration compared to the RPGRIP1 KO mice23. The loss of the outer nuclear layer was initiated by postnatal day (P) 12 and nearly complete by P28. The rod outer segments were rarely formed, while the cone outer segments were initially formed but degenerated rapidly23. A naturally occurring dog model has a 44-nucleotide insertion in exon 2 of the Rpgrip1 gene, resulting in an altered open reading frame and introduction of a premature stop codon11. RPGRIP1 deficient dogs exhibited loss of cone function by 6 weeks of age consistent with shortened inner and outer segments. Rod photoreceptor function appeared normal by 10 weeks of age, but by 23 weeks of age rod cells grossly degenerated with a significantly thinned outer nuclear layer. Only a few photoreceptors remained by 45 weeks of age24.
Here we report a new zebrafish model of RPGRIP1 generated by ethyl nitrosourea (ENU) mutagenesis. Homozygous rpgrip1 mutant fish exhibited severely affected rod cells at an early stage, followed by progressive degeneration of cone cells. Rod outer segments were absent and rhodopsin mislocalization was apparent by 5 days post fertilization (dpf) of age, the earliest time point examined, as were trafficking defects of some ciliary proteins. Our data support the hypothesis that Rpgrip1 is required for rod outer segment formation possibly through regulating ciliary transport processes.
Results
Identification of zebrafish rpgrip1 mutants
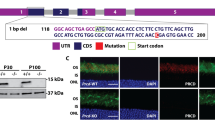
Zebrafish rpgrip1 encodes an open reading frame of 1342 amino acids and contains at least 28 exons, spanning approximately 28 kb on chromosome 24. Zebrafish Rpgrip1 protein has similar functional domains to human RPGRIP1with conserved protein identities of 24.93%, 36.08% and 31.34% for SMC/CC, C2 and RID domains, respectively (Fig. 1A). Zebrafish rpgrip1 expression significantly increased during embryogenesis (Supplementary Material, Fig. S 1A and B), and in adult tissues rpgrip1 was detected predominantly in the eye (Supplementary Material, Fig. S 1C and D). A large library of ENU-mutagenized fish were screened by TILLING25 for mutations in zebrafish ciliopathy genes, a C → T transversion at nucleotide 2206 in exon 18 of the rpgrip1 gene was identified, resulting in a premature stop codon (Q736X) and loss of a BbvI restriction site, which was subsequently used for genotyping rpgrip1 mutants (Fig. 1B,C). We predicted that the nonsense mutation would lead to either a truncated 735 amino acids polypeptide and/or nonsense-mediated RNA decay (NMD). Quantitative real-time PCR assay found rpgrip1 mRNA was decreased by about 85% in 5 dpf rpgrip1 −/− embryos when compared to wildtype siblings (Supplementary Material, Fig. S2), confirming that NMD was most likely occurring. Both heterozygous and homozygous rpgrip1 mutant fish exhibited normal external morphology and were fertile. The mutant larvae also developed a normal swim bladder, an indirect sign of fish health. We phenotypically characterized the mutant fish, all the examined homozygous rpgrip1 fish showed same phenotypes, suggesting a complete penetrance. There is no variable expressivity. Because zebrafish rpgrip1 is a large gene, we could not clone the full-length cDNA, so we did not performed any rescue experiment.
Genetic identification of rpgrip1 mutant zebrafish. (A) Schematic structure of human RPGRIP1, zebrafish Rrpgrip1 (zRPGRIP1) and truncated zebrafish rpgrip1 mutant. (B) Sequence validation of the C to T conversion in homozygous zebrafish, which resulted in a nonsense mutation and changing glutamine codon into stop codon (CAG to TAG, indicated with a box). (C) Gel picture of BbvI digested PCR products amplified using wild type and heterozygous fish genomic DNAs as templates. The PCR fragment was 99 bp and BbvI cut the fragment amplified from wildtype zebrafish to 59 bp and 40 bp, but could not cut the fragment amplified from mutant zebrafish. Wildtype zebrafish (rpgrip1 +/+) showed only one band because both bands (59 bp and 40 bp) are too close and could not be well separated. The heterozygous zebrafish, rpgrip1 +/−, showed two bands: the upper band is for the mutant PCR product, the lower band is BbvI-cut PCR product (59 bp and 40 bp). The homozygous mutant zebrafish, rpgrip1 −/−, showed one band, the 99 bp fragment, which BbvI could not cut. UD, undigested PCR product; D, BbvI-digested fragments.
Rod outer segment development was defective in rpgrip1 mutants
Previous reports in mice suggested RPGRIP1 participates in rod outer segment formation and disk morphogenesis21,23. We first examined photoreceptors in wildtype, heterozygous and homozygous rpgrip1 mutant larvae at 5 and 7 dpf by hematoxylin and eosin staining. Photoreceptor outer/inner segments of wildtype, heterozygous and homozygous siblings at 5 and 7 dpf were visible by light microscopy of retinal sections(the rpgrip1 +/− larvae were indistinguishable from wildtype siblings - data not shown); however, outer/inner segments were significantly shorter in rpgrip1 −/− retina compared to wildtype siblings (Fig. 2A and Supplementary Material, Fig. S 3). It is well known that rod outer segments are longer, thinner and more cylindrical in shape compared to cone outer segments which are shorter and conical shaped26,27,28. Rod outer segments are surrounded by plasma membrane; whereas cone outer segments have parallel membranes called lamellae, which are partially closed on one side by plasmalemma26,27,28. To determine whether the significantly decreased length of outer/inner segments in rpgrip1 mutant larvae was caused by the defect of rod outer segment development, we examined outer segment ultrastructure in wildtype and rpgrip1 mutant siblings using transmission electron microscopy. At 5dpf rod and cone outer segments were well formed in wildtype siblings; whereas in rpgrip1 −/− mutants rod outer segments were not observed, however, cone outer segments were normal (Fig. 2B). Anti-rhodopsin antibody (4D2) labelling of rod outer segments by immunofluorescent analysis demonstrated stained outer segments visible in wildtype but absent in rpgrip1 −/− siblings. Rhodopsin was mislocalized in rod cells through to the synapses (Fig. 2C).
Rod outer segments were not developed in rpgrip1 −/− zebrafish. (A) Photoreceptor outer segments and inner segments were shorter in the retinas of rpgrip1 −/− zebrafish at 5 and 7dpf shown by haematoxylin & eosin staining. Double-headed yellow arrow shows the outer/inner segments; the measurement of outer segment length was shown in Supplementary Materials Fig. S3. Inserts show a 3.5 × magnification of the selected regions. (B) Ultrastructural analysis using transmission electron microscopy revealed both rod and cone outer segments were formed in the retinas of 5dpf wildtype siblings but only cone outer segments were presented in the mutants; rod outer segments were not observed in the retinas of 5 examined RPGRIP1 −/− mutants at 5dpf. (C) Immunostaining with anti-rhodopsin antibody (4D2) further confirmed rod outer segments were not formed in the retinas of rpgrip1 −/− mutants at 5dpf; rhodopsin was mislocalized in whole rod cell body. Nuclei were shown in blue using DAPI. COS, cone outer segments; GCL, ganglion cell layer; INL, inner nuclear layer; M, mitochondria; MEL, melanosome; ONL, outer nuclear layer; RPE, retinal pigment epithelium; ROS, rod outer segments.
Early retinal degeneration in rpgrip1 mutants
Previous reports show that RPGRIP1 mutations can cause LCA, juvenile RP and cone-rod dystrophy in humans7,8,9,10 and early-onset retinal degeneration in mice21,23. We examined retinal degeneration in heterozygous and homozygous fish at different ages using histological methods. Heterozygous fish did not present with photoreceptor abnormalities, even as late as 24 months of age (data not shown). The outer nuclear layers of rpgrip1 −/− retinas at 5 and 7 dpf were the same as that of wildtype siblings (Fig. 2A); however, some rod cells displayed an abnormal morphology with condensed and rounded-up nuclei at 2 weeks of age. By age of 3 weeks, degeneration of rod cells was evident in the mutant fish (data not shown). Only some rod nuclei remained at 1 month post-fertilization (mpf) when compared to age-matched controls (Fig. 3). Zebrafish retina development continues into adult stages29, as observed in 3 mpf controls, where approximately four layers of rod nuclei were visible, but in the rpgrip1 mutant, only a few rod nuclei were present. This is consistent with the loss of photoreceptor cells, and in agreement we detected significantly increased cell death in rpgrip1 mutant retinas at 14 dpf, 1 mpf and 3 mpf using TUNEL assays (Fig. 4). At 3 mpf, cone cells presented normal morphology, though they were not tightly aligned together as in wildtype siblings (Fig. 3), possibly due to the degeneration of the rod cells resulting in structural changes to the cone cell layer. At 6 mpf, the cone cells appeared disorganized and loss of cone cells was much more evident in rpgrip1 −/− mutants. At 13 mpf, the number of cone cells was further decreased (Fig. 3). Transmission electron micrographs of the retinal sections of rpgrip1 −/− mutants at 13 mpf showed that only single cones with normal morphology remained (Supplementary Material, Fig. S4). At 18 mpf single cones were also affected: the morphology of these cells was abnormal and some were degenerated (Fig. 3); At 23 mpf very few single cone cells were evident (Fig. 3).
Haematoxylin & eosin staining of retinal sections of wild type and rpgrip1 −/− zebrafish at different ages, showing progressive retinal degeneration. WT, wildtype; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium. The scale bars are 10 µm.
TUNEL assay showed significantly increased cell death in rpgrip1 −/− mutant retinas. Retina cryosections of wildtype (WT) and rpgrip1 −/− zebrafish at 14 dpf, 1 mf and 3 mpf were subjected to TUNEL assay. (A) TUNEL-positive cells were present in the outer nuclear layer (ONL) of rpgrip1 −/− mutant retinas. TUNEL-positive cells were barely observed in ONL of wildtype retina at three age points. (B) Graph indicating significant cell death (12%, 9% and 8% respectively) in the mutant retina compared to the wildtype retina at each age point. The data were collected by counting positive cells from 5 retinal sections of wildtype and rpgrip1 mutant respectively and shown as mean ± SD. SD: standard deviation. **p < 0.01.
Immunofluorescence labelling was used to examine both rod and cone cells. Rhodopsin was mislocalized to the inner segments, outer nuclear layer and synapses of rod cells at all time points examined (Fig. 2C and Supplementary Material, Fig. S5). The rod cells appeared to be disorganized at 14 dpf. The fluorescence signal (labelled by 4D2 antibody) was remarkably reduced by 1 mpf, suggesting significant degeneration of rod cells. At 3 mpf, only a weak fluorescence signals were detected, possibly because all the rod cells had degenerated. At 6 mpf, only a little background fluorescence signals were presented (Supplementary Material, Fig. S5). Immunostaining with ZPR1 antibody which labels red and green cone cells (double cones) showed those cells were morphologically normal at 3, 4, 5 dpf and 1 mpf (Fig. 5 and data not shown). At 6 mpf, the cone cells were markedly disorganized and the outer segments were obviously shorter. By 12 mpf, only a few fragmented double cone cell bodies were observed (Fig. 5). As cone opsins are partially mislocalized in the cell bodies of rpgrip1mutant mice21,23, we examined cone opsin localization in the retinas of rpgrip1 −/− and wildtype siblings at 14 dpf using antibodies which target to red, green or UV opsins. Red, green and UV opsins were localized in cone outer segments in the wildtype retina; however, the three types of opsins were partially mislocalized in the outer nuclear layer of rpgrip1 −/− mutants (Fig. 6).
Immunostaining with Zpr1 antibody exhibited red/green double cones that were normal at early ages and degenerated at later ages. Nuclei were shown in blue using DAPI. Double cones of rpgrip1 −/− mutants were morphologically normal at 2wpf and 1 mpf, shown here. While at 3mpf, double cones were disorganized, the number of double cones was markedly reduced at 6 mpf and only some double cone cell bodies remained at 12 mpf.
Partial mislocalization of cone opsins in rpgrip1 −/− retinas. Green, red and UV opsins were localized in cone outer segments of 14 dpf wildtype zebrafish retinas. In rpgrip1 mutant zebrafish retinas, green, red and UV opsins were localized in outer segments and partially mislocalized in cone cell bodies (arrow) and synapses (arrowhead). GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; OPL, outer plexiform layer; RPE, retinal pigment epithelium.
Visual function was impaired in RPGRIP1 mutants
We used electroretinography (ERG) recordings to evaluate rpgrip1 −/− fish for abnormal visual function. Zebrafish cone cells mature earlier than rod cells, so the ERG response of larvae at 5 and 7 dpf is determined by cones30,31. 7dpf zebrafish were dark-adapted for at least 30 minutes before ERG recordings. Compared with wildtype siblings, the scotopic b-wave amplitudes were significantly reduced by approximately 30% (p < 0.01) and the OFF response (d-wave) was absent (Fig. 7A,B), suggesting dysfunction of cone cells, possibly due to the partial mislocalization of cone opsins (Fig. 6). We also evaluated whether rpgrip1 −/− fish are functionally responsive to light by examining the visual background adaption (VBA) response in 7dpf fish. VBA, a neuroendocrine response, is controlled through the retino-thalamic tract32 and blind fish fail to display this response33. When fish are placed on a dark background, the pigment granules (melanosome) are dispersed within pigmented skin cells; in contrast when fish are placed on a light background, the melanosomes are aggregated to form small granules32. 7 dpf wildtype and rpgrip1 −/− larvae were dark-adapted for 30 mins then exposed to bright light for 15 mins. The wildtype larvae exhibited small pigment granules as expected, however rpgrip1 −/− larvae displayed larger diffuse pigment granules, suggesting visual impairment (Fig. 7C).
Impairment of visual function in rpgrip1 −/− zebrafish. (A) Representative ERG traces from wildtype and rpgrip1 −/− zebrafish at 7dpf. D-wave was absent in the mutant zebrafish. (B) b-wave amplitudes of rpgrip1 −/− zebrafish (n = 10) were significantly reduced when compared to wildtype siblings (n = 11). The result is shown as mean ± SD. SD: standard deviation. **p < 0.01. (C) Visual background adaption for wildtype (n = 10) and rpgrip1 −/− zebrafish (n = 10) at 7dpf responding to dark adaption and bright light stimulation. Representative images showing that melanin of wildtype zebrafish was aggregated into small granules after light adaption, while rpgrip1 −/− zebrafish displayed larger diffused melanin granules.
Rpgrip1 mutants exhibited defects in ciliary protein trafficking
Mammalian RPGRIP1 directly or indirectly interacts with different proteins and possibly functions in ciliary protein trafficking17. RPGRIP1 physically interacts with RPGR through its C-terminal RPGR-interacting domain3,4,5. Localization of RPGR to the connecting cilium of mouse retina is dependent on RPGRIP1, and RPGR was absent in the connecting cilium of RPGRIP1 knock-out mouse21. We examined whether Rpgr localization in rpgrip1 −/− zebrafish retina was affected. Immunostaining with an antibody, previously used to localize RPGR to connecting cilia and outer segments of adult zebrafish photoreceptors34, revealed that Rpgr was localized to photoreceptor inner segments of 14 dpf rpgrip1 −/− zebrafish retina, while Rpgr was localized to outer segments and connecting cilia of wildtype sibling photoreceptors. Rpgr signal was also significantly decreased in rpgrip1 mutant retinas (Supplementary Material, Fig. S 6A).
Previous work showed RPGR directly interacted with a small GTPase Rab8 and functioned as a guanine nucleotide exchange factor (GEF) for Rab835. Knock-down of RPGR reduced Rab8 targeting to primary cilia in vitro 35. Since RPGR localization was disrupted in RPGRIP1 knockout mouse21, we asked whether Rab8 localization in rpgrip1 −/− fish photoreceptors was affected. Immunofluorescence assays were performed using an anti-Rab8 polyclonal antibody raised against a human Rab8 peptide from the C-terminal end that is highly conserved between human and zebrafish Rab8 (86% identity, data not shown). Rab8 was mainly localised to the ciliary base of photoreceptor cells, though weak signal was also detected in outer segments of photoreceptor cells (Fig. 8A), consistent with early reports of Rab8 localization in zebrafish, Xenopus and mouse photoreceptors36,37,38,39. However in rpgrip1 −/− siblings, the Rab8 signal at the ciliary base was significantly decreased, and signals were also detected in the inner segments of cones, the localization of Rab8 to the outer segments of photoreceptor cells was also lost, indicating RPGRIP1 is partially required for the Rab8 localization in photoreceptors (Fig. 8A,D).
Mislocalization of ciliary proteins in rpgrip1 mutant fish. (A) Rab8 was mainly localized to the connecting cilium of rod cells and some weak fluorescence signals were present in the outer segments of photoreceptor cells in wildtype (WT) zebrafish at 2 wpf; it was mislocalized in inner segments of photoreceptor cells and the localization to the outer segments of photoreceptor cells was lost in rpgrip1 mutant zebrafish at 2 wpf. (B,C) Transducin and GRK1 exhibited abnormal distribution in cone outer segments and loss of localization to rod outer segment, because the rod outer segments were absent in rpgrip1 mutant zebrafish. INL, inner nuclear layer; ONL, outer nuclear layer. (D) Quantification of fluorescence intensity of Rab8, GNAT1 and GRK1 signals. The fluorescence intensity was measured within three outer segment areas (25 × 25 µm²/area) in each section (four sections from four individual retinas were used) by Zen (Zeiss) at the condition of same laser intensity and master gain, and the data was analysed by t test with Graph Prism. The result is shown as mean ± SD. SD: standard deviation. p = 0.0027 (Rab8); p = 0.0084 (GNAT1); p = 0.0164 (GRK1).
Transducin has been reported to be mislocalized in Rpgrip1 nmf247 mutant mice23. We wondered whether transducin was mislocalized in rpgrip1 −/− zebrafish. We examined transducin localization in zebrafish retina by immunostaining with anti-Gnat1 (the alpha subunit of rod transducin) antibody, which was used for immunofluorescence assay in rp2 knock-out zebrafish40. At 14 dpf, Gnat1 was localized at the outer segments of wildtype photoreceptors. However, the fluorescence signal of Gnat1 was mainly observed in the inner segments of rod cells of rpgrip1 −/− mutants, possibly because rod outer segments were absent, and the fluorescence signals were also significantly reduced (Fig. 8B,D). The localization of G-protein-coupled receptor kinases (GRK1 and GRK7) in wildtype and rpgrip1 mutant retinas was examined using anti-human GRK1 and anti-zebrafish GRK7a antibodies41,42. GRK1 was detected in outer segments of both rod and cone cells, probably because the anti-human GRK1 antibody could recognize both zebrafish GRK1a and GRK1b, which were localized to rod and cone outer segments respectively42. However, GRK1 signals were localized at inner segments of rod cells and most GRK1 fluorescence signals were significantly reduced from cone outer segments (Fig. 8C,D). Additionally, the localization of GRK7a (the cone opsin kinase) was not affected in rpgrip1 −/− mutants (Supplementary Material, Fig. S6B,C). Since the fluorescence signals of RPGR, rab8, GNAT1, GRK1 were decreased in rpgrip1 mutant retinas at 14 dpf, we examined the level of these proteins and found expression of all proteins to be significantly decreased (Supplementary Material, Fig. S7).
Discussion
In this study, we characterized a newly identified zebrafish mutant, which carried a nonsense mutation in the rpgrip1 gene. Rpgrip1 −/− zebrafish did not form rod OS and displayed rhodopsin mislocalization. However, cone cells developed normally, although cone opsins were partially mislocalized, suggesting RPGRIP1 may have a major role in rod cells. Mice with complete depletion of rpgrip1 also do not form rod OS, but cone OS was initially formed and abnormal23. In contrast to zebrafish and mouse mutants, rpgrip1 mutant dogs appeared to form both rod and cone OS24. The vertebrate photoreceptor OS is a specialized compartment, comprising stacks of disk membranes. It contains the visual pigment molecules essential for phototransduction. Rhodopsin is the most abundant molecule contained within OS membranes and appears to be required for OS elaboration. Mice carrying homozygous mutations in the Rho gene do not form OS43. Depletion of the transcription factor, CRX, which regulates expression of many retinal genes including the Rho gene, also results in a failure of OS formation in the mouse model44.
Rhodopsin is synthesized in the endoplasmic reticulum and undergoes glycosylation in the Golgi complex. After sorting and exiting from the Golgi complex, rhodopsin reaches the trans-Golgi network (TGN) membranes and incorporates into rhodopsin transport carriers (RTCs), where it binds the activated small GTPase Arf4 through its C-terminal VxPx ciliary targeting signal to initiate the assembly of the rhodopsin ciliary targeting complex. Rab8 is activated by Rabin8 during budding of RTCs and in turn this regulates the tethering of RTCs to the periciliary plasma membrane and fusion with the ciliary membranes. Once rhodopsin is in the ciliary membranes, it is further transported to rod OS through motor proteins and intraflagellar transport machinery45,46. Defects in any step of the trafficking procedures will result in rhodopsin mislocalization, abnormal rod OS formation and retinal degeneration. Rab8 is a major participant of rhodopsin-bearing vesicle trafficking and play a critical role for the delivery of rhodopsin-containing post-Golgi vesicles to the base of the connecting cilium. Early data showed Rab8 was enriched in post-Golgi vesicles, co-localized with rhodopsin-containing post-Golgi vesicles at the base of OS, and also localized at the ciliary stalk of OS. Rab8 was also associated and partially colocalized with microfilaments37. Transgenic Xenopus carrying the canine Rab8 dominant negative (T22N) form display rhodopsin mislocalization and accumulation of rhodopsin-bearing vesicles near the periciliary ridge complex, indicating Rab8 is required for docking the post-Golgi rhodopsin-containing membrane to the plasma membrane38. However, recent data demonstrated that mice with deletion of Rab8a only or with Rab8a deletion and overexpression of Rab8b dominant-negative form had normal outer segment structure and correct distribution of rhodopsin and other phototransduction proteins, suggesting Rab8 is not required for mouse rhodopsin targeting to outer segments47. However, the current findings in our study indicate that RPGRIP1 is required for Rab8 ciliary localization and is involved in rhodopsin ciliary vesicle transport in zebrafish.
The mechanisms underlying the role of RPGRIP1 for Rab8 ciliary localization remains elusive. It is possible that this is through RPGR or other RPGRIP1 interacting partners. CEP290, a component of the RGRIP1 protein complex and a ciliopathy-causing gene, is also required for Rab8 primary cilium localization48. CC2D2A, a CEP290 interactor, also has a role in Rab8 mediated ciliary vesicle transport, and deficiency of Cc2d2a in zebrafish leads to mislocalization of Rab8 in cone cells and abnormal OS development36. Depletion of another ciliopathy-causing gene, Ahi1, also a CEP290 interactor, in mice resulted in decreased Rab8 at the base of cilium39. The Bardet-Biedl syndrome (BBS) protein complex (BBSome) mediates ciliogenesis by recruiting Rab8 to the primary cilium through BBS1 physically interacting with Rabin849. The ciliopathies are a group of highly genetically heterogeneous disorders and the ciliopathy-associated proteins form a protein complex/network, e.g. BBSome and NPHP-JBTS-MKS, for their interlinked function49,50. It is highly possible that the ciliopathy protein complex/network may share some common molecular pathways, one of which is Rab8 mediated post-Golgi ciliary vesicle trafficking.
Rhodopsin mislocalization is a common phenotypic feature in humans and animal models with retinal ciliopathy. RPGRIP1 deficiency in mouse, dog and zebrafish causes rhodopsin mislocalization (Fig. 2C, Supplementary Material, Fig. S5, and ref.21,23,24). Similarly, depletion of two RPGRIP1 physically interacting partners, NPHP4 and SPATA7, in mice also results in mislocalization of rhodopsin18,20. It has been proposed that mislocalized rhodopsin contributes to photoreceptor cell death, that the level of mislocalized rhodopsin most likely correlates to the speed of retinal degeneration and that the reduction of mislocalized rhodopsin can ameliorate rod cell death18,51,52. Mislocalized rhodopsin triggered the apoptosis of rod cells in the retinas of Spata7 and Kif3a mutant mice18,52. It is probably that rod death in rpgrip1 −/− zebrafish is via apoptosis, which was confirmed by TUNEL assay (Fig. 4). How mislocalized rhodopsin triggers rod cell apoptosis remains to be determined, although an in vitro study has suggested rhodopsin mislocalization stimulates G proteins that increase the activity of adenylate cyclase thereby enhancing generation of cAMP53. Previous studies have reported that the cAMP level was increased in some RP animal models and, as a result, it has been proposed to be a promoter of photoreceptor cell apoptosis54.
In RP and LCA patients and animal models, loss of rod cells leads to secondary death of cones (1). Rpgrip1 −/− zebrafish recapitulate the clinical presentations of LCA and juvenile RP patients by displaying early rapid rod cell degeneration followed by secondary cone cell death (Fig. 3 and Supplementary Material, Fig. S4). Zebrafish have a cone-rich retina with four types of cones (red, green, blue and UV) that provide an excellent model to study the mechanisms of cone cell death. In rpgrip1 mutant zebrafish long cones (red and green cones) were fully degenerated by 13 months of age; short cones (blue and UV cones), however, appeared to be morphologically normal at that age (Supplementary Material, Fig. S4). Short cones were abnormally organized at 18 months of age and most single cones were degenerated by the age of 23 months. The mechanisms underlying selective cone cell death at different time points require further investigation.
In summary, the present study describes the cellular and molecular pathology of a novel rpgrip1 mutant zebrafish model. This model will provide an opportunity for future investigation into the cellular function of RPGRIP1 and elucidation of the underlying disease mechanisms as well as enabling development of drug candidates for the treatment of conditions caused by RPGRIP1 mutations.
Materials and Methods
Animals
An ENU (N-Nitroso-N-ethylurea) mutagenized library of F1 male zebrafish was prepared and ENU-induced heterozygous mutations were identified using CEL1-based TILLING (Targeting Induced Local Lesions IN Genomes)25. Zebrafish containing mutations were maintained for further characterization. All experiments using zebrafish were carried out in accordance with the UK home office animal care guidelines and approved by the Animal Ethics and Welfare Committee, Department of Life Sciences, Glasgow Caledonian University; a minimal number of animals were used for this study. Zebrafish larvae and adult were maintained at 28 °C on a 14/10 h light/dark cycle.
Bioinformatic analysis
Peptide sequences of human and zebrafish RPGRIP1 were obtained from Ensembl.org database and were aligned using Clustal Omega alignment tool (http://www.ebi.ac.uk/Tools/msa/clustalo/). Conserved sequences were boxshaded using BoxShade 3.21 (http://www.ch.embnet.org/software/BOX_form.html) and primers were designed using NCBI primer design tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
Gene expression analysis
Total RNA was extracted from different tissues of adult zebrafish and embryos at various stages of development using Trizol (Invitrogen) and cDNAs were synthesized using Trancriptor high fidelity cDNA synthesis kit (Roche). End-point PCR analysis was carried out using NEB standard Taq polymerase system and quantitative PCR was performed using platinum SYBR green qPCR supermix (Life technologies). The sequences of all the primers will be provided as requested.
Genotyping
The zebrafish were genotyped using DNA extracted from the fin clip. Individual fish were anaesthetised by immersing in 0.016% Tricaine for 30–45 seconds, then a small portion of the tail fin was clipped and placed in a sterile 0.2 ml PCR tube. Once the fin was clipped the fish was immediately placed in system water to recover. To extract DNA from the fin clip, 25 µl of methanol was added and incubated at room temperature for 5 minutes. 15 µl of TE-Tween20 (10 mM Tris, 1 mM EDTA (pH 8.0), 0.33% Tween 20) and 0.5 µl of 10 mg/ml proteinase K were added to the sample and incubated at 56 °C for at least 2 hours in a thermal cycler machine. The tube was then incubated at 98 °C for 10 minutes to inactivate the proteinase K. 135 µl of dH2O was added to the sample and mixed well. 2 µl of the extracted DNA was used as template to amplify the DNA fragment using primers GT211: AAAAATGTACAAAAACTAAGGCCTACC and GT212: AAGAGCGAGGATCTCGTTGATGGAAGCACTG. The PCR products were subjected to sequencing or restriction digestion using BbvI restriction enzyme. 3 µl of the PCR sample was added to 1 µl of 10X NEB buffer 2, 0.5 µl BbvI (10U/µl) and 5.5 µl dH2O, and incubated at 37 °C for 3 hours and the samples were subjected to electrophoresis on 3% agarose gel at 70 V for 90 minutes.
Histology and immunostaining
Enucleated eyes were fixed in 4% paraformaldehyde overnight. For haematoxylin & eosin staining and immunohistochemistry analysis, fixed eyes were dehydrated with series ethanol treatment and embedded in paraffin. For immunohistostaining, fixed eyes were washed in PBS twice (5 minutes each) and cryoprotected in 5%, 15% and 20% sucrose each for 2 hours (h) at room temperature (RT) and moulded in OCT medium (VWR) and 10 µm sections were cut in cryostat. 7 µm paraffin sections were deparaffinised and blocked in 2% BSA/PBS for 30 minutes at RT and then incubated in primary antibodies at RT for 2 h (Anti-rhodopsin (1:500; 4D2 from Abcam). After washing in PBS twice, sections were incubated at RT for 2 h in anti-FITC mouse (1:500; Sigma) diluted in 2% BSA/PBS. For immunostaining with anti-RPGR (1:500; Sigma, HPA001593), anti-Zpr1 (1:500; ZIRC), anti-Rab8 (1:500; Sigma), anti-GRK1 (1:50; Abcam), anti-GRK7a (1:50; gift from Prof. Stephan Neuhauss, University of Zurich) and anti-GNAT1 (1:50; Abcam) antibodies, 10 µm cryosections were thawed at RT for 30 minutes, blocked in 5% sheep serum, 0.3% Triton X-100 in PBS for 1 h and incubated in primary antibody diluted in diluting solution (1% BSA/PBS, 0.3% Triton X-100) for 2 h at RT. Then sections were washed with PBS thrice (5 minutes each) and then incubated in secondary antibodies for 2 h at RT (anti-FITC mouse, 1:500; Sigma), AlexaFluor 594-conjugated anti-rabbit (1:500; Invitrogen)). After incubating sections in the secondary antibody, the sections were washed with PBS twice (5 minutes each) and mounted with vectashield mounting medium containing DAPI, which stains the nuclei. All the sections were imaged using LSM 510 confocal microscopy (Carl Zeiss).
TUNEL assay
Cell death in the retinas of rpgrip1 −/− mutant zebrafish at 14 dpf, 1 mpf and 3 mpf was detected by the DeadEnd™ Fluorometric TUNEL System (Promega) following the manufacturer’s instruction. The cryosections of zebrafish eyes were washed with 1 × PBS for 5 min before being fixed again by 4% PFA/PBS. After permeabilisation with 20 μg/ml proteinase K solution, the sections were labelled with rTdT reaction mix for 1 hour at 37 °C and the reaction was stopped with 2 × SSC. Slides were mounted with DAPI and images were captured using ZEISS LSM 800 confocal microscopy. In order to quantify cell death, the number of TUNEL positive cells in a sample of 500 cells of the outer nuclear layer was counted from five slides (100 cells in each slide from each fish sample) of wildtype and mutant retinas respectively.
Electron microscopy
Anaesthetised zebra fish (5 days and 13 months old) were fixed in 4% Glutaraldehyde/1% Paraformaldehyde/0.1 M Sodium Cacodylate Buffer for 1.5hrs at room temperature. The specimens were washed twice in 0.1 M Sodium Cacodylate and then post-fixed in 1% Osmium Tetroxide/0.1 M Sodium Cacodylate for 1hr. The Osmium Tetroxide was washed off with three 15mins-changes in distilled water then specimens were stained with 0.5% Uranyl Acetate for 1hr and placed in the dark. The zebrafish were dehydrated through a series of ethanol 30, 50, 70 and 90% for 15 mins each then 100% Ethanol four 10 mins changes and finally dried in 100% Ethanol four 10 mins changes. The specimens were given four 10mins-changes of Propylene Oxide before leaving in a 1:1 PO:EPON resin mix overnight. Over the next day specimens were given 2–3 changes of pure EPON 812 Araldite then freshly embedded in moulds and polymerised at 60 °C for 24–48hrs. Ultrathin sections of 50–60 nm thickness were cut using a Leica UTC Ultratome, then collected on 100 mesh formvar coated copper grids and contrast stained with 2% Uranyl Acetate and Reynolds Lead Citrate for 5 mins each. The sections of zebrafish eyes were viewed on a Tecnai T20 TEM running at 200 kV and DM3/Tiff images captured using Gatan Digital imaging software.
Electroretinography (ERG)
ERG recording for the zebrafish was carried out according to methods described by Makhankov et al.55. 7dpf old larvae were dark-adapted for at least 30 minutes before ERG recording. Animals were anaesthetised in tricaine (MS222, Sigma) and immobilized using 0.8 mg/ml Esmeron (Sigma), then placed on top of a piece of wet filter tissue. A straight silver wire was placed beneath the larvae as the reference electrode. To hold the fish down and to provide oxygenation, a small piece of wet tissue was placed on the fish trunk. A glass suction electrode with a ~10 micron tip opening was positioned onto the fish cornea using an MX763OR manipulator from SD Instruments under a Nikon E600FN microscope. ERG recordings were made using Axon Multiclamp 700B amplifier with a gain of 2000, digitized with CED Power 1401 mkII and sampled with Signal 5 software (Cambridge Electronic Design, Cambridge) at 10 kHz. Signals were band-filtered between 3 and 100 Hz. Illumination was provided by a customised battery-powered white LED (50k mcd), controlled using TTL pulses generated through Power 1401 mkII. Test LED illumination was applied after 30 minutes of dark adaption.
Visual background adaption (VBA) assay
VBA assay was performed as previously described33. Briefly, Wildtype and rpgrip1 −/− embryos were incubated at 28 °C till 7dpf. Embryos were then put in Petri dishes and underwent a dark adaption for more than 30 min. Then, the Petri dishes were placing on the stand of the microscope and a bright light was shone on the embryos from above. Images were taken immediately when the light was switched on. The light remained on for 15 minutes and again images were taken.
Western blotting
14dpf wildtype and rpgrip1 mutant zebrafish heads (with eyes) were homogenised by T-PERTM tissue protein extraction reagent (Thermo Fisher Scientific, UK) with Complete™ protease inhibitor (Roche). Protein concentrations were determined Bio-Rad DCTM protein assay. The equal amounts of total proteins were separated by SDS-PAGE (NuPAGE; 10% gels) which was transferred into nitrocellulose membranes. The membranes were blocked in 5% non-fat milk in TBST at room temperature. Then, the target protein were detected by incubation with primary antibodies including RPGR (1:500, Sigma, HPA001593), GRK1 (1:500, Abcam), GNAT1 (1:500, Gene Tex), Rab8 (1:1000, Sigma) and acetylated tubulin (1:5000, Sigma). After washing, the membranes were incubated with fluorescence IRDye 600–800 RD-conjugated secondary antibodies in TBST. The specific bands were visualized and quantified using Li-cor Odyssey FC and Image Studio software.
Statistical analysis
All the statistical analysis was carried out using Graphpad Prism (Version 5) (GraphPad Software, www.graphpad.com). An unpaired two tail t test was carried out to determine statistically significant difference between wildtype and rpgrip1 −/− groups for rpgrip1 mRNA analyses and for ERG analyses. For the spatial and temporal gene expression analysis of rpgrip1 at various developmental stages and different tissues one-way ANOVA with Tukey’s multiple comparison tests was performed using GraphPad prism.
References
Wright, A. F., Chakarova, C. F., Abd El-Aziz, M. M. & Bhattacharya, S. S. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet 11, 273–284 (2010).
den Hollander, A. I., Roepman, R., Koenekoop, R. K. & Cremers, F. P. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 27, 391–419 (2008).
Boylan, J. P. & Wright, A. F. Identification of a novel protein interacting with RPGR. Hum Mol Genet 9, 2085–2093 (2000).
Roepman, R. et al. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet 9, 2095–2105 (2000).
Hong, D. H., Yue, G., Adamian, M. & Li, T. Retinitis pigmentosa GTPase regulator (RPGR)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J Biol Chem 276, 12091–12099 (2001).
Shu, X. et al. RPGR mutation analysis and disease: an update. Hum Mutat 28, 322–328 (2007).
Dryja, T. P. et al. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet 68, 1295–1298 (2001).
Gerber, S. et al. Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis. Eur. J Hum Genet 9, 561–571 (2001).
Booij, J. C. et al. Identification of mutations in the AIPL1, CRB1, GUCY2D, RPE65 and RPGRIP1 genes in patients with juvenile retinitis pigmentosa. J. Med. Genet. 42, e67 (2005).
Hameed, A. et al. Evidence of RPGRIP1 genemutations associated with receive cone-rod dystrophy. J. Med. Genet. 40, 616–619 (2003).
Mellersh, C. S. et al. Canine RPGRIP1 mutation establishes cone-rod dystrophy in miniature longhaired dachshunds as a homologue of human Leber congenital amaurosis. Genomics 88, 293–301 (2006).
Mavlyutov, T. A., Zhao, H. & Ferreira, P. A. Species-specific subcellular localization of RPGR and RPGRIP isoforms: implications for the phenotypic variability of congenital retinopathies among species. Hum. Mol. Genet. 11, 1899–1907 (2002).
Castagnet, P., Mavlyutov, T., Cai, Y., Zhong, F. & Ferreira, P. RPGRIP1s with distinct neuronal localization and biochemical properties associate selectively with RanBP2 in amacrine neurons. Hum. Mol. Genet. 12, 1847–1863 (2003).
Shu., X. et al. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum. Mol. Genet. 14, 1183–1197 (2005).
Coene, K. L. et al. The ciliopathy-associated protein homologs RPGRIP1 and RPGRIP1L are linked to cilium integrity through interaction with Nek4 serine/threonine kinase. Hum. Mol. Genet. 20, 3592–3605 (2011).
Arts, H. H., Cremers, F. P., Knoers, N. V. & Roepman, R. Focus on molecules: RPGRIP1. Exp. Eye Res. 88, 332–333 (2009).
Patnaik, S. R., Raghupathy, R. K., Zhang, X., Mansfield, D. & Shu, X. The role of RPGR and its interacting proteins in ciliopathies. J. Ophthalmol. 2015, 414781 (2015).
Eblimit, A. et al. Spata7 is a retinal ciliopathy gene critical for correct RPGRIP1 localization and protein trafficking in the retina. Hum. Mol. Genet. 24, 1584–1601 (2015).
Wang, H. et al. SPATA7 cause Leber congenital amaurosis and juvenile retinitis pigmentosa. Am. J. Hum. Genet. 84, 380–387 (2009).
Roepman, R. et al. Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations. Proc. Natl. Acad. Sci. USA 102, 18520–18525 (2005).
Zhao, Y. et al. The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc. Natl. Acad. Sci.,USA 100, 3965–3970 (2003).
Patil, H. et al. Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons. Cell Death Dis. 3, e355 (2012).
Won, J. et al. RPGRIP1 is essential for normal rod photoreceptor outer segment elaboration and morphogenesis. Hum. Mol. Genet. 18, 4329–4339 (2009).
Turney, C. et al. Pathological and electrophysiological features of a canine cone-rod dystrophy in the miniature longhaired dachshund. Invest Ophthalmol Vis. Sci. 48, 4240–4249 (2007).
Draper, B. W., McCallum, C. M., Stout, J. L., Slade, A. J. & Moens, C. B. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 77, 91–112 (2004).
Corless, J. M. Cone outer segments: a biophysical model of membrane dynamics, shape retention, and lamella formation. Biophys J. 102, 2697–2705 (2012).
Mustafi, D., Engel, A. H. & Palczewski, K. Structure of cone photoreceptors. Prog Retin Eye Res. 28, 289–302 (2009).
Kennedy, B. & Malicki, J. What drives cell morphogenesis: a look inside the vertebrate photoreceptor. Dev Dyn. 238, 2115–2138 (2009).
Cerveny, K. L., Varga, M. & Wilson, S. W. Continued growth and circuit building in the anamniote visual system. Dev Neurobiol. 72, 328–345 (2012).
Brockerhoff, S. E. et al. Light stimulates a transducing-independent increase of cytoplasmic Ca2+ and suppression of current in cones from the zebrafish mutant nof. J. Neurosci. 23, 470–480 (2003).
Stearns, G., Evangelista, M., Fadool, J. M. & Brockerhoff, S. E. A mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in zebrafish. J Neurosci. 27, 13866–13874 (2007).
Balm, P. H. & Groeneveld, D. The melanin-concentrating hormone system in fish. Ann. N.Y. Acad. Sci. 839, 205–209 (1998).
Neuhauss, S. C. et al. Genetic disorders of vision revealed by a behavioral screen of 400 essential loci in zebrafish. J. Neurosci. 19, 8603–8615 (1999).
Shu, X. et al. Zebrafish Rpgr is required for normal retinal development and plays a role in dynein-based retrograde transport processes. Hum. Mol. Genet. 19, 657–670 (2010).
Murga-Zamalloa, C. A., Atkins, S. J., Peranen, J., Swaroop, A. & Khanna, H. Interaction of retinitis pigmentosa GTPase: implications for cilia dysfunction and photoreceptor degeneration. Hum. Mol. Genet. 19, 3591–3598 (2010).
Bachmann-Gagescu, R. et al. The ciliopathy gene cc2d2a controls zebrafish photoreceptor outer segment development through a role in Rab8-dependent vesicle trafficking. Hum. Mol. Genet. 20, 4041–4055 (2011).
Deretic, D. et al. Rab8 in retinal photoreceptors may participate in rhodopsin transport and in rod outer segment disk morphogenesis. J. Cell Sci. 108(Pt 1), 215–224 (1995).
Moritz, O. L. et al. Mutant rab8 impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol. Bol. Cell 12, 2341–2351 (2001).
Westfall, J. E. et al. Retinal degeneration and failure of photoreceptor outer segment formation in mice with targeted deletion of the Joubert syndrome gene, Ahi1. J. Neurosci. 30, 8759–8768 (2010).
Liu, F. et al. Knockout of RP2 decreases GRK1 and rod transducin subunits and leads to photoreceptor degeneration in zebrafish. Hum. Mol. Genet. 24, 4648–4659 (2015).
Rinner, O., Makhankov, Y. V., Biehlmaier, O. & Neuhauss, S. C. Knockdown of cone-specific kinase GRK7 in larval zebrafish leads to impaired cone response recovery and delayed dark adaptation. Neuron 47, 231–242 (2005).
Wada., Y., Sugiyama, J., Okano, T. & Fukada, Y. GRK1 and GRK7: unique cellular distribution and widely different activities of opsin phosphorylation in the zebrafish rods and cones. J. Neurochem. 98, 824–837 (2006).
Humphries, M. M. et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 15, 216–219 (1997).
Morrow, E. M., Furukawa, T., Raviola, E. & Cepko, C. L. Synaptogenesis and outer segment formation are perturbed in the neural retina of Crx mutant mice. BMC. Neurosci. 6, 5 (2005).
Wang, J. & Deretic, D. Molecular complexs that direct rhodopsin transport to primary cilia. Prog Retin Eye Res. 38, 1–19 (2014).
Insinna, C. & Besharse, J. C. Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev. Dyn. 237, 1982–1992 (2008).
Ying, G. et al. Small GTPases Rab8a and Rab11a Are Dispensable for Rhodopsin Transport in Mouse Photoreceptors. PLoS One 11, e0161236 (2016).
Kim, J., Krishnaswami, S. R. & Gleeson, J. G. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 17, 3796–3805 (2008).
Nachury, M. V. et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 (2007).
Sang, L. et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528 (2011).
Louie, C. M. et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat. Genet 42, 175–180 (2010).
Lopes, V. S. et al. Dysfunction of heterotrimeric kinesin-2 in rod photoreceptor cells and the role of opsin mislocalization in rapid cell death. Mol. Biol. Cell 21, 4076–4088 (2010).
Alfinito, P. D. & Townes-Anderson, E. Activation of mislocalization opsin kills rod cells: a novel mechanism for rod cell death in retinal diseases. Proc.Natl.Acad.Sci.,USA 99, 5655–5660 (2002).
Traverso, V., Bush, R. A., Sieving, P. A. & Deretic, D. Retinal cAMP levels during the progression of retinal degeneration in rhodopsin P23H and S334ter transgenic rats. Invest. Ophthalmol. Vis. Sci. 43, 1655–1661 (2002).
Makhankov, Y. V., Rinner, O. & Neuhauss, S. C. An inexpensive device for non-invasive electroretinography in small aquatic vertebrates. J. Neurosci. Methods 135, 205–210 (2004).
Acknowledgements
We thank Prof. Stephan C.F. Neuhauss (University of Zurich) for the kind gift of the zebrafish GRK7a antibody. We also thank Prof. John A. Craft and Daphne L McCulloch (Glasgow Caledonian University) for advice and suggestions. We would like to thank the Royal Society of London, the National Eye Research Centre, the Visual Research Trust, Fight for Sight, the W.H. Ross Foundation, the Rosetrees Trust, and the Glasgow Children’s Hospital Charity for supporting this work. This work was also supported by the Deanship of Scientific Research at King Saud University for funding this research (Research Project) grant number ‘RGP – VPP – 219’. The maintenance of GCU Zebrafish Facility was funded by the EU INTERREG NEW noPILLS programme.
Author information
Authors and Affiliations
Contributions
Xinhua Shu designed the work. Rakesh K. Raghupathy, Xun Zhang, Fei Liu, Reem H. Alhasani, Lincoln Biswas, Saeed Akhtar and Wenchang Li carried out the experiments. Luyuan Pan and Cecilia B. Moens provided rpgrip1 mutant zebrafish. Wenchang Li, Mugen Liu, Breandan N. Kennedy and Xinhua Shu analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Raghupathy, R.K., Zhang, X., Liu, F. et al. Rpgrip1 is required for rod outer segment development and ciliary protein trafficking in zebrafish. Sci Rep 7, 16881 (2017). https://doi.org/10.1038/s41598-017-12838-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-12838-x
This article is cited by
-
Novel mutations of RPGR in Chinese families with X-linked retinitis pigmentosa
BMC Ophthalmology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.