Abstract
Somatic embryogenesis receptor kinases (SERKs) belong to a small gene family of receptor-like kinases involved in signal transduction. A total of 54 genes were shortlisted from the wheat genome survey sequence of which 5 were classified as SERKs and 49 were identified as SERK-like (SERLs). Tissue- specific expression of TaSERKs at major developmental stages of wheat corroborates their indispensable role during somatic and zygotic embryogenesis. TaSERK transcripts show inherent differences in their hormonal sensitivities, i.e. TaSERK2 and TaSERK3 elicits auxin- specific responses while TaSERK1, 4 and 5 were more specific towards BR-mediated regulation. The ectopic expression of TaSERK1, 2, 3, 4 and 5 in Arabidopsis led to enhanced plant height, larger silique size and increased seed yield. Zygotic embryogenesis specific genes showed a differential pattern in TaSERK Arabidopsis transgenics specifically in the silique tissues. Elongated hypocotyls and enhanced root growth were observed in the overexpression transgenic lines of all five TaSERKs. The inhibitory action of auxin and brassinosteroid in all the TaSERK transgenic lines indicates their role in regulating root development. The results obtained imply redundant functions of TaSERKs in maintaining plant growth and development.
Similar content being viewed by others
Introduction
Somatic embryogenesis (SE) is the developmental reprogramming of somatic cells towards the embryogenic pathway which forms the basis of cellular totipotency in higher plants1,2. This unique developmental pathway involves a plethora of characteristic events viz., cellular dedifferentiation, cell division activation, reorganization of physiology and regulation of gene expression patterns3. Several genes involved in embryogenic competence have been studied in Arabidopsis such as SERK4,5, LTP2, BBM6,7, LEC8,9, PKL10,11, CLV12, WUS13, AGL–1514 and LEC1–LIKE15. Interestingly, a SERK related gene, functioning in ancestral conjugate algae, may have been recruited with a novel function similar to SE during evolution from unicellular algae to multicellular plant organisms16. In wheat, earlier reports manifested 2,4–D induced SE in the leaf base region17 which was further demonstrated to be mediated by Ca2+–CaM pathway18,19, providing necessary insight into the process of plant embryogenesis.
SOMATIC EMBRYOGENESIS RECEPTOR LIKE KINASE (SERK) first isolated from carrot (Daucus carota) embryogenic cells, is considered a characteristic molecular marker for SE in carrot, Dactylis glomerata and Arabidopsis 4,5,20. Since it is expressed in somatic cultures exhibiting close homology with animal and plant receptor kinases, it was named as somatic embryogenesis receptor kinase (SERK) gene. SERK genes belong to a small receptor like kinase family (RLKs) identified in many plant species with five members in Arabidopsis 5, three in Zea mays 21, five in Medicago truncatula 22, four in Helianthus annus 23, two in Oryza sativa 24,25, three in Vitis vinifera 26, three in Phoenix dactylifera 27 and at least three in Triticum aestivum 28. In addition, SERK-like genes have also been reported in Poa pratensis and rice, with eight29 and nine members30 respectively. SERK gene expression in D. carota appears in embryogenic competent cell cultures and continues to the globular stage of embryos while no expression is detected in non-embryogenic cultures4. In D. glomerata, expression of SERK was reported in leaf segments and continues in shoot apical meristems20. Ectopic expression of AtSERK1 results in enhanced embryogenic cell formation5. AtSERK1 and AtSERK2 function redundantly in maintaining the development of the male gametophyte31. In P. pratensis, PpSERK1 expression was high during premeiosis and decreased during meiosis and post-meiotic stages, whereas expression in PpSERK2 was high from premeiosis to anthesis29. Contrastingly in Z. mays, SERK expression was reported in both embryogenic and non-embryogenic callus cultures21. In O. sativa, OsSERK1 is expressed in phytohormone sensitive tissues where it mediates defense signal transduction while OsSERK2 is expressed in all other plant organs24,25. Recently in P. notatum, PnSERK2 was correlated with the onset of apomixis as it showed expression in nucellar cells at the meiosis stage of the apomictic genotype32. These studies substantiate that the SERK genes play a crucial role during embryogenesis and have functional relevance in other facets of plant growth and development.
The present study was undertaken to gain insight into the expression and functional significance of SERK genes in wheat, T. aestivum. To achieve this, we cloned and characterised five TaSERKs and raised the overexpression (OE) transgenics in Arabidopsis. Here, we report their sequence analysis, structural organisation, phylogenetic relationship and expression analysis in different zygotic and somatic tissues of wheat. We also demonstrate the effect of auxin and brassinosteroid on root growth in TaSERK OE transgenic lines. Differential expression analysis of other embryogenesis related genes in OE transgenics demonstrates the possible role of TaSERKs in embryogenesis and seed development. Constitutive expression of TaSERKs in Arabidopsis results in enhanced hypocotyl length, plant height, altered silique size and seed yield.
Experimental Procedures
Plant material and growth conditions
Triticum aestivum
Seeds of T. aestivum var. PBW343 were surface sterilised with 4% sodium hypochlorite for 30 min and inoculated on water soaked cotton bed and covered with Klin wrap for maintaining humidity. The seeds were grown under culture room conditions at 28 °C, with a daily photoperiodic regime of 16 h light and 8 h dark cycle where light was provided by fluorescent tubes (Philip TL 40 W/ 54) at a fluence rate of 80–100 µmol m−2s−1, as per experimental requirements. 13–d–old wheat seedling tissues were used for detailed experiments according to the protocol described earlier17,19. The zygotic tissue of wheat was raised, collected from field–grown plant, and immediately frozen in liquid nitrogen and stored at −80 °C until use. Embryogenic and non–embryogenic calli were raised as described previously33 and for auxin (2,4–D) and brassinosteroid (epi–BL) leaf base induction treatment, experimental method was carried out as described earlier34.
Arabidopsis thaliana
To raise OE transgenic lines in Arabidopsis thaliana ecotype Col–0, plants were grown in pots containing Soilrite (Kelpirite, Bangalore; 1:1:1 ratio of Vermiculite, Perlite and Sphagnum moss) supplemented with OS medium35 in a culture room under 80–100 µmol m−2 s−1 at 22 ± 1 °C with 16 h /8 h light and dark photoperiod regime. TaSERKs: pMDC32 was transformed in Agrobacterium and transgenic plants were generated as described previously36.
Genome wide analysis of SERKs in wheat
To identify homologues of SERK in wheat (T. aestivum), the National Centre for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi), the Arabidopsis information resource (TAIR, http://www.arabidopsis.org/Blast/index.jsp) and Rice Genome Annotation Project (RGAP, http://rice.plantbiology.msu.edu/analyses_search_blast.shtml) databases were used. The deduced amino acid sequences of the known SERK proteins was employed to search for other homologues in wheat by using the TBLASTN program. The redundant sequences were removed using CLC main workbench software. The search was based on the presence of the characteristic features of SERKs, i.e. presence of SPP motif and the C–terminal domain. Additionally, we also made an attempt to identify the SERK homologues from wheat genome survey sequences. For this, the wheat genome sequences were downloaded from URGI sequence repository (http://wheat-urgi.versailles.inra.fr/) which was then Blast searched (blastn version 2.2.6) using CDS sequences of the already known wheat SERKs as a query. Sequences obtained from the BLAST were then utilised for protein prediction using GENSCAN version 1.037. Protein sequences were retrieved and analysed using TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) for the identification of transmembrane helices also. The sequences were then aligned for the search of SPP motif to identify SERKs in wheat.
In silico analysis of TaSERKs
The nucleotide and protein sequences of cloned TaSERKs were analysed using Gene Runner Program 3.04 (http://www.genenames.com). Deduced protein sequences were used to decipher domain organization using SMART (http://smart.embl/heidelberg.de/). The nucleotide and amino acid sequence were searched to obtain homologues from other plants as well (using NCBI database BLAST program). Phylogenetic tree of TaSERKs was generated using the neighbor-joining (NJ) method in MEGA (version 6) software program.
RNA Isolation and cDNA synthesis
RNA from wheat embryogenic calli and overexpression Arabidopsis transgenics were isolated by RNeasy Plant mini kit (Qiagen, Germany) according to the manufacturer’s instructions followed by DNase–I treatment for removal of genomic DNA contamination. For cDNA synthesis, 2 µg RNA was used for the amplification and the PCR conditions was followed according to the manufacturer’s instructions using Superscript III one–step RT–PCR (Invitrogen, USA). The cDNA synthesised was used as a template for further amplification of TaSERKs. For real–time expression analysis, cDNA was prepared from 2 µg RNA using High capacity cDNA archive kit (Applied Biosystems, USA)38. Primers used for real–time PCR analysis are listed in Supplementary Table S2.
Isolation of full–length cDNA of TaSERKs (TaSERK1, 2, 3, 4, 5)
For the amplification of full length cDNA of TaSERKs, RNA was isolated from the wheat embryogenic calli and cDNA prepared by one step RT–PCR using Superscript® III First–Strand Synthesis System RT–PCR kit (Invitrogen, USA) was used as a template for the amplification of TaSERK genes using Phusion High Fidelity Taq polymerase (Finnzymes). The thermal cycling condition was as follows: initial denaturation at 98 °C for 30 s followed by amplification for 35 cycles at 98 °C for 10 s, annealing at 62 °C for 30 s, extension at 72 °C for 1 min and a final extension at 72 °C for 7 min. Each of the full length amplified products of all TaSERKs obtained were then cloned individually in pDRIVE vector (PCR cloning kit, Qiagen, Germany) and entry vector pENTRTM/D–TOPO (Invitrogen Inc. USA) as described previously39. Primers used for above cloning were listed in Supplementary Table S2.
Hypocotyl assay
For hypocotyl assay, seeds were germinated on half-strength MS medium supplemented with 2% sucrose and 0.8% agar in Petri plates kept in growth room at 22 ± 1 °C. After 7 d of growth, the hypocotyl length of ten seedlings each from the WT and TaSERKs overexpression (TaSERKs-OE) lines were measured.
Root growth assay
For examining the effect of brassinosteroid (epi–BL) and auxin (2,4–D) on root growth assay, seedlings were grown on half-strength MS medium supplemented with 2% sucrose and 0.8% agar in plates kept vertically for 3 d at 22 ± 1 °C. Seedlings were then transferred on to a fresh MS medium supplemented with 24–epibrassinolide and 2,4–D (Sigma, St Louis, MO, USA) at different working concentrations and placed vertically under normal culture conditions for 4 d. The root length was measured on the fifth day of transfer and was compared to control seedlings. All experiments were done in triplicates, and the values presented in the data are mean of these experiments, and standard error was calculated.
Statistical analysis
Data of 10–15 seedlings for root growth measurement, 20–25 seedlings for hypocotyl elongation assay, 10–20 plants for morphological and phenotypic evaluation from WT and transgenic plants were collected. Student’s t-test was calculated for significant differences between WT and transgenic lines. A p-value of 0.05 was considered significant.
Results
Sequence analysis of TaSERK genes and relationship with other family members
Our previous study (Singla et al.)28 had identified three TaSERKs from T. aestivum, one of which was specifically isolated from an auxin induced cDNA library40 (TaSERK3). In the present study, we identify additional SERK genes in the wheat genome from sequence analysis through BlastN, Blastp, TBlastX search of ESTs and cDNA clones using NCBI, RGAP, KOME databases and analysed the sequences after multiple sequence alignment by the CLC main workbench program. From the above sequence analysis, we identified five SERKs which were named as TaSERK1 (Accession no. AK333001), TaSERK2 (Accession no. AK3336771), TaSERK3 (Accession no. BT009223), TaSERK4 (Accession no. Ta76279_4565) and TaSERK5 (Accession no. BT009426). The sequences were then confirmed by amplifying full length cDNAs and verified by sequencing (Supplementary Fig. S1).
TaSERK1 harbours a 168 bp 5′UTR and 306 bp 3′UTR; TaSERK2 has an 110 bp and 307 bp long 5′ and 3′UTR; TaSERK3 has a 133 bp and 244 bp 5′ and 3′UTR; TaSERK4 has a 397 bp and 272 bp 5′ and 3′UTR and TaSERK5 contains a 130 bp 5′UTR and 284 bp 3′UTR, respectively. TaSERK1, TaSERK2, TaSERK3, TaSERK4 and TaSERK5 encode proteins of 628, 623, 624, 474 and 628 amino acids with predicted molecular weight of 68.94 kDa, 68.48 kDa, 68.76 kDa, 52 kDa and 69.07 kDa, respectively. Sequence analysis revealed that among the TaSERK members, TaSERK1 shows the highest identity with TaSERK4 (92%) and TaSERK5 (98%). Amongst Arabidopsis AtSERK members, TaSERK1 is closest to AtSERK2 (85%). TaSERK2 shows closest identity with TaSERK3 (91%) and with AtSERK2 (85%). In addition, TaSERK3 was found to be closest to TaSERK4 and TaSERK5, TaSERK4 with TaSERK1 and TaSERK5, and TaSERK5 with TaSERK1 and TaSERK4, respectively.
Multiple sequence alignment of the deduced amino acid sequences of the TaSERKs and OsSERKs gene family from rice (Supplementary Fig. S2) indicated that TaSERKs are similar to OsSERK1 and OsSERK2, sharing characteristic domain features of RLKs, including five leucine–rich repeats (LRR), a SPP (ser–pro–pro) motif (a hallmark feature of SERK gene family), a transmembrane domain and a serine/threonine kinase domain at the carboxyl terminus responsible for phosphorylating downstream proteins30. Detailed domain analysis of TaSERK proteins displayed the presence of a leucine zipper region (Supplementary Fig. S3). The leucine zipper sequence is represented from position 37–58 in TaSERK1, from 29–50 in TaSERK2, from 29–50 in TaSERK3 and from 37–58 in TaSERK5. Only TaSERK4 was found to lack this domain. Additionally, a putative protein kinase ATP–binding site is present in the kinase domain at position 311–333 in TaSERK1, 305–327 in TaSERK2, 304–326 in TaSERK3, 156–178 in TaSERK4 and 311–333 in TaSERK5. A Ser/Thr kinase active–site signature in subdomain VI at position 428–440 in TaSERK1, 422–434 in TaSERK2, 424–436 in TaSERK3, 273–285 in TaSERK4 and 428–440 in TaSERK5 is indicative of serine/threonine kinases.
Phylogenetic analysis revealed that TaSERKs clustered together with SERKs in other monocot species, with SERKs in dicot plants clustering separately (Fig. 1). It is evident from the tree that TaSERK1 and TaSERK5 are closest to OsSERK1 (AK103038), TaSERK4 is closest to ZmSERK1 (CAC37640), and TaSERK2 and TaSERK3 are closely related to OsSERK2 (AK099777).
Differential expression of TaSERK genes
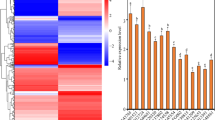
Expression profile of TaSERK genes in vegetative tissues (root and shoot) and zygotic tissues viz., spike, anther, ovary, milky stage of seed (MSS), developing seed (DS) and mature seed (MS) revealed a vast range of expression patterns in wheat. The expression of TaSERK1 and TaSERK4 up-regulated by 2-fold in shoots compared to the root tissue whereas expression in other zygotic tissues was not significantly increased (Fig. 2). The nearly similar expression profile of TaSERK1 and TaSERK4 suggests that they may be functionally overlapping and redundant in action. TaSERK2 expression was up-regulated in the ovaries by 15-fold followed by an 8-fold change in the anther and MSS as compared to the root tissue suggests its predominant role during zygotic embryogenesis. High expression of TaSERK3 was observed in the shoot (18-fold) and zygotic tissues such as anthers by 20-fold change, followed by an 8-fold change in MS, then 5-fold in MSS, spike (4-fold), as compared to the roots. The significant differential expression of TaSERK3 indicates its role during both somatic and zygotic embryogenesis. Expression of TaSERK5 up-regulated in a vegetative tissue, shoots by 4-fold as well as in zygotic tissue, spikes by 9-fold whereas a low level of expression in other zygotic tissues indicates that TaSERK5 might play a rather specific role during zygotic embryogenesis. Therefore, differential expression of TaSERK gene indicates a higher complexity of this gene family in the functional aspects of plant development.
Expression profiles of TaSERK1, 2, 3, 4 and 5 by qRT-PCR. cDNAs normalized to housekeeping gene, ACTIN, in different tissues. The error bars represent mean ± SD of two biological replicates, each analysed with three technical replicates. Asterisks above error bars represent the significance levels (Students t-test; *p value ≤ 0.05).
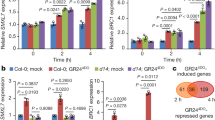
The expression patterns of TaSERK1, 2, 3, 4 and 5 were examined in wheat embryogenic callus (EC) and non-embryogenic callus (NEC) grown under dark and light culture conditions (Fig. 3). TaSERK1 (6-fold), TaSERK2 (7-fold), followed by TaSERK4 (4-fold) and TaSERK3 (2.5-fold) was found to be up-regulated significantly in EC grown under light conditions relative to NEC. Under light conditions, TaSERK5 was found to be down-regulated in EC and up-regulated only in NEC (2.5-fold), however, under dark culture conditions except for TaSERK2 and TaSERK3 which showed up-regulation in EC, all other TaSERK1, 3 and 5 was down-regulated in EC relative to NEC.
Expression analysis of TaSERK1, 2, 3, 4 and 5 in wheat embryogenic and non-embryogenic callus. cDNAs normalized to housekeeping gene, ACTIN, in different tissues grown under dark and light culture conditions, respectively. The error bars represent mean ± SD of two biological replicates, each analysed with three technical replicates. Asterisks above the bars represent the significance levels (Students t-test; *p value ≤ 0.05).
Hormonal sensitivity of TaSERKs
The expression levels of TaSERK1 and TaSERK5 (Supplementary Fig. S4) showed BR-mediated up-regulation by 3-fold in treated leaf base explants followed by TaSERK4 and TaSERK2 which was up-regulated by 2 fold whereas lower expression was observed in TaSERK3. In the presence of 2,4–D only TaSERK2 was up-regulated ≥2.5 fold as compared to other TaSERKs. Therefore, this data suggests that TaSERK1, TaSERK4 and TaSERK5 are preferentially BR-regulated while TaSERK2 is preferentially responsive to 2,4-D.
Generation of overexpressing TaSERK1, 2, 3, 4 and 5 Arabidopsis transgenics
To decipher the functional role of TaSERK genes in planta, each of the five TaSERK cDNAs were independently fused in the OE Gateway vector pMDC32 under the control of CaMV 35 S promoter to generate overexpressing transgenic lines of TaSERK1, 2, 3, 4 and 5 in Arabidopsis. All TaSERK OE transgenics were confirmed by PCR using hptII and (GSP) gene-specific primers (Supplementary Fig. S5) and selected lines were grown to the homozygous stage as described earlier36. Transcript levels of selected transgenic lines of TaSERK1, 2, 3, 4 and 5 were examined by real-time PCR analysis which exhibited variation in the expression level with respect to WT (Supplementary Fig. S6). Three independent lines from each TaSERK transgenic were selected for further analysis on the basis of transcript levels and sufficient seed availability.
Hormone responsive root growth of TaSERKs OE transgenics
The root phenotype of OE TaSERK transgenic and WT Arabidopsis plants in the presence of different plant hormones was examined. The results showed that with increasing concentration of auxin (2,4-D), the root length of TaSERK1 transgenic lines (L5, L11 and L14) gradually decreased (Supplementary Fig. S7) whereas under control conditions, root growth of TaSERK1 was significantly longer than WT (Fig. 4A). Similar trends were also observed in other TaSERKs 2, 3, 4 and 5 transgenics in the presence of auxin (Fig. 4B–E).
Root growth elongation assay of TaSERK1, 2, 3, 4 and 5 OE plants under 2,4-D treatment. Histograms represent the root length of WT and TaSERKs-OE 7-d old seedlings at different concentrations of auxin supplemented medium. Values are mean ± SE for 10 seedlings each. The asterisks (*p ≤ 0.05) indicate statistically significant differences between WT and transgenic lines.
We also examined the effect of brassinosteroid (24 epi-BL) on the root growth of TaSERK transgenics compared to WT Arabidopsis seedlings (Supplementary Fig. S8). Here the root length of TaSERK1 transgenic lines (L5, L11 and L14) increased at 1 nM 24 epi-BL whereas higher concentrations reduces the root length (Fig. 5A). Similarly, the root length of the other TaSERK transgenics (TaSERK 2, 3, 4 and 5) also exhibited same response to different concentrations of epi-BL (Fig. 5B–E). Thus, the above result shows that TaSERK-OE plants are sensitive to the inhibitory effect of auxin and BR in a dose- dependent manner.
Root growth elongation assay of TaSERK1, 2, 3, 4 and 5 OE plants under epi-BL treatment. Histograms represent the root length of WT and TaSERKs-OE 7-d old seedlings at different concentrations of BR supplemented medium. Values are mean ± SE for 10 seedlings each. The asterisks (*p ≤ 0.05) indicate statistically significant differences between WT and transgenic lines.
Enhanced hypocotyl elongation upon OE TaSERKs
Arabidopsis TaSERK transgenic and WT seedlings grown under control culture conditions of 16 h light and 8 h dark photoperiod for seven days showed enhanced hypocotyl length in all the 1, 2, 3, 4 and 5 transgenic lines (Fig. 6A). Measurement and statistical analysis of the hypocotyl length (Fig. 6B) indicated significant differences between all five TaSERK transgenic seedlings compared to the WT. The above results clearly demonstrate that OE of TaSERKs in Arabidopsis promotes hypocotyl elongation under light conditions.
Hypocotyl elongation assay. (A) Phenotype of hypocotyl elongation between OE TaSERK transgenics and WT Arabidopsis seedlings grown on MS medium for seven days. (B) Histogram representing the hypocotyl length of WT and TaSERK transgenics. Graph plotted taking ± SE of twenty seedlings for each Arabidopsis lines. The asterisks (*p ≤ 0.05) indicate statistically significant differences between WT and transgenic lines.
Constitutive expression of TaSERK genes enhance plant growth and seed yield
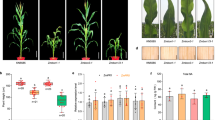
In the present study, constitutive expression of TaSERKs in Arabidopsis resulted in an overall increase in plant growth and productivity. The effect of OE of TaSERKs on plant growth was monitored during the course of development. The TaSERK1 OE transgenic lines (L5, L11 and L14) showed increased plant height compared to the wild–type (Fig. 7A) after 30 days of germination. Elongation continues in the TaSERK1-OE lines whereas it ceases in WT after 40 days. The ectopic expression of TaSERK1 in Arabidopsis also results in larger silique size as well as an increase in the number of siliques per plant with respect to WT (Fig. 7B). This difference is further reflected in the seed weight which showed a significant increase in seed yield per plant in different transgenic lines (Fig. 7C). However, no significant difference was observed in the size of the seed (Supplementary Table S1) between WT and transgenics, implying that the enlarged silique size was due to a greater number of seeds per plant. The difference were also observed in the leaf morphology and rosette leaf numbers, with rosette leaf numbers being higher in WT compared to the transgenics (Supplementary Table S1). Morphological differences of the other TaSERK OE transgenics were also measured (Supplementary Figs S9 and S10 and Supplementary Table S1). Here, we observed that all the TaSERK transgenic plants demonstrate an overall increase in plant height, larger siliques, an increased number of siliques per plant and increased seed yield when compared to the WT. No appreciable differences, however, was observed in the length of the siliques in TaSERK5 transgenics; therefore the increase in seed yield here could be attributed to an increase in the number of siliques per plant examined (Supplementary Table S1). Thus, the above similarity in morphometric analyses for all the TaSERK transgenic plants can be attributed to the high sequence similarity and functional redundancy between the five TaSERKs.
Morphological analysis of TaSERK1 OE lines in Arabidopsis thaliana. (A) TaSERK1 showed increase in plant height as compared to the Col-0 WT grown under 16 h light and 8 h dark culture condition. (B) Two month old TaSERK lines had larger siliques than WT. (C) Graphical representation of silique length, total number of siliques per plant and seed weight per individual plant (N = 10). Data represents mean ± SE. The asterisks (*p ≤ 0.05) indicate statistically significant differences between WT and transgenics.
Expression of various zygotic genes in TaSERK -OE transgenics
To gain insight into the expression profile of known embryogenesis related genes in TaSERK OE Arabidopsis plants, real-time quantitative PCR was performed to examine the transcript levels of LEC1, WUS, BBM and AGL–15 zygotic genes. Interestingly, while no expression of LEC1 gene was observed in either the seedling tissues or in the flower tissues of TaSERK -OE plants, the expression of these genes was significantly higher in silique tissues of TaSERK transgenics. The LEC1 gene was markedly up-regulated in siliques, with TaSERK2-OE and TaSERK3-OE plants exhibiting ≥20–30 fold change, and 15-fold-change in TaSERK1–OE plants in comparison to WT (Fig. 8A) however, TaSERK4–OE and TaSERK5–OE plants showed lower levels of expression as compared to TaSERK1, 2 and 3. A significant change in the expression of the WUS gene was observed in the silique tissues compared to seedling and flower tissues of TaSERK -OE plants with respect to WT (Fig. 8B). In contrast to the expression of LEC1, WUS expression was detected in flower tissues of all TaSERK transgenic plants compared to the WT. The expression level analysis of BBM (Fig. 8C) and AGL–15 (Fig. 8D) in different tissues of TaSERK-OE transgenic plants, displayed higher expression of both these genes in the silique tissues compared to their meager increase in seedling and flower tissues (Fig. 8C,D). BBM expression was highest in TaSERK2–OE followed by TaSERK1 and TaSERK3–OE with only small differences observed in TaSERK4 and TaSERK5–OE plants. AGL–15 expression levels increased significantly in silique tissues of OE lines of TaSERK1, 2, 3 and 4 Arabidopsis plants, but were reduced in the TaSERK5–OE line. Unlike LEC2, WUS and AGL–15 the level of expression of BBM was drastically reduced. The above results thus suggest that TaSERKs-OE in Arabidopsis alters the expression of zygotic genes specifically in the siliques, implying the coordination of somatic and zygotic genes during seed development.
Expression profile of zygotic genes (A) LEC (B) WUS (C) BBM (D) AGL-15 in seedlings, siliques and flower tissue of WT and TaSERKs-OE Arabidopsis plants. Values were normalised to Arabidopsis ACTIN gene expression. The error bars represent mean ± SE of two biological replicates, each analysed with three technical replicates. The asterisks (*p ≤ 0.05) indicate statistically significant differences between WT and transgenics.
Discussion
Structural similarity of TaSERK genes
The plant RLKs form a large gene family comprises of more than 600 members in Arabidopsis and almost 300 LRR–RLKs in rice41,42. Amongst the SERK proteins identified so far, the most extensively studied was the Arabidopsis AtSERK1 which plays a pleiotropic role5,43,44,45,46. Sequence and structural analyses indicated that TaSERK1, 2, 3, 4 and 5 encode a typical SERK protein belonging to the LRR–RLK family47. The predicted domain structure of TaSERKs consists of a signal peptide, leucine zipper (LZ) region (absent in TaSERK4), five LRR domains, a characteristic SPP motif, a single transmembrane domain, serine/threonine kinase domain and a highly conserved C–terminal domain which are very similar to other characterized SERKs including AtSERK15, OsSERK1 and OsSERK230, and ZmSERK121. The five LRR repeats in the SERK domain structure form a horseshoe-shaped cavity predicted to be involved in protein-protein interactions during molecular recognition processes in animals and plants48. TaSERK1, 2, 3, 4 and 5 were recently found to interact with TaBRI1 at the plasma membrane49. The hallmark of SERK proteins is the presence of a SPP motif that acts as a hinge providing flexibility to the extracellular part of the receptor5. The sequence similarity of TaSERK1, TaSERK2, TaSERK3, TaSERK4 and TaSERK5 with AtSERK1 and AtSERK2 implies that the corresponding TaSERKs may play similar functional roles during morphogenesis, cell signaling and plant development. The C–terminal leucine rich domain highly conserved in SERK proteins plays a key role in protein–protein interactions4. Alignment of the five TaSERKs reveals differential amino acid sequences present in the signal peptide, with TaSERK2, TaSERK3 and TaSERK4 lacking some amino acid residues compared to the TaSERK1 and TaSERK5. Some differences in amino acids were also located in the SPP region of TaSERK3 and TaSERK4. Differences in the C–terminal domain may be responsible for differential expression and function. Phylogenetic studies reveal that TaSERK1 and TaSERK5 may perform a similar function as they clustered together. TaSERK2 and TaSERK3 also cluster together while TaSERK4 was found to be more closely associated with ZmSERK121, indicating that TaSERKs may play a redundant role in SE and have various functional aspects as observed for other plant species. The sequence analyses and phylogenetic tree reveal that the conservation of TaSERKs is very extensive within a subgroup encompassing the monocot OsSERK1, OsSERK2, ZmSERK1, ZmSERK2 and ZmSERK3 and the dicot AtSERK1, AtSERK2, PpSERK1, PpSERK2, MtSERK2 and MtSERK5.
Expression of TaSERKs correlates in different tissues and organs
There are various reports on the probable role of SERKs in different aspects of plant development. In the present study, we analysed the expression profile of TaSERKs revealing their participation both in somatic as well as zygotic embryogenesis. Amongst all five TaSERK members, the expression pattern analyses of TaSERK1 and TaSERK4 appears to implicate them specifically in SE rather than in zygotic tissues, thus suggesting specificity to SE similar to the OsSERK1 and OsSERK2 30. Interestingly, TaSERK2 and TaSERK5 are highly expressed in ovary tissues and spikes in contrast to TaSERK3 which showed high abundance of transcript in anthers but low abundance in ovaries, respectively implies that these may play a significant role during zygotic embryogenesis, consistent with the results of AtSERK1, AtSERK2, MtSERK1 andCitSERK1 22,31,50,51. The high expression level of TaSERK1, TaSERK2 and TaSERK4 in embryogenic callus specifically under light culture conditions with low expression in non–embryogenic callus both under light and dark conditions, suggest a functional similarity to ZmSERK1 and ZmSERK2 genes which were reported to be expressed both in embryogenic and non–embryogenic callus cultures21. The SERK expression patterns28 were also reported in, callus tissues of Dactylis glomerata 20 and Arabidopsis thaliana 5, validating the indispensable role of SERKs during SE in these plants. Thus, the above expression patterns corroborate that the function of the wheat SERK gene family members is not only restricted to embryogenesis but they play a crucial role in other organs at major developmental stages.
The expression of the five TaSERKs was also monitored under the influence of phytohormones, auxin and BR. The transcript level of TaSERK1 and TaSERK2 showed upregulation upon induction with 2,4–D as compared to TaSERK3, 4 and 5. In contrast, TaSERK1 and TaSERK5 were induced significantly to a high extent in the presence of BR. These results suggest that the TaSERK2 response is highly auxin mediated compared to other TaSERKs, while TaSERK1 and TaSERK5 may be mediated by the BR signaling pathway, consistent with AtSERK1, 2, 3 and 4 52.
Constitutive expression of TaSERKs regulate root and hypocotyl growth in Arabidopsis
Earlier reports and results from the present study clearly indicate that SERKs also regulate root development. It was reported that auxins play a key role in root development of Arabidopsis plants53,54,55 and together with BRs also participate in regulating the growth largely through mediating control over essential genes56,57,58,59. Evidence from genetic mutant analysis validates the role of SERKs regulating root development primarily via a BR–independent pathway as the triple mutant serk2bak1bkk1 exhibited shortened root phenotype that was rescued by the expression of SERK1, BAK1/SERK3 and BKK1/SERK4, respectively60. OE of each of the five TaSERKs enhanced primary root growth in Arabidopsis and also elicited sensitivity to the inhibitory action of exogenously supplied auxin and brassinosteroid hormones in a dose-dependent manner, suggesting their functional role in the development and an enhancement of root growth at lower concentrations of BR in an auxin-independent manner. Nonetheless, it is still unclear how the TaSERKs modulate root development when induced in combination with auxin and BR.
The OE of TaSERK 1, 2, 3, 4 and 5 led to the diverse changes in the growth and morphology of Arabidopsis. The major alterations were observed in plant height, silique size and seed yield. In contrast, the constitutive expression of AtSERK1 did not show any altered phenotype; however, seedlings which overexpressed AtSERK1 initiated SE with higher efficiency5. Transgenic plants harbouring TaSERK1, TaSERK2, TaSERK3, TaSERK4 and TaSERK5 independently displayed common alterations in plant height, rosette and leaf development, silique morphology, seed yield, and seed size. The phenotypic observations suggest that enhanced expression of TaSERK family members exhibit overlapping functions in Arabidopsis transgenics.
TaSERK enhances the expression of zygotic genes in Arabidopsis
A small set of transcription factor (TF) specific genes reported from earlier experimental studies which play an essential role in the embryogenic development includes LEAFY COTYLEDON(LEC)8,61, WUSCHEL (WUS)13, BABY BOOM (BBM)6 and AGAMOUS–LIKE15 (AGL15)62. The LEC genes are presumed to link the maturation phase of zygotic embryogenesis (ZE) and initiation of SE via the establishment of a suitable environment for cell differentiation63. LEC1 expression in Arabidopsis embryogenesis culture is also involved in differentiation and development apart from somatic embryo induction64. Arabidopsis LEC1 was found to be expressed at higher levels in siliques during early embryo development compared to maturing embryos65. In our study, LEC1 transcript expression correlated with a significant increase in siliques on transgenic TaSERK2, TaSERK3 and TaSERK1 plants, suggesting a link between SERK gene family members and LEC in regulating gene expression during the maturation phase of embryo development. The WUS gene in Arabidopsis promotes the transition from vegetative to embryonic phase in all tissues and organs13. Here, we show that the ectopic expression of TaSERKs in Arabidopsis results in increased expression of WUS in siliques and lower expression in flowers, indicating that constitutively expressing TaSERK1, 2 and 3 modulate zygotic embryo development. Further, some studies have reported the expression of BBM during zygotic and pollen–derived SE6. In our study, we show that TaSERK1, 2 and 3 transgenic plants exhibit higher expression of BBM specifically in siliques, suggesting ectopic expression of TaSERK1, 2 and 3 in Arabidopsis may regulates zygotic embryo development. We also examined the expression pattern of AGL–15, a MADS domain TF reported to play a role in embryogenesis. AGL–15 was found to be expressed during early zygotic embryogenesis66,67 and its ectopic expression upregulated AtSERK1 expression62. The higher expression of AGL–15 in TaSERK1, 2, 3 and 4 Arabidopsis transgenics suggests a correlation between SERK and zygotic genes. TaSERK5 transgenics, however, do not participate in the regulation of zygotic embryo related gene expression in plant development. Nevertheless, the expression levels of LEC1, WUS, BBM, and AGL-15 were significantly affected in silique tissues of TaSERK transgenic plants, indicating that they may be involved in the regulation of seed development.
Conclusion
TaSERK gene family members are functionally redundant based on phenotypic observations as well as gene expression patterns in transgenic lines. TaSERKs display sequence conservation and exhibit differential expression in zygotic and somatic tissues of wheat. The effect of auxin and brassinosteroid on tissue specific expression and in root growth suggests the involvement of TaSERKs in hormonal regulation pathway. Taken together, the findings from the present study confirms that TaSERKs exert a profound effect on embryogenesis, plant growth and development, thus suggesting wide- range function of TaSERK genes which were earlier restricted to their role in SE.
References
Chugh, A. & Khurana, P. Gene expression during somatic embryogenesis–recent advances. Curr Sci. 83, 715–730 (2002).
Zeng, F. et al. Isolation and characterization of genes associated to cotton somatic embryogenesis by suppression subtractive hybridization and macroarray. Plant Mol Biol. 60, 167–183 (2006).
Yang, X. & Zhang, X. Regulation of somatic embryogenesis in higher plants. Crit Rev Plant Sci. 29, 36–57 (2010).
Schmidt, E. D. L., Guzzo, F., Toonen, M. A. J. & de Vries, S. C. A leucine–rich repeat containing receptor–like kinase marks somatic plant cells competent to form embryos. Dev. 124, 2049–2062 (1997).
Hecht, V. et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–16 (2001).
Boutilier, K. et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 14, 1737–1749 (2002).
Srinivasan, C. et al. Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta. 225, 341–351 (2007).
Stone, S. L. et al. LEAFY COTYLEDON 2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98, 11806–11811 (2001).
Gaj, M. D., Zhang, S., Harada, J. J. & Leamux, P. G. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta. 222, 977–988 (2005).
Ogas, J., Cheng, J. C., Sung, Z. R. & Somerville, C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 277, 91–94 (1997).
Li, H. C. et al. PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44, 1010–1022 (2005a).
Mordhorst, A. P. et al. Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics. 149, 549–563 (1998).
Zuo, J., Niu, Q. W., Frugis, G. & Chua, N. H. The WUSCHEL gene promotes vegetative–to–embryonic transition in Arabidopsis. Plant J. 30, 349–359 (2002).
Rounsley, S. D., Ditta, G. S. & Yanofsky, M. F. Diverse roles for MADS Box genes in Arabidopsis development. Plant Cell. 7, 1259–1269 (1995).
Kwong, R. M. et al. LEAFY COTYLEDON1–LIKE defines a class of regulators essential for embryo development. Plant Cell. 15, 5–18 (2003).
Sasaki, G. et al. Multiple receptor–like kinase cDNAs from liverwort Marchantia polymorpha and two charophycean green algae, Closterium ehrenbergii and Nitella axillaris: extensive gene duplications and gene shufflings in the early evolution of streptophytes. Gene. 401, 135–144 (2007).
Mahalakshmi, A., Khurana, J. P. & Khurana, P. Rapid induction of somatic embryogenesis by 2,4–D in leaf base cultures of wheat (Triticum aestivum L). Plant Biotech. 20, 267–273 (2003).
Patnaik, D. & Khurana, P. Identification of a phosphoprotein expressed during somatic embryogenesis in wheat leaf base cultures. J Plant Biochem Biotechnol. 14, 149–154 (2005).
Mahalakshmi, A., Singla, B., Khurana, J. P. & Khurana, P. Role of calcium–calmodulin in auxin–induced somatic embryogenesis in leaf base cultures of wheat (Triticum aestivum var. HD 2329). Plant Cell Tissue Organ Cult. 88, 167–174 (2007).
Somleva, M. N., Schmidt, E. D. L. & de Vries, S. C. Embryogenic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Rep. 19, 718–726 (2000).
Baudino, S. et al. Molecular characterization of two novel maize LRR receptor–like kinases, which belong to the SERK gene family. Planta. 213, 1–10 (2001).
Nolan, K. E., Irwanto, R. R. & Rose, R. J. Auxin up–regulates MtSERK1 expression in both Medicago truncatula root–forming and embryogenic cultures. Plant Physiol. 133, 218–230 (2003).
Thomas, C., Meyer, D., Himber, C. & Steinmetz, A. Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis. Plant Physiol Biochem. 42, 35–42 (2004).
Ito, Y., Takaya, K. & Kurata, N. Expression of SERK family receptor–like protein kinase genes in rice. Biochim Biophys Acta. 1730, 253–258 (2005).
Hu, H., Xiong, L. & Yang, Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta. 222, 107–117 (2005).
Schellenbaum, P. et al. Characterization of VvSERK1, VvSERK2, VvSERK3 and VvL1L genes and their expression during somatic embryogenesis of grapevine (Vitis vinifera L.). Plant Cell Rep. 27, 1799–1809 (2008).
Rekik, I., Elleuch, A., Kriaa, W. & Drira, N. Molecular cloning and in silico analysis of three somatic embryogenesis receptor kinase mRNA from date palm. Genetika. 45, 837–853 (2013).
Singla, B., Khurana, J. P. & Khurana, P. Characterization of three somatic embryogenesis receptor kinase genes from wheat. Triticum aestivum. Plant Cell Rep. 27, 833–843 (2008).
Albertini, E. et al. SERK and APOSTART, candidate genes for apomixis in Poa pratensis. Plant Physiol. 138, 2185–2199 (2005).
Singla, B., Khurana, J. P. & Khurana, P. Structural characterization and expression analysis of the SERK/SERL gene family in rice (Oryza sativa). Int J Plant Genomics. 539402 (2009).
Colcombet, J., Boisson–Dernier, A., Ros–Palau, R., Vera, C. E. & Schroeder, J. I. Arabidopsis somatic embryogenesis receptor kinases1 and 2 are essential for tapetum development and microspore maturation. Plant Cell. 17, 3350–3361 (2005).
Podio, M. et al. Characterization and expression analysis of SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) genes in sexual and apomictic Paspalum notatum. Plant Mol Biol. 84, 479–495 (2014).
Khurana, J., Chugh, A. & Khurana, P. Regeneration from mature and immature embryos and transient gene expression via Agrobacterium–mediated transformation in emmer wheat (Triticum dicoccum Schuble). Indian J Exp Biol. 40, 295–303 (2002).
Singla, B., Khurana, J. P. & Khurana, P. An early auxin–responsive Aux/IAA gene from wheat (Triticum aestivum) is induced by epibrassinolide and differentially regulated by light and calcium. J Exp Bot. 57, 4059–4070 (2006).
Somerville, C. R. & Ogren, W. L. Isolation of photorespiration mutants in Arabidopsis thaliana. In M. Edelman, R. B. Hollick & N. H. Chua, eds, methods in chloroplast molecular biology. Elsevier Biomedical Press, New York, 129–139 (1982).
Checker, V. G. & Khurana, P. Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Rep. 32, 1729–1741 (2013).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 268, 78–94 (1997).
Khurana, N., Chauhan, H. & Khurana, P. Expression analysis of a heat–inducible, Myo–inositol–1 phosphate synthase (MIPS) gene from wheat and the alternatively spliced variants of rice and Arabidopsis. Plant Cell Rep. 31, 237–251 (2012).
Singh, A. & Khurana, P. Molecular and functional characterization of a wheat B2 protein imparting adverse temperature tolerance and influencing plant growth. Front Plant Sci. 7, 642 (2016).
Singla, B., Tyagi, A. K., Khurana, J. P. & Khurana, P. Analysis of expression profile of selected genes expressed during auxin–induced somatic embryogenesis in leaf base system of wheat (Triticum aestivum) and their possible interactions. Plant Mol Biol. 65, 677–692 (2007).
Shiu, S. H. & Bleecker, A. B. Receptor–like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98, 10763–10768 (2001).
Shiu, S. H. et al. Comparative analysis of the receptor–like kinase family in Arabidopsis and rice. Plant Cell. 16, 1220–1234 (2004).
Shah, K., Gadella, T. W. Jr., van Erp, H., Hecht, V. & de Vries, S. C. Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J Mol Biol. 309, 641–655 (2001).
Shah, K., Vervoort, J. & de Vries, S. C. Role of threonines in Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J Biol Chem. 276, 41263–41269 (2001).
Shah, K., Schmidt, E. D., Vlak, J. M. & de Vries, S. C. Expression of the Daucus carota somatic embryogenesis receptor kinase (DcSERK) protein in insect cells. Biochimie. 83, 415–421 (2001).
Shah, K., Russinova, E., Gadela, T. W. J. Jr., Willemse, J. & de Vries, S. C. The Arabidopsis kinase–associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev. 16, 1707–1720 (2002).
Walker, J. C. Structure and function of the receptor–like protein kinases of higher plants. Plant Mol Biol. 26, 1599–1609 (1994).
Kobe, B. & Deisenhofer, J. The leucine–rich repeat: a versatile binding motif. Trends Biochem Sci. 19, 415–421 (1994).
Singh, A. & Khurana, P. Wheat Brassinosteroid-Insensitive1 (TaBRI1) interacts with members of TaSERK gene family and causes early flowering and seed yield enhancement in Arabidopsis. PLoS One 11, e0153273 (2016).
Albrecht, C., Russinova, E., Hecht, V., Baaijens, E. & de Vries, S. The Arabidopsis thaliana somatic embryogenesis receptor–like kinases 1 and 2 control male sporogenesis. Plant Cell. 17, 3337–3349 (2005).
Shimada, T. et al. Isolation and characterization of the somatic embryogenesis receptor–like kinase gene homologue (CitSERK1) from Citrus unshiu Marc. Sci Hortic. 103, 233–238 (2005).
Gou, X. et al. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8, e1002452 (2012).
Wei, H. et al. Research progresses on auxin response factors. J Integr Plant Biol. 48, 622–627 (2006).
Bennett, T. & Scheres, B. Root development–Two meristems for the price of one. Curr Top Dev Biol. 91, 67–102 (2010).
Overvoorde, P., Fukaki, H. & Beeckman, T. Auxin control of root development. Cold Spring Harb Perspect Biol. 2, a001537 (2010).
Bao, F. et al. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134, 1624–1631 (2004).
Li, L., Xu, J., Xu, Z. & Xue, H. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell. 17, 2738–2753 (2005b).
Mouchel, C. F., Osmont, K. S. & Hardtke, C. S. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 443, 458–461 (2006).
Kim, T. W. et al. Elongation and gravitropic responses of Arabidopsis roots are regulated by brassinolide and IAA. Plant Cell Environ. 30, 679–689 (2007).
Du, J. et al. Somatic embryogenesis receptor kinases control root development mainly via brassinosteroid– independent actions in Arabidopsis thaliana. J Integr Plant Biol. 54, 388–399 (2012).
Ledwoń, A. & Gaj, M. D. Leafy Cotyledon2 gene expression and auxin treatment in relation to embryogenic capacity of Arabidopsis somatic cells. Plant Cell Rep. 28, 1677–1688 (2009).
Harding, E. W., Tang, W. N., Nichols, K. W., Fernandez, D. E. & Perry, S. E. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS–Like 15. Plant Physiol. 133, 653–663 (2003).
Braybrook, S. A. & Harada, J. J. LECs go crazy in embryo development. Trends Plant Sci. 13, 624–630 (2008).
Ledwoń, A. & Gaj, M. D. LEAFY COTYLEDON1, FUSCA3 expression and auxin treatment in relation to somatic embryogenesis induction in Arabidopsis. Plant Growth Regul. 65, 157–167 (2011).
Lotan, T. et al. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 93, 1195–1205 (1998).
Perry, S. E., Nichols, K. W. & Fernandez, D. E. The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell. 8, 1977–1989 (1996).
Perry, S. E., Lehti, M. D. & Fernandez, D. E. The MADS–domain protein AGAMOUS–like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol. 120, 121–129 (1999).
Acknowledgements
This work was financially supported by the Department of Biotechnology, Government of India and partially by Indo-Swiss Collaboration in Biotechnology (ISCB). A.S. acknowledges the Council of Scientific and Industrial Research (CSIR) for the award of research fellowships.
Author information
Authors and Affiliations
Contributions
A.S. and P.K. conceived and designed the study. A.S. performed the experiments. A.S. and P.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Singh, A., Khurana, P. Ectopic expression of Triticum aestivum SERK genes (TaSERKs) control plant growth and development in Arabidopsis . Sci Rep 7, 12368 (2017). https://doi.org/10.1038/s41598-017-10038-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-10038-1
This article is cited by
-
Identification of a novel promoter region responsible for the embryo-specific expression of SERK1 in pineapple
Horticulture, Environment, and Biotechnology (2023)
-
Epigenetic modifications and miRNAs determine the transition of somatic cells into somatic embryos
Plant Cell Reports (2023)
-
Genome-Wide Identification and Analysis of the TaSERK Gene Family in Bread Wheat Triticum aestivum L. and TaSERK8 Overexpression Study in Rice
Journal of Plant Growth Regulation (2023)
-
Greater morphological and primary metabolic adaptations in roots contribute to phosphate-deficiency tolerance in the bread wheat cultivar Kenong199
BMC Plant Biology (2021)
-
Genes, proteins and other networks regulating somatic embryogenesis in plants
Journal of Genetic Engineering and Biotechnology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.