Abstract
We report a simple single-molecule fluorescence imaging method that increases the temporal resolution of any type of array detector by >5-fold with full field-of-view. We spread single-molecule spots to adjacent pixels by rotating a mirror in the detection path during the exposure time of a single frame, which encodes temporal information into the spatial domain. Our approach allowed us to monitor fast blinking of an organic dye, the dissociation kinetics of very short DNA and conformational changes of biomolecules with much improved temporal resolution than the conventional method. Our technique is useful when a large field-of-view is required, for example, in the case of weakly interacting biomolecules or cellular imaging.
Similar content being viewed by others
Introduction
A camera-based detector has been widely used to study many biological systems with single-molecule fluorescence techniques because of its high sensitivity and high-throughput1, 2. However, in spite of remarkable technical advancements over the last decade, the maximum frame rate of the detector, i.e. electron multiplying charge-coupled device (EMCCD) or scientific complementary metal-oxide semiconductor (sCMOS) camera is still at 50–100 frames per second. Cropping the field-of-view (FOV) is the simplest way to improve temporal resolution, however, it decreases the number of molecules that can be observed simultaneously3. This is especially disadvantageous for studying weakly interacting biomolecules because the complex formation will occur very rarely4. For example, assuming that the binding rate is 106 M−1 s−1 and the accessible concentration of fluorophore-labeled biomolecules is 10 nM in TIRF microscopy, the rate of the complex formation amounts to ~10−2 s−1. Alternatively, stroboscopic illumination could be used, but it loses the dynamic information during which the excitation pulses are turned off5. Thus, a technique that uses a full chip but provides higher temporal resolution than a given frame rate is still lacking.
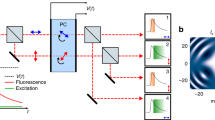
Interestingly, high-speed photography overcame frame rate problem long time ago by means of a rapidly rotating mirror that spreads images to multiple cameras at different time points6. Inspired by that technique, we devised a simple method that encodes temporal information into the spatial domain by exploiting the sparsity of single-molecule samples similar to spatially encoding a spectrum7 or an axial position of single molecules8. To do so, we used an objective-type total internal reflection fluorescence microscope with EMCCD, in which a beam-splitter and a single-axis galvo mirror were installed between two relay lenses (Fig. 1a, Supplementary Fig. 1). During the exposure period (∆t = 20 ms), we swept the galvo mirror in N steps (for instance, N = 5), which resulted in N equally spaced single-molecule spots produced on the image plane with higher temporal resolution (∆t/N = 4 ms) (Fig. 1b,c, Supplementary Fig. 2). The number of steps and the distance between the spots were controlled by a waveform generator (Supplementary Fig. 3). The beam splitter was used to obtain unmodified images either for comparison or for determination of an initial point of the event.
Spatially encoded fast single molecule spectroscopy with full field-of-view. (a) Schematic of spatially encoding temporal information in single-molecule imaging. The galvo mirror was rotated during the camera exposure time. BS, beam splitter; GM, galvo mirror. (b,c) Single-molecule images of Atto647N attached to single-stranded DNA obtained by conventional (left) and high temporal resolution imaging (right) at 2 mM Trolox. Average of ten frames in (c). (d) Blinking dynamics of Atto647N at 50 µM Trolox. Single-molecule raw images (top) and fluorescence time traces (bottom) at 4-ms time resolution. Scale bars, 1 µm (b,d) and 2 µm (c).
Results
As a proof-of-principle experiment, we first recorded fluorescence transients of Atto647N labeled to a single-stranded DNA on the surface. Photoblinking happens when the fluorophore intermittently switches back and forth between a bright fluorescent state and a dark nonfluorescent state on a timescale that ranges from microseconds to minutes, depending on different dark states9. However, an addition of Trolox with oxygen removal effectively prevents photoblinking and photobleaching of many fluorophores10 due to a combinatorial effect of its reducing and oxidizing properties11. For Atto647N, the on-count and the off-time are dependent on the concentration of the reducer (Trolox) and oxidizer (a quinone derivative of Trolox), respectively, and thus, its blinking dynamics could be readily tuned by changing the concentration of Trolox. As shown in Fig. 1d, our method enabled us to observe clearly the blinking dynamics of the fluorophore at low concentration of Trolox, whereas the temporal resolution of 20 ms by the conventional method did not allow measuring the blinking dynamics (Supplementary Fig. 4). We used auto-correlation analysis to determine the off-time12, 13, which was 4.6 ± 0.8 ms and 3.5 ± 0.5 ms at 50 μM and 200 μM Trolox, respectively (Supplementary Fig. 5). Both methods exhibited steady fluorescence intensity at high concentration of Trolox (Supplementary Fig. 6). Note that the fast blinking dynamics with millisecond time scale has been observed generally through confocal microscopy with a single pixel detector11.
Since signal-to-noise ratio (SNR) is a crucial criterion in fast single-molecule measurements, we analyzed it using fluorescence trajectories of Atto647N at 2 mM of Trolox (n > 50). Here we defined SNRB = (I S − I B)/σ B and SNRS = (I S − I B)/σ s, where I S is the mean of signal intensity, I B is the mean of background intensity, σ s and σ B are their respective standard deviations. The measured SNRB and SNRS were 34.0 ± 5.2 and 11.5 ± 3.1 at 20 ms resolution whereas they were 8.3 ± 1.4 and 4.1 ± 0.6 at 4 ms resolution (Supplementary Fig. 7). At given illumination intensity, the average photobleaching time was 69.7 s.
Next, we measured the dissociation kinetics of short double-stranded DNA (n < 8 nt) as an example of weakly interacting biomolecules. The binding (association) and unbinding (dissociation) of double-stranded DNA (dsDNA) through base-pairing is an essential process in DNA replication and repair14, and those processes are also used for point accumulation imaging in nanoscale topography (DNA-PAINT), leading to super-resolution fluorescence imaging for fixed samples15. However, the kinetics of extremely short dsDNA are still elusive due to the fact that (i) the dissociation rate is fast (k off > 20 s−1)15, and (ii) the association rate (104~106 M−1s−1) limits the number of observable binding events unless the molecules are confined to the vesicle14 or trapped electro-kinetically16. A previous study reported at least seven contiguous base pairs were required for fast annealing14 and the unbinding rate k off (1/τon) of 9-bp DNA with a mismatched sequence was ~5 s−1. Here, our method allowed us to observe the rare binding events of the 7-bp DNA strand with a 1-bp mismatch at one end of the duplex (7-1 bp) labeled with Cy3B (Fig. 2a, Supplementary Fig. 8), exhibiting the on-time (τon = 1/k off) of 7.7 ± 0.5 ms. We excluded the possibility that on/off fluorescence signals were due to the photoblinking of Cy3B (Supplementary Fig. 9). Our results were in good agreement with the base pair length dependence of the on-time as previously reported (Fig. 2b)15. Compared to cropping FOV, our method yielded 4.5-fold higher throughput (Fig. 2c) and allowed for the detection of more molecules for imaging densities up to 0.13 molecules/μm2 (Fig. 2d and Supplementary Fig. 10). But at higher imaging density, overlapping between the swept spots results in diminished throughput.
Binding and unbinding kinetics of short double-stranded DNA. (a) Dwell time histogram of 7-1 bp over five movies of 2,000 frames. Inset: Single-molecule raw images of binding and unbinding events of Cy3B-labeled single-stranded DNA. (b) Dependence of the on-time (τon) on the length of DNA duplexes from five movies. The on-time of 7-1, 7 and 9 bp were 7.7 ± 0.5 ms, 43.5 ± 2 ms and 530 ± 12 ms, respectively. Data are the mean ± standard deviation from >6,000 binding events. (c) The number of binding events of 7-1 bp with sweeping mirror or cropping FOV from five movies. Scale bars, 1 µm. (d) The number of resolvable molecules at different imaging densities.
Lastly, we monitored the conformational dynamics of biomolecules using single-molecule FRET where we slightly modified the detection setup (Supplementary Fig. 1b). In the presence of magnesium ions, a four-way DNA structure known as Holliday junction (HJ) folds itself into two possible stacked structures (Fig. 3a)17. Previous studies showed that the transition rates were highly dependent on DNA sequences, concentrations of counter ions and temperature17. Particularly, the transition rate decreases dramatically with an increase of the concentration of magnesium. In order to observe fast dynamics of HJ, we used a low magnesium concentration during the experiments. By hidden Markov model analysis, we observed fast two-state conformational changes of single HJ molecules (Fig. 3b and Supplementary Fig. 11). Two distinct FRET populations were markedly revealed at 5-ms time resolution as contrasted with a broad FRET distribution obtained at 20-ms time resolution (Fig. 3c). The transition rate at 2 mM of magnesium ions was 61.1 ± 4.7 s−1 (Fig. 3d) which is in agreement of with the literature18.
Holliday junction dynamics. (a) Cartoon of Holliday junction dynamics. (b) Single-molecule fluorescence intensity (top) and FRET time traces (bottom) of Holliday junction showing rapid conformational changes at 2 mM Mg2+. In FRET time trace (solid gray line), a red solid line is a fit with hidden Markov model. (c) Single-molecule FRET histograms of HJ dynamics with 20-ms and 5-ms time resolution from ~750 and ~3,000 data points, respectively. (d) Transition rates of Holliday junctions at different magnesium concentrations. More than 30 molecules from 5 movies were taken.
Discussion
Our simple method demonstrated 5-fold higher temporal resolution in single-molecule fluorescence spectroscopy, which enables the study of 100–200 molecules simultaneously. Further improvement in temporal resolution can be realized with increase of the step number N up to 10. In addition, our method could be used at even higher imaging densities with many overlapped spots taking advantage of that the peak positions of each spot are pre-determined by the applied voltage. Note that it is applicable to not only a larger detector such as sCMOS19 for improved throughput, but also to improve the temporal resolution of zero-mode waveguides20. Our technique will be particularly useful when cropping FOV is not feasible, for example, in the case of cellular imaging.
Methods
Single-molecule TIRF microscope
Single-molecule imaging was carried out on an objective-type total internal reflection fluorescence (TIRF) microscope built around Olympus IX73 (Supplementary Fig. 1a) similar to previous work3. Combined via a polarizing beam splitter, a green laser (Cobolt 06-DPL,532 nm), for excitation of Cy3B) and a red laser (Cobolt 06-MLD, 638 nm) with a laser clean-up filter were coupled to a single mode fiber. Collimated beams from the fiber were focused onto the back focal plane of an objective lens (UPlanSApo, 100x/1.40 oil, Olympus) by a focusing lens (f = 300 mm) and reflected by a multiband dichroic mirror (Di03-R405/488/532/635, Semrock). Fluorescence emission was collected by the same objective and filtered through a double notch filter (ZET532/638 m, Chroma). Then it was passed through the tube lens (fTL = 180 mm) and two relay lenses (f3 = 100 mm, f4 = 120 mm), and detected by EMCCD (iXon Ultra 897, Andor). An adjustable slit was located at the intermediate image plane to limit the final image to half of the EMCCD. A single-axis galvo mirror (GVS211, Thorlabs) placed between the relay lenses was rotated in a step-wise manner by a function generator (DG1032Z, Rigol) and it was synchronized with an output of EMCCD (Supplementary Fig. 1c). The step response time of the galvo mirror was 0.6 ms. The time lag between the steps was negligible. For the measurements of dye blinking and short stranded DNA binding/unbinding events, a beam splitter (BSS10R, 30:70 (R:T), Thorlabs) was installed in front of the galvo mirror to separate the fluorescence beam into two channels, i.e. a conventional image with 20-ms time resolution and a spatially encoded high temporal resolution image. For the single-molecule FRET experiments, the fluorescence beam was directly swept by the galvo mirror and then split into donor and acceptor channels through a dichroic beam splitter (FF640-FDi01, Semrock) (Supplementary Fig. 1b). In both cases, the separated beams were focused onto the left and right side of the EMCCD with 512 × 512 pixels corresponding to an imaging area of 68 µm × 68 µm. The exposure time was 20 ms and the lag between images was 1.7 ms. In smFRET experiments, a short-pass filter was additionally used to suppress auto-fluorescence from the coverslip.
Temporal calibration
We generated N-step function by raising the voltage from 0 to V in V/(N − 1) millivolt increments, where N is the number of spots and V is the voltage amplitude of the final spot (Supplementary Fig. 1c). While the generated waveform was fed into the galvo mirror, fluorescent crimson beads of 200-nm diameter (F8806, Invitrogen) sparsely immobilized on the coverslip were imaged by the TIRF microscope (Supplementary Fig. 3a). The total sweeping distance (L) between the first and the last spots increased linearly with the voltage applied to the galvo mirror, exhibiting the slope of 0.25 pixel/mV (Supplementary Fig. 3b). For unambiguous measurements, we set a criterion that the distance between two adjacent spots, 0.25 V/(N−1) in pixel is approximately double the full width at half maximum (FWHM) of single molecule spot. For example, since the FWHM of single Atto647N was 2.56 ± 0.55 pixels, corresponding to 340 ± 73 nm, all spots were well separated when the sweeping voltage was 96 mV (24 pixels) in 5 steps case.
Data analysis
Similar to smFRET analysis1, 21, we pre-selected single-molecule spots on the left side and then identified the corresponding spots on the right side by the mapping coefficients which were calculated using the fluorescent beads with V = 0 mV. In blinking dynamics and short dsDNA kinetics, we summed the fluorescence intensity in 7 × 7 pixels area around the peak. For the spots generated by sweeping mirror on the right side, as sliding the area horizontally with 6 pixels, we concatenated those intensities either for building time traces or for binding event judging. The signal-to-noise ratio (SNR) of our method was assessed by using fluorescence time traces of Atto647N at 2 mM Trolox for each sweeping step. We obtained average auto-correlation curves from fluorescence time trajectories (n > 30) and fitted them with (k off/k on)exp[−(k on + k off)t], where we assumed a two-state system made of a bright and a dark state with transition rates of k on and k off 12, 13. For DNA binding/unbinding experiments we used either a threshold (I = I B + 3 × σ B) or hidden Markov analysis22 to determine the binding time; however, we mainly used the former that was simple and robust to assign the binding and unbinding events. In HJ experiments, we used the first ten frames without the mirror sweeping for mapping and selecting single-molecules, and extracted the high-temporal fluorescence intensity from the mirror swept image as mentioned above. We used vbFRET software23 to determine the number of states and the transition rates of the acquired time traces in HJ dynamics. To avoid any interference among the sweeping spots, it was important to select good single molecule spots that do not overlap each other within the range of sweeping distance. All time traces in Figs 1, 3 and Supplementary Figures were background-subtracted. The FRET efficiency in Fig. 3 was approximated as I A /(I D + I A ), where I D and I A are fluorescence intensities of the donor and acceptor, respectively, with background and leakage-correction.
Preparation of flow chamber
We used polyethylene glycol (PEG)-coated flow chambers as described elsewhere1, 3 except the experiments of Atto647N blinking dynamics.
DNA samples
All oligonucleotides were purchased from IDT. Amino-modified oligonucleotides were labeled with Cy3B NHS (PA63101, GE Healthcare) or Atto647N NHS (18373, Sigma) in 0.1 M borate buffer (pH 8.5) and purified twice through ethanol precipitation. Please see Supporting Table 1 for complete sequence information.
Blinking dynamics of Atto647N
A flow chamber was passivated with 1 mg/mL of biotin-labeled bovine serum albumin (A8549, Sigma) followed by rinsing and an introduction of Neutravidin (31000, Thermo Fisher). Then single-stranded DNA labeled with Atto647N (ssDNA-A647N) was immobilized to the coverslip. We used an imaging buffer consisted of 20 mM Tris (pH 8.0), 50 mM NaCl, an enzymatic oxygen scavenger system (0.8% (w/v) dextrose, 1 mg/mL glucose oxidase (G2133, Sigma), 0.04 mg/mL catalase (2190015MU, EMD Millipore)) and Trolox (sc-200810, Santa Cruz) with various concentrations. In order to increase the Trolox-quinone concentration, Trolox was prepared at least one week before the experiment. An average power of the red laser was ~12 mW at the back focal plane of the objective lens. Single molecule movies were analyzed using our custom made MATLAB scripts as described earlier3.
Binding and unbinding kinetics of short double-stranded DNA
We introduced 50 µL neutravidin (20 µg/mL) to the flow chamber coated with PEG and biotin-PEG. After washing, we incubated the capture DNA (10 nM) for 1 min followed by an injection of 50 µL Cy3B labeled ssDNA (1 nM) in the imaging buffer (1 × PBS, 600 mM NaCl, 10 mM NiCl2, 0.8% (w/v) dextrose, 1 mg/mL glucose oxidase, 0.04 mg/mL catalase). Here we used Ni2+ instead of Trolox to improve the photostability of Cy3B24. The excitation power of 532 nm laser was ~10 mW. The voltage applied to the galvo mirror was 112 mV in 5 steps and the total sweeping distance was approximately 28 pixels. More than ten thousand independent events were recorded to build the dwell time histogram of τon, exhibiting 7.7 ± 0.5 ms, 43.5 ± 2.0 ms and 530 ± 12 ms for the 7-1, 7 and 9 bp DNA strands, respectively. The total number of binding/unbinding events of the 7–1 bp DNA was ~12,500 during 2,000 frames. The number of non-specific binding events was ~800 during the same imaging time so its contribution was negligible (Fig. 2c, Supplementary Fig. 8c). We examined the possibility that on/off fluorescence signals might be attributed to photophysics. To this end, we measured fluorescence trajectories of 9 bp dsDNA labeled with Cy3B which has τon = 530 ± 12 ms. During the ON time, the blinking events of Cy3B were rarely observed at 4 ms time resolution among ~6,600 time traces. SNRB and SNRS of Cy3B were 8.2 ± 1.6 and 4.0 ± 0.6.
Holliday junction dynamics
The Holliday junction sample was prepared by mixing Seq. 1,2,3 and 4 with a molar ratio of 1:0.9:1:1 (final concentration, ~5 μM each) in 10 mM Tris-HCl (pH 8.0) and 50 mM NaCl followed by slowly cooling down from 95 °C to room temperature for ~2 h. HJ molecules were immobilized on the PEG-coated coverslip and imaged with an imaging buffer (20 mM Tris (pH 8.0), 2–20 mM MgCl2, 50 mM NaCl, 2 mM Trolox, 0.8% (w/v) dextrose, 1 mg/mL glucose oxidase, 0.04 mg/mL catalase). We applied 96 mV to the galvo mirror (4 steps), resulting to 5-ms time resolution (Supplementary Fig. 11b). For each concentration of magnesium, more than 30 time traces of different molecules (from more than 5 movies) were used to calculate the transition rates.
References
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008).
Gorman, J. & Greene, E. C. Visualizing one-dimensional diffusion of proteins along DNA. Nat. Struct. Mol. Biol. 15, 768–774 (2008).
Heo, P. et al. A chemical controller of SNARE-driven membrane fusion that primes vesicles for Ca2+-triggered millisecond exocytosis. J. Am. Chem. Soc. 138, 4512–4521 (2016).
Holzmeister, P., Acuna, G. P., Grohmann, D. & Tinnefeld, P. Breaking the concentration limit of optical single-molecule detection. Chem. Soc. Rev. 43, 1014–1028 (2014).
Farooq, S. & Hohlbein, J. Camera-based single-molecule FRET detection with improved time resolution. Phys. Chem. Chem. Phys. 17, 27862–27872 (2015)
Ray, S. F. High speed photography and photonics. 167–180 (Focal press, 1997).
Zhang, Z., Kenny, S. J., Hauser, M., Li, W. & Xu, K. Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy. Nat. Methods 12, 935–938 (2015).
Shechtman, Y., Weiss, L. E., Backer, A. S., Lee, M. Y. & Moerner, W. E. Multicolour localization microscopy by point-spread-function engineering. Nat. Photon. 10, 590–594 (2016).
Ha, T. & Tinnefeld, P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 63, 595–617 (2012).
Rasnik, I., McKinney, S. A. & Ha, T. Nonblinking and longlasting single-molecule fluorescence imaging. Nat. Methods 3, 891–893 (2006).
Cordes, T., Vogelsang, J. & Tinnefeld, P. On the mechanism of Trolox as antiblinking and antibleaching reagent. J. Am. Chem. Soc. 131, 5018–5019 (2009).
Kim, H. D. et al. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl. Acad. Sci. USA 99, 4284–4289 (2002).
Han, K. Y. et al. Dark state photophysics of nitrogen-vacancy centres in diamond. New J. Phys. 14, 123002 (2012).
Cisse, I. I., Kim, H. & Ha, T. A rule of seven in Watson-Crick base-pairing of mismatched sequences. Nat. Struct. Mol. Biol. 19, 623–627 (2012).
Jungmann, R. et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 10, 4756–4761 (2010).
Wang, Q. & Moerner, W. E. Single-molecule motions enable direct visualization of biomolecular interactions in solution. Nat. Methods 11, 555–558 (2014).
McKinney, S. A., Declais, A. C., Lilley, D. M. J. & Ha, T. Structural dynamics of individual Holliday junctions. Nat. Struct. Biol. 10, 93–97 (2003).
Joo, C., McKinney, S. A., Lilley, D. M. J. & Ha, T. Exploring rare conformational species and ionic effects in DNA Holliday junctions using single-molecule spectroscopy. J. Mol. Biol. 341, 739–751 (2004).
Juette, M. F. et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Hua, B. et al. An improved surface passivation method for single-molecule studies. Nat. Methods 11, 1233–1236 (2014).
McKinney, S. A., Joo, C. & Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006).
Bronson, J. E., Fei, J., Hofman, J. M., Gonzalez, R. L. Jr. & Wiggins, C. H. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 97, 3196–3205 (2009).
Glembockyte, V., Lincoln, R. & Cosa, G. Cy3 photoprotection mediated by Ni2+ for extended single-molecule imaging: old tricks for new techniques. J. Am. Chem. Soc. 137, 1116–1122 (2015).
Acknowledgements
We thank Benjamin Croop for carefully reading manuscript, Boyang Hua and Taekjip Ha (Johns Hopkins University) for Holliday junction samples in the initial experiments, and Harold Kim (Georgia Tech) for fruitful discussions. This work was supported by CREOL at University of Central Florida.
Author information
Authors and Affiliations
Contributions
J.T., Y.S., S.P. and K.Y.H. conceived the project. J.T. designed and performed the experiments, analyzed the data. J.T. and K.Y.H. wrote the manuscript with feedback from other authors.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tang, J., Sun, Y., Pang, S. et al. Spatially encoded fast single-molecule fluorescence spectroscopy with full field-of-view. Sci Rep 7, 10945 (2017). https://doi.org/10.1038/s41598-017-10837-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-10837-6
This article is cited by
-
Transcriptional mutagenesis of α-synuclein caused by DNA oxidation in Parkinson’s disease pathogenesis
Acta Neuropathologica (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.