Abstract
Several protein tyrosine phosphatase non-receptor 22 (PTPN22) single-nucleotide polymorphisms (SNPs) have been significantly related with rheumatoid arthritis (RA) susceptibility. Nevertheless, its potential influence on PTPN22 expression in RA has not been completely elucidated. Furthermore, PTPN22 binds to C-Src tyrosine kinase (CSK) forming a key complex in autoimmunity. However, the information of CSK gene in RA is scarce. In this study, we analyzed the relative PTPN22 and CSK expression in peripheral blood from 89 RA patients and 43 controls to determine if the most relevant PTPN22 (rs2488457, rs2476601 and rs33996649) and CSK (rs34933034 and rs1378942) polymorphisms may influence on PTPN22 and CSK expression in RA. The association between PTPN22 and CSK expression in RA patients and their clinical characteristics was also evaluated. Our study shows for the first time a marked down-regulation of PTPN22 expression in RA patients carrying the risk alleles of PTPN22 rs2488457 and rs2476601 compared to controls (p = 0.004 and p = 0.007, respectively). Furthermore, CSK expression was significantly lower in RA patients than in controls (p < 0.0001). Interestingly, a reduced PTPN22 expression was disclosed in RA patients with ischemic heart disease (p = 0.009). The transcriptional suppression of this PTPN22/CSK complex may have a noteworthy clinical relevance in RA patients.
Similar content being viewed by others
Introduction
Mutations in the protein tyrosine phosphatase non-receptor 22 (PTPN22) gene are associated with numerous autoimmune diseases1, 2. In this regard, several PTPN22 single-nucleotide polymorphisms (SNPs) have been significantly related with susceptibility to rheumatoid arthritis (RA)3,4,5,6,7. Moreover, it has been suggested that PTPN22 modulation, at a transcriptional level, may influence on inflammatory processes associated with RA8, 9. In particular, the expression of a PTPN22 isoform was correlated with RA activity in whole peripheral blood from RA patients10. Accordingly, PTPN22 is considered as the main non-HLA genetic risk factor involved in RA pathogenesis11, and, in fact, PTPN22 expression profiles have been proposed as biomarkers of RA8,9,10. Despite these evidences, the influence of PTPN22 genetic variants on PTPN22 expression in RA has not been completely elucidated and contradictory results have been published. In this regard, whereas a study did not find any polymorphism associated with the expression of PTPN22 splice forms in peripheral blood cells of RA patients8, a possible trend for association between PTPN22 rs2488457 SNP and PTPN22 gene expression was described in RA patients from China12.
Since PTPN22 is an intracellular tyrosine phosphatase that mainly inhibits T-cell receptor (TCR) signaling pathway, it is critically involved in the development of autoimmune diseases1, 2. This function is strengthened by the interaction with C-Src tyrosine kinase (CSK), also a negative regulator of TCR signaling13, 14. In this regard, two well-known CSK SNPs (rs34933034 and rs1378942) have been linked to systemic lupus erythematosus and systemic sclerosis15, 16. However, to the best of our knowledge, the information of CSK gene in RA is scarce.
A recent work from Walsh et al. reported some PTPN22 and CSK polymorphisms as potential expression quantitative trait loci (eQTLs) in whole blood from RA patients17 (information retrieved from additional files included in that manuscript). However, this study was not specially focused on the implication of PTPN22 and CSK in the pathogenesis of RA.
Taking all these considerations into account, in this study we determined if the most relevant PTPN22 (rs2488457, rs2476601 and rs33996649) and CSK (rs34933034 and rs1378942) polymorphisms may influence on PTPN22 and CSK expression in whole peripheral blood of RA patients when compared to healthy controls. The association between PTPN22 and CSK expression in RA patients and their clinical characteristics was also studied.
Results
Influence of PTPN22 genetic variants on PTPN22 expression
In a first step, we compared the PTPN22 mRNA expression between healthy controls and patients with RA (4.66 ± 1.52 vs 4.07 ± 1.57, p = 0.04). However, these differences were not statistically significant after adjustment (p = 0.22) (Fig. 1a).
Relative PTPN22 (a) and CSK (b) mRNA expression in healthy controls and patients with RA. PTPN22 and CSK expression was normalized to beta-actin and GAPDH as housekeeping genes. P-values were adjusted for sex, age, and cardiovascular risk factors. Horizontal bars indicate mean value of each study group. ns indicates not significant differences.
To determine if PTPN22 rs2488457, rs2476601 and rs33996649 polymorphisms may influence on PTPN22 expression, we assessed the PTPN22 expression in RA patients and controls carrying the reference and the risk allele for each polymorphism (Table 1). After checking that PTPN22 rs2488457, rs2476601 and rs33996649 were in HWE, we observed a significant down-regulation of PTPN22 in patients with RA carrying PTPN22 rs2488457 risk allele G and rs2476601 risk allele A compared to healthy controls (p = 0.004 and 0.007, respectively) (Fig. 2a,b and Table 1 ). A decreased PTPN22 mRNA expression was also disclosed between RA patients and healthy controls carrying the haplotype GA, composed of both risk alleles (Supplementary Table 1). No statistical analysis could be carried out to compare PTPN22 expression between patients and controls carrying the PTPN22 rs33996649 polymorphism risk allele T, since only one control subject carried this allele (Table 1). According to individuals carrying the reference allele, no significant differences in PTPN22 mRNA expression were found between patients with RA and controls in any of the cases (Table 1).
Relative PTPN22 and CSK mRNA expression in healthy controls and patients with RA according to PTPN22 and CSK polymorphisms. PTPN22 and CSK expression was normalized to beta-actin and GAPDH as housekeeping genes. P-values were adjusted for sex, age, and cardiovascular risk factors. (a and b) Differential PTPN22 expression between controls and patients with RA stratified according to their reference and risk allele of PTPN22 rs2488457 (a) and PTPN22 rs2476601 (b) polymorphisms. (c and d) Differential CSK expression between controls and patients with RA stratified according to their reference and risk allele of CSK rs34933034 (c) and CSK rs1378942 (d) polymorphisms. Horizontal bars indicate mean value of each study group.
Influence of CSK genetic variants on CSK expression
Firstly, we found a significant decrease in CSK mRNA expression in RA patients compared to controls (2.50 ± 0.77 vs 3.28 ± 1.02, p < 0.0001) (Fig. 1b). Then, we compared the CSK expression between these two study groups according to their reference and risk allele for CSK rs34933034 and rs1378942 (Table 2). Both polymorphisms were in HWE. We disclosed a decreased expression of CSK in patients with RA respect to controls, independently on whether they carried the CSK rs34933034 and rs1378942 reference or risk allele (Fig. 2c,d and Table 2 ). This decrease in CSK expression was statistically significant in all the cases (p < 0.05), except for individuals who carried CSK rs34933034 risk allele A, that showed a marginally significant decrease (p = 0.059). Similar results were obtained when haplotype analysis was performed (Supplementary Table 1).
Association of PTPN22 and CSK mRNA expression with clinical characteristics in RA patients
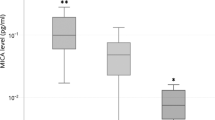
Interestingly, a significant association between PTPN22 expression and ischemic heart disease (IHD) was observed in patients with RA, after and adjustment for sex, age at time of study, and CV risk factors (p = 0.009) (Fig. 3). In this regard, RA patients with IHD had a lower PTPN22 expression (3.02 ± 1.11) than those without IHD (4.32 ± 1.57) (Fig. 3). In addition, an inverse correlation was disclosed between CSK expression in patients with RA and their C-reactive protein (CRP) levels at time of study (r = −0.26, p = 0.019). No significant differences were found between the PTPN22 and CSK mRNA expression of patients with RA and other clinical characteristics, including disease duration, rheumatoid factor, anti-cyclic citrullinated peptide antibodies status, erosions, extra-articular manifestations, and CRP levels and erythrocyte sedimentation rate at RA onset.
Decrease PTPN22 mRNA expression in RA patients with IHD. Box plot showing the PTPN22 mRNA expression, normalized to beta-actin and GAPDH expression, in patients with rheumatoid arthritis (RA) stratified according to the presence (n = 16) or absence of ischemic heart disease (IHD) (n = 71). P-value was adjusted for sex, age, and cardiovascular risk factors.
PTPN22 and CSK serum levels in RA and controls
The measurement of circulating levels of PTPN22 and CSK was assessed to provide supplementary reliable data between the expression of mRNA and the corresponding protein levels. No significant differences were found between patients with RA and controls in PTPN22 and CSK protein levels after adjustment by potential confounding factors. In this regard, PTPN22 levels were 450.59 ± 328.76 pg/ml in RA patients and 285.80 ± 259.70 pg/ml in controls (p = 0.20). Similarly, CSK levels were 39.02 ± 26.10 pg/ml in RA patients and 32.35 ± 24.53 in controls (p = 0.76).
Discussion
Since an association between some PTPN22 genetic variants and RA susceptibility was described3,4,5,6,7, functional analyses of PTPN22 have been considered necessary to further understand the role of this gene in RA and other autoimmune diseases1, 9. Our study disclosed for the first time that two PTPN22 genetic variants, rs2488457, located in the promoter region, and rs2476601, located in exon 14, down-regulate the transcription and function of PTPN22 in patients with RA compared to controls, who have higher PTPN22 expression. This was observed both when the PTPN22 SNPs were analyzed individually or forming haplotypes. Our results confirm the trend found between PTPN22 rs2488457 and PTPN22 expression in Chinese RA patients12. Furthermore, Zhang et al. had previously shown that this SNP was associated not only with the development of an inflammatory disease known as Vogt-Koyanagi-Harada, but also with a decreased PTPN22 expression, only assessed in healthy controls18. A potential influence between rs2488457 and ulcerative colitis was also described19. Regarding PTPN22 rs2476601, it has been clearly shown that it is the main variant involved in the susceptibility to develop autoimmune diseases1,2,3, and due to its functional effect it is able to decrease the interaction between PTPN22 and CSK14, 20. No association between this SNP and PTPN22 modulation had previously been shown8, 21, 22. However, PTPN22 rs2476601 and also PTPN22 rs2488457 have been recently proposed as potential cis-eQTLs in whole blood from RA patients17. This finding is in keeping with our results, supporting that these genetic variants may play an important role in the regulatory region of PTPN22. Based on our data, we feel that the decreased PTPN22 expression in patients carrying the risk alleles of PTPN22 rs2488457 and rs2476601 may lead to a pro-inflammatory status in RA, since T cells activation would not be inhibited by this gene. In fact, PTPN22 deficiency significantly increased the severity of disease in a mouse model of RA23.
An important implication of PTPN22 in the development of atherosclerosis has previously been proposed24. In this regard, we disclosed for the first time a marked down-regulation of PTPN22 gene in RA patients with IHD. Considering a previous study from our group which showed that PTPN22 rs2476601 polymorphism was not directly associated with an increased risk of CV disease in RA patients25, we think that this association may be SNP-independent and could constitute an useful tool to identify RA patients with a higher CV risk.
We also demonstrated a significant decrease of CSK gene in peripheral blood from RA patients. A down-regulation of CSK, which acts as a tumor suppressor gene, has previously been associated with colon cancer26. However, the role of CSK gene in RA remains to be elucidated, even though it has been proposed as an autoimmunity risk factor16. Since a study demonstrated a decrease of inflammation in a rat model of RA injected with Csk virus27, we consider that the down-regulation of CSK may cause a high inflammatory response in our patients, contributing to RA progression. Finally, we observed that CSK rs34933034 and rs1378942 did not influence on CSK expression since its reduction in RA patients as compared to controls was independent of the polymorphisms. Therefore, the lower CSK mRNA expression shown in our RA patients when compared to healthy controls may be more related to the disease itself. The negative association of CSK mRNA expression with CRP, a systemic marker of inflammation, reinforces our hypothesis.
In summary, our study reveals for the first time that the risk alleles of PTPN22 rs2488457 and PTPN22 rs2476601 influence on the down-regulation of PTPN22 in RA. This confirms the important role of PTPN22 in RA. Furthermore, it indicates that CSK may also be considered as a genetic risk factor for the development of RA. The transcriptional suppression of the PTPN22/CSK complex, as a key regulator of the immune response, may have a noteworthy clinical relevance in patients with RA, playing an important role in disease progression. Compensatory mechanisms through other biologic pathways may be crucial to control RA.
Subjects and Methods
Patients and controls
For experiments involving humans and the use of human blood samples, all the methods were carried out in accordance with the approved guidelines and regulations, according to the Declaration of Helsinki. All experimental protocols were approved by the local Ethics Committees of Clinical research of Cantabria for Hospital Universitario Marqués de Valdecilla in Santander and of Madrid for Hospital Universitario de La Princesa. An informed consent was obtained from all subjects before being enrolled.
Peripheral blood samples were collected from 89 patients with RA who met the 1987 American College of Rheumatology (ACR) and also the 2010 ACR/European League Against Rheumatism criteria for RA28, 29 and 43 healthy controls recruited from Hospital Universitario Marqués de Valdecilla (Santander, Spain) and Hospital Universitario de La Princesa (Madrid, Spain). Supplementary Table 2 shows the main demographic and clinical characteristics of subjects enrolled in this study.
Expression profiles of PTPN22 and CSK genes
Total RNA from peripheral blood was isolated using NucleoSpin RNA Blood Midi kit (Macherey-Nagel) according to the manufacturer’s protocol, as we previously described30, 31. 1 μg of total RNA was reverse-transcribed using iScriptTM Advanced cDNA Synthesis Kit for RT- qPCR (Bio-Rad, Hercules, CA, USA). 20 ng of cDNA was used for quantitative real-time PCR (qPCR), using SsoAdvancedTM Universal SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA). Primers for amplification of PTPN22 (135 bp, UniGene ID Hs.535276), CSK (73 bp, UniGene ID Hs.77793), beta-actin (62 bp, UniGene ID Hs.520640) and GAPDH (117 bp, UniGene ID Hs. 544577) genes were acquired from PrimePCR Assays, Bio-Rad, Hercules, CA, USA. All samples were examined in triplicate and controls were included in each reaction. qPCRs were performed in a 7900 HT real-time instrument (Applied Biosystems, Foster City, CA, USA) following the conditions previously reported31. Amplicons of the different genes and a 50 base pairs DNA ladder (ThermoScientific) were electrophoresed on 2% agarose gel stained with GelGreen Nucleic Acid Gel stain (Biotium) to check the appropriate size of each amplicon (Supplementary Figure 1). CSK gene was expressed successfully in all samples. However, PTPN22 was not detected in 2 out of the 89 patients enrolled. The relative PTPN22 and CSK mRNA (messenger RNA) expression was analyzed by the comparative threshold cycle method, using beta-actin and GAPDH as housekeeping genes30. Normalized values were obtained for each sample and mean values were determined for each study group.
PTPN22 and CSK genotyping
Genomic DNA was extracted from peripheral blood samples using NucleoSpin Blood Kit (Macherey-Nagel) according to the manufacturer’s recommendations. Genotyping of the PTPN22 rs2488457 (C > G), PTPN22 rs2476601 (G > A), PTPN22 rs33996649 (C > T), CSK 34933034 (G > A) and CSK rs1378942 (A > C) was performed using TaqMan probes in a 7900 HT real-time instrument (Applied Biosystems, Foster City, CA, USA), as previously reported32. Negative controls were included to check the accuracy of genotyping. The linkage disequilibrium pattern of the PTPN22 and CSK polymorphisms is displayed in Supplementary Figure 2.
PTPN22 and CSK serum levels by enzyme-linked immunosorbent assay (ELISA)
PTPN22 and CSK serum levels were determined using commercial kits acquired from CUSABIO (Catalog Number: CSB-EL019036HU and CSB-EL006056HU, respectively), according to the manufacturer’s instructions. All samples were analyzed in duplicate.
Statistical analysis
First, Student’s t test was used to compare differences in PTPN22 and CSK mRNA expression between: patients and controls, patients and controls stratified according to their reference or risk allele for each SNP, as well as patients and controls stratified according to PTPN22 and CSK haplotypes. Differences in PTPN22 and CSK serum levels between RA patients and controls were also assessed by Student’s t test. Results were expressed as mean ± standard deviation for each study group. Next, covariance analysis was performed in order to adjust the results by potential confounding factors, including sex, age at time of study, and cardiovascular (CV) risk factors (hypertension, dyslipidemia, smoking, diabetes and obesity)33. The association between PTPN22 and CSK mRNA expression in RA patients and their clinical characteristics was evaluated using Student’s t test or Pearson partial correlation coefficients (r) as required, after adjusting for the confounding factors commented above. In all cases, p-values < 0.05 were considered statistically significant. Regarding genotyping, Hardy-Weinberg equilibrium (HWE) was checked using Χ2 test. The statistical power of the study is displayed in Supplementary Table 3. All these analysis were performed with STATA statistical software 12.0 (Stata Corp., College Station, TX, USA).
References
Stanford, S. M. & Bottini, N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol. 10, 602–611 (2014).
Gregersen, P. K. Gaining insight into PTPN22 and autoimmunity. Nat Genet. 37, 1300–1302 (2005).
Jiang, Y. et al. Meta-analysis of 125 rheumatoid arthritis-related single nucleotide polymorphisms studied in the past two decades. PLoS One. 7, e51571 (2012).
Begovich, A. B. et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 75, 330–337 (2004).
Martín, J. E. et al. Evidence for PTPN22 R620W polymorphism as the sole common risk variant for rheumatoid arthritis in the 1p13.2 region. J Rheumatol. 38, 2290–2296 (2011).
Rodríguez-Rodríguez, L. et al. The PTPN22 R263Q polymorphism is a risk factor for rheumatoid arthritis in Caucasian case-control samples. Arthritis Rheum. 63, 365–372 (2011).
Orozco, G. et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 52, 219–224 (2005).
Ronninger, M. et al. The balance of expression of PTPN22 splice forms is significantly different in rheumatoid arthritis patients compared with controls. Genome Med. 4, 2 (2012).
Díaz-Gallo, L. M. & Martín, J. PTPN22 splice forms: a new role in rheumatoid arthritis. Genome Med. 4, 13 (2012).
Chang, H. H. et al. PTPN22.6, a dominant negative isoform of PTPN22 and potential biomarker of rheumatoid arthritis. PLoS One. 7, e33067 (2012).
Messemaker, T. C., Huizinga, T. W. & Kurreeman, F. Immunogenetics of rheumatoid arthritis: understanding functional implications. J Autoimmun. 64, 74–81 (2015).
Huang, J. J. et al. A PTPN22 promoter polymorphism −1123G > C is associated with RA pathogenesis in Chinese. Rheumatol Int. 32, 767–771 (2012).
Cloutier, J. F. & Veillette, A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med. 189, 111–121 (1999).
de la Puerta, M. L. et al. The autoimmunity risk variant Lyp-W620 cooperates with CSK in the regulation of TCR signaling. PLoS One. 8, e54569 (2013).
Manjarrez-Orduño, N. et al. CSK regulatory polymorphism is associated with systemic lupus erythematosus and influences B-cell signaling and activation. Nat Genet. 44, 1227–1230 (2012).
Martín, J. E. et al. Identification of CSK as a systemic sclerosis genetic risk factor through genome wide association study follow-up. Hum Mol Genet 21, 2825–2835 (2012).
Walsh, A. M. et al. Integrative genomic deconvolution of rheumatoid arthritis GWAS loci into gene and cell type associations. Genome Biol. 17, 79 (2016).
Zhang, Q. et al. A functional variant of PTPN22 confers risk for Vogt-Koyanagi-Harada syndrome but not for ankylosing spondylitis. PLoS One 9, e96943 (2014).
Chen, Z. et al. Association of PTPN22 gene (rs2488457) polymorphism with ulcerative colitis and high levels of PTPN22 mRNA in ulcerative colitis. Int J Colorectal Dis. 28, 1351–1358 (2013).
Vang, T. et al. LYP inhibits T-cell activation when dissociated from CSK. Nat Chem Biol. 8, 437–446 (2012).
Viken, M. K. et al. The PTPN22 promoter polymorphism −1123G > C association cannot be distinguished from the 1858C > T association in a Norwegian rheumatoid arthritis material. Tissue Antigens. 70, 190–197 (2007).
Machado-Contreras, J. R. et al. Distribution of PTPN22 polymorphisms in SLE from western Mexico: correlation with mRNA expression and disease activity. Clin Exp Med. 16, 399–406 (2016).
Maine, C. J., Marquardt, K., Cheung, J. & Sherman, L. A. PTPN22 controls the germinal center by influencing the numbers and activity of T follicular helper cells. J Immunol. 192, 1415–1424 (2014).
Saccucci, P. et al. Atherosclerosis and PTPN22: a study in coronary artery disease. Cardiology. 119, 54–56 (2011).
Palomino-Morales, R. et al. Lack of association of PTPN22, STAT4 and TRAF1/C5 gene polymorphisms with cardiovascular risk in rheumatoid arthritis. Clin Exp Rheumatol. 28, 695–701 (2010).
Kunte, D. P. et al. Down-regulation of the tumor suppressor gene C-terminal Src kinase: an early event during premalignant colonic epithelial hyperproliferation. FEBS Lett. 579, 3497–3502 (2005).
Takayanagi, H. et al. Suppression of arthritic bone destruction by adenovirus-mediated csk gene transfer to synoviocytes and osteoclasts. J Clin Invest. 104, 137–146 (1999).
Arnett, F. C. et al. The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 31, 315–224 (1988).
Aletaha, D. et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 62, 2569–2581 (2010).
Remuzgo-Martínez, S. et al. Decreased expression of methylene tetrahydrofolate reductase (MTHFR) gene in patients with rheumatoid arthritis. Clin Exp Rheumatol. 34, 106–110 (2016).
Remuzgo-Martínez, S. et al. Expression of osteoprotegerin and its ligands, RANKL and TRAIL, in rheumatoid arthritis. Sci Rep. 6, 29713 (2016).
López-Mejías, R. et al. Role of PTPN22 and CSK gene polymorphisms as predictors of susceptibility and clinical heterogeneity in patients with Henoch-Schönlein purpura (IgA vasculitis). Arthritis Res Ther. 17, 286 (2015).
Gonzalez-Gay, M. A. et al. HLA-DRB1 and persistent chronic inflammation contribute to cardiovascular events and cardiovascular mortality in patients with rheumatoid arthritis. Arthritis Rheum. 57, 125–132 (2007).
Acknowledgements
We wish to thank all the patients with RA and controls who participated to make this study possible. This study was supported by European Union FEDER funds and “Fondo de Investigación Sanitaria” (grant PI12/00060 and PI15/00525) from “Instituto de Salud Carlos III” (ISCIII, Health Ministry, Spain). This work was also partially supported by RETICS Programs RD12/0009 (RIER) from ISCIII (Spain), and in part by grants from the European IMI BTCure Programme. SR-M is supported by funds from the RETICS Program (RIER) from the ISCIII, Spain (RD16/0012/0009). FG is a recipient of a Sara Borrell post-doctoral fellowship from the ISCIII (CD15/00095). RL-M is supported by funds of the Miguel Servet type I programme from the ISCIII (CP16/00033). BU is supported by funds from the RETICS Program (RIER) from the ISCIII (RD12/0009/0013).
Author information
Authors and Affiliations
Contributions
S.R.-M. and F.G. carried out the conception and design of the study, expression assays, statistical analysis and interpretation of data and drafted the manuscript. S.C. and A.C. participated in the acquisition of clinical data and samples, interpretation of data, and helped to draft the manuscript. P.M.-F., B.U., V.M., V.P., J.G.-V., T.P., G.O.-V., J.I.-V., R.B. and J.M. participated in acquisition of clinical data, expression and genotyping assays, and revised critically the manuscript. J.L. was involved in the statistical analysis, interpretation of data and helped to draft the manuscript. R.L.-M. and M.A.G.-G. made substantial contributions to the conception and study design, interpretation of data, coordination, and contributed substantially in the manuscript drafting. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Remuzgo-Martínez, S., Genre, F., Castañeda, S. et al. Protein tyrosine phosphatase non-receptor 22 and C-Src tyrosine kinase genes are down-regulated in patients with rheumatoid arthritis. Sci Rep 7, 10525 (2017). https://doi.org/10.1038/s41598-017-10915-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-10915-9
This article is cited by
-
Selection signatures for local and regional adaptation in Chinese Mongolian horse breeds reveal candidate genes for hoof health
BMC Genomics (2023)
-
PTPN22 +788 G>A (R263Q) Polymorphism is Associated with mRNA Expression but it is not a Susceptibility Marker for Rheumatoid Arthritis Patients from Western Mexico
Biochemical Genetics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.