Abstract
Current interest in Foxp3+ T-regulatory (Treg) cells as therapeutic targets in transplantation is largely focused on their harvesting pre-transplant, expansion and infusion post-transplantation. An alternate strategy of pharmacologic modulation of Treg function using histone/protein deacetylase inhibitors (HDACi) may allow more titratable and longer-term dosing. However, the effects of broadly acting HDACi vary, such that HDAC isoform-selective targeting is likely required. We report data from mice with constitutive or conditional deletion of HDAC11 within Foxp3+ Treg cells, and their use, along with small molecule HDAC11 inhibitors, in allograft models. Global HDAC11 deletion had no effect on health or development, and compared to WT controls, Foxp3+ Tregs lacking HDAC11 showed increased suppressive function, and increased expression of Foxp3 and TGF-β. Likewise, compared to WT recipients, conditional deletion of HDAC11 within Tregs led to long-term survival of fully MHC-mismatched cardiac allografts, and prevented development of transplant arteriosclerosis in an MHC class II-mismatched allograft model. The translational significance of HDAC11 targeting was shown by the ability of an HDAC11i to promote long-term allograft allografts in fully MHC-disparate strains. These data are powerful stimuli for the further development and testing of HDAC11-selective pharmacologic inhibitors, and may ultimately provide new therapies for transplantation and autoimmune diseases.
Similar content being viewed by others
Introduction
Histone/protein deacetylase (HDAC) enzymes developed in metazoans prior to the evolution of histones1, 2, consistent with current knowledge that HDACs play important roles in regulating the acetylation and function of many non-histone proteins3,4,5, in addition to regulating chromatin accessibility and nucleosome remodeling by deacetylating lysine residues within histone tails6. Studies of the protein acetylome are continuing to advance such knowledge7, 8. Our prior work, showing that small molecule pan-HDAC inhibitors (HDACi) can increase Foxp3 acetylation and DNA binding and thereby enhance Foxp3+ T-regulatory (Treg) cell production and suppressive activity9, led us to analyze the roles of individual HDACs in Foxp3+ Treg cells. This is a necessary step since pan-HDACi can have undesirable effects that are thought to likely curtail their use beyond oncology10. Moreover, while gene deletion and/or pharmacological targeting of some HDACs, e.g. HDAC611 and HDAC99, 12, boosts Treg function, deletion of others, e.g. HDAC313, ablates Treg suppressive function and leads to lethal autoimmunity. In the course of these studies, we have analyzed the biology of HDAC11 in Treg cells.
HDAC11 is the sole member of the class IV family of zinc-dependent HDACs, sharing sequence similarity to both class I and II HDAC proteins, and is the most recently identified and, at 39-kilodaltons, the smallest known HDAC14. HDAC11 is highly conserved between species, and is relatively tissue-specific in that high expression is limited to kidney, heart, brain, skeletal muscle, and testis14. However, along with HDAC10, HDAC11 is one of the least studied HDAC family members6. Recently, HDAC11 was identified as a transcriptional repressor of Il10 expression in mouse and human antigen-presenting cells (APCs)15, and in a separate study, shown to associate with the survival of motor neurons (SMN) complex and regulate mRNA splicing8. Another HDAC, HDAC6, can physically associate with HDAC11 in both the cytoplasm and nuceli14. The association of HDAC6 and HDAC11 in APCs promotes Il10 expression16. HDAC6 and HDAC11 also interact at the vitamin D receptor to regulate expression of MYC17, providing a second example of gene regulation by dynamic complexes that contain both HDAC6 and HDAC11 enzymes. We now report genetic and pharmacologic data as to the role of HDAC11 in Foxp3+ Tregs, and show that HDAC11 targeting can enhance Treg function and induce Treg-dependent suppression of allograft rejection.
Results
HDAC11 can co-associate with, and deacetylate, Foxp3
Foxp3 plays a central role in controlling Treg development and functions by regulating the expression of several hundred target genes that collectively determine the phenotype and suppressive activity of Treg cells18. In addition, the binding of Foxp3 to target genes is regulated by acetylation and deacetylation9. These considerations led us to first assess whether Foxp3 could physically interact with HDAC11. After transfection of 293 T cells with Flag-tagged Foxp3 and Myc-tagged HDAC11 constructs, cells were lysed, proteins immunoprecipitated with anti-Myc (HDAC11) antibody, and probed with anti-mouse Foxp3 and anti-Flag antibodies. We found that immunoprecipitation of HDAC11 led to co-precipitation of Foxp3 (Fig. 1A, Supplementary Fig. S1). Comparable results were found when proteins were immunoprecipitated with anti-Flag (Foxp3) antibody, and probed with anti-Myc (HDAC11) antibody (Fig. 1B, (Supplementary Fig. S2). Hence, Foxp3 and HDAC11 can co-associate.
HDAC11 can associate with, and deacetylate, Foxp3. (A) Co-immunoprecipitation of Foxp3 protein when HDAC11 is immunoprecipitated from 293 T cells transfected with FLAG-tagged Foxp3 and Myc-tagged Hdac11; original uncropped gels are shown in Supplementary Fig. 1. (B) Co-immunoprecipitation of HDAC11 protein when Foxp3 is immunoprecipitated from 293 T cells transfected with FLAG-tagged Foxp3 and Myc-tagged HDAC11; original uncropped gels are shown in Supplementary Fig. 2. (C) Western blots showing that the acetylation of lysine-31 (K31) of Foxp3 by p300 (lane 3) is reversed by the presence of HDAC11 (lane 5); original uncropped gels are shown in Supplementary Fig. 3. (D) Detection of HDAC11 (nucleus and cytoplasm) and Foxp3 (nucleus) in resting Tregs, whereas CD3/CD28 mAb activation promotes their nuclear co-localization (original magnifications ×400).
Next, we tested whether HDAC11 could deacetylate Foxp3. Consistent with our previous report19, co-transfection of 293 T cells with Foxp3 and p300 led to a marked increase in Foxp3 acetylation (Fig. 1C, lane 3, and Supplementary Fig. S3). Cotransfection of 293 T cells with Foxp3 and HDAC11 led to decreased expression of Foxp3 (Fig. 1C, lane 4), consistent with knowledge that the binding of acetylated Foxp3 to a conserved noncoding sequence (CNS) within a Foxp3 intronic site (CNS2) is required for optimal and stable expression of Foxp3 protein20. Importantly, compared to the presence of Foxp3 and p300 (Fig. 1C, lane 3), the addition of HDAC11 decreased both Foxp3 expression and Foxp3 acetylation (Fig. 1C, lane 5). Hence, HDAC11 can bind to Foxp3 and promote its deacetylation.
Lastly, we undertook immunofluorescent localization of HDAC11 and Foxp3 in murine Treg cells. In freshly isolated, unstimulated cells, HDAC11 was detected within both cytoplasmic and nuclear regions, whereas Foxp3 was located almost entirely within the nuclei of Treg cells (Fig. 1D). However, activation with CD3/CD28 monoclonal antibodies (mAbs) led to the nuclear translocation of HDAC11, and co-localization of HDAC11 and Foxp3 within the nuclei of murine Treg cells (Fig. 1D). Hence, in vitro studies suggest that HDAC11 might contribute to the regulation of Foxp3 expression and function in Treg cells.
Deletion of HDAC11 in Foxp3+ Tregs enhances Treg suppressive function
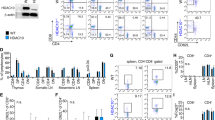
We conditionally deleted HDAC11 in Treg cells by crossing Hdac11 fl/fl and Foxp3YFP-Cre mice (Supplementary Fig. S4). The resultant (hereafter HDAC11−/−) mice developed normally and had no obvious phenotypic abnormalities. Flow cytometric analysis of T cell populations from the thymus, spleen or lymph nodes showed no significant differences between HDAC11−/− and WT mice (Fig. 2A). However, compared to WT Treg cells, Foxp3+ Treg cells lacking HDAC11 had an increased ability to inhibit the proliferation in vitro of conventional CFSE-labeled CD4 + CD25- T-effector (Teff) cells (Fig. 2B).
Conditional HDAC11 deletion promotes Foxp3+ Treg suppressive function. (A) Deletion of HDAC11 within developing Foxp3+ Tregs did not affect the proportions of thymic, splenic or LN Treg cells; data are from 4 to 6 weeks old male mice, and are representative of results in 4 mice/group. (B) In vitro Treg suppression assay of the proliferation of CFSE-labeled WT Teff cells using Tregs from WT vs. conditional HDAC11−/− mice, with percentage of proliferating cells shown in each panel, and representative of 4 separate experiments. (C) Gene expression (qPCR) by WT vs. conditional HDAC11−/− Tregs at rest or activated with plate-bound CD3/CD28 mAbs for 24 h; normalized gene expression data are shown as mean ± SD, 4 mice/group; *p < 0.05, **p < 0.01, ***p < 0.005. (D) ChIP assays comparing levels of Foxp3 binding at the IL-2 and TGF-β promoters in WT and HDAC11−/− Treg cells. (E) Enhanced Treg suppressive function in vitro is due, at least in part, to increased TGF-β production, as shown by normalization of the suppressive function of conditional HDAC11−/− Treg cells when incubated in the presence of a neutralizing anti-TGF-β mAb; the percentage of proliferating cells is shown in each panel and representative of 3 separate experiments.
Analysis of gene expression by real-time quantitative PCR (qPCR) showed that compared to WT Treg cells, freshly isolated HDAC11−/− Tregs had increased expression of Foxp3 (p < 0.05) and TGF-β (b < 0.001) (Fig. 2C). Upon cell activation, in addition to increased Foxp3 and TGF-β expression, HDAC11−/− Tregs showed increased expression of CTLA4 and GITR (p < 0.01), whereas their expression of IL-10 was not significantly different from WT cells under resting or activating conditions (Fig. 2C). ChIP assays showed that the enrichment of Foxp3 at the IL-2 and TGF-β promoters was markedly increased in HDAC11−/− vs. WT Treg cells (Fig. 2D). These data led us to test whether the increased TGF-β production by HDAC11−/− vs. WT Tregs contributed to their greater in vitro suppressive function. The addition of a neutralizing anti-TGF-β mAb largely erased the suppressive function of both WT and HDAC11−/− Tregs (Fig. 2E). Hence, HDAC11 deletion enhances expression of multiple Treg-associated genes, including Foxp3, and increases Treg suppressive function in vitro through a TGF-β dependent mechanism.
Deletion of HDAC11 promotes chromatin remodeling and Treg gene expression
Foxp3 gene expression reflects the contributions of three evolutionarily conserved non-coding sequences (CNS) within Foxp3 intronic regions that function as enhancers, as well as the actions of the Foxp3 promoter itself. ChIP studies assessing acetylated H3-associated chromatin remodeling showed that deletion of Hdac11 increased chromatin accessibility at the Foxp3 promoter and at all 3 Foxp3 enhancer regions (Fig. 3A). Reporter assays using a luciferase-linked Foxp3 promoter showed that HDAC11 decreased the signal driven by NFκB/p65, alone or in the presence of transfected Foxp3, and impaired luciferase activity driven by p65, Foxp3 and the HAT enzyme, p300 (Fig. 3B). The inhibitory effects of HDAC11 at the Foxp3 promoter were reversed by use of JB3-22, an HDAC11 pharmacologic inhibitor (HDAC11i, IC50 0.235 µM) (Fig. 3B). JB3-22 also partially reversed the inhibitory effect of HDAC11 on p300-induced Foxp3 acetylation at lysine 31 (Supplementary Fig. S5), consistent with the ability of HDAC11 to directly regulate acetylation of Foxp3.
Conditional HDAC11 deletion promotes chromatin remodeling at the Foxp3 locus and Treg gene expression. (A) ChIP studies of the Foxp3 promoter and 3 enhancer regions in WT vs. conditional HDAC11−/− Treg cells, showing increased acetyl-H3 binding at all 4 sites in HDAC11−/− vs. WT Treg cells. (B) Luciferase reporter assays showing the impact of overexpression, and also inhibition, of HDAC11 in 293 T cells that were transiently transfected with murine Foxp3-promoter-Luc vector plus expression vectors for p65, HDAC11, Foxp3, and p300, as specified, or empty vector (EV) along with 50 ng of pRL-TK Renilla luciferase internal control vector; EV was added to ensure equal amounts of DNA per reaction. After 48 h, cells were treated with PMA (6 ng/ml) and ionomycin (1 μM) for 5 to 6 h, and 1 μM HDAC11i where specified, and cell lysates were harvested for luciferase assays. Firefly luminescence data was normalized to Renilla luminescence and analyzed by ANOVA (*p < 0.05, **p < 0.005); data are representative of the results of 3 or more independent experiments. (C) Heat-map of selected genes that were upregulated (red) or downregulated (blue) in conditional HDAC11−/− vs. WT Treg cells. (D) qPCR evaluation of WT vs. conditional HDAC11−/− Treg cells, showing increased expression of Jun, Junb, Fos and Fosb in HDAC11−/− Tregs; normalized data are shown as mean ± SD, 4 mice/group, and *p < 0.05, **p < 0.01.
We next undertook microarray analyses of HDAC11−/− and WT Tregs to assess HDAC11-dependent global changes in gene expression (Fig. 3C and Supplementary Fig. S6). HDAC11 deletion led to the induction of a series of genes previously linked with DNA replication licensing control, including Cdt1, geminin and at least 6 of the genes associated with the hexameric minichromosome maintenance (MCM) DNA helicase21, 22. HDAC11 deletion also decreased expression of additional genes known to interact with, or to be regulated, by HDAC11, including E4f1 and Tblx18, 17. With regard to genes relevant to Treg biology, HDAC11 deletion decreased expression of Rorc and multiple cytokine receptor genes and increased expression of Fos, Fosb, Jun and Junb, whose protein products dimerize to form the AP-1 transcription factor which binds to the Foxp3 promoter23, CNS124 and CNS225 regions. Increased expression of Fos, Fosb, Jun and Junb genes in HDAC11−/− vs. WT Tregs was confirmed by qPCR (Fig. 3D). Hence, HDAC11 deletion in Foxp3+ Tregs promotes expression of genes important to protection against oxidative stresses22 and important to the development and maintenance of the Treg lineage23.
HDAC11 deletion promotes Treg-dependent allograft survival
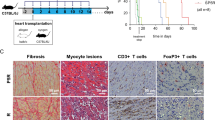
We used 3 murine allograft models to assess the effects of HDAC11 gene deletion in vivo. First, we performed cardiac allografting using fully MHC-disparate BALB/c donors (H-2d) and C57BL/6 recipients (H-2b), comparing survival in WT mice versus those lacking HDAC11 within their Foxp3+ Treg cells. Recipients were treated with a sub-therapeutic regimen of 14 days of rapamycin (RPM, 0.2 mg/kg/d, i.p.), beginning on the day of engraftment, that dampens conventional T cell responses but preserves Foxp3+ Treg cell function9. In contrast to cardiac allografts that were acutely rejected in WT mice, mice whose Tregs lacked HDAC11 but were otherwise comparable maintained their grafts long-term (>100 days, p < 0.01) (Fig. 4A). Compared to grafts harvested at day 7 post-transplant from WT recipients that showed diffuse mononuclear cell infiltration and extensive myocytolysis, grafts from mice with HDAC11−/− Tregs showed only small focal mononuclear infiltrates and had well preserved myocardia and vessels (Fig. 4B).
Effects of conditional or global HDAC11 deletion on cardiac allograft survival. (A) Compared to the acute rejection of BALB/c cardiac allografts in WT recipients, about two thirds of allografts in mice with conditional deletion of HDAC11 within their Tregs survived for >100 days (p < 0.01, n = 6 allografts/group). (B) Histology (H&E) of corresponding cardiac allograft groups as shown in panel (A) when harvested at day 7 post-transplant; note the absence of graft infiltration and myocyte necrosis in HDAC11−/− hosts compared to WT controls. (C) Histology of Bm12 cardiac allografts harvested at 30 days post-transplant from WT mice or mice with conditional deletion of HDAC11 within their Foxp3+ Treg cells, with a marked difference in the development of transplant arteriosclerosis (p < 0.001), myocyte injury and mononuclear cell infiltration (see Results for quantitative data on vessel scores for the 2 groups). (D) Compared to the acute rejection of BALB/c allografts in WT recipients, about two thirds of cardiac allografts in mice with global deletion of HDAC11 within their Tregs survived for >100 days (p < 0.01, n = 10 allografts/group).
In a second model, WT mice or mice lacking HDAC11 within their Foxp3+ Treg cells were transplanted with class IIa-disparate Bm12 cardiac allografts; recipients received no additional therapy. This class II MHC-mismatched strain combination is widely used to model the development of transplant arteriosclerosis26, 27, a major and persistent problem in clinical transplantation28. Allografts harvested from WT recipients at 30 d post-transplant showed severe intimal proliferation within muscular arteries (mean vessel score of 2.3 ± 0.3), as well as focal infarcts and dense interstitial mononuclear cell infiltrates, whereas lack of HDAC11 within recipient Tregs led to preservation of myocardial tissues and an absence of intimal proliferation (mean vessel score of 0.2 ± 0.1, p < 001) (Fig. 4C).
In a third model, we tested whether constitutive deletion of HDAC11 affected alloresponsiveness, using the fully MHC disparate BALB/c- > C57BL/6 combination plus 14 d of low-dose RPM therapy (RPM, 0.2 mg/kg/d, i.p.). As with conditional deletion in in Tregs (Fig. 4A), global deletion of HDAC11 protected mice against the acute rejection seen in WT recipients, with 60% surviving long-term (>100 d, p < 0.01) (Fig. 4D). These data show that conditional deletion of HDAC11 within Foxp3+ Tregs can markedly prolong allograft survival across a full MHC disparity, and can protect against the development of transplant arteriosclerosis. The beneficial effects of global deletion in allograft recipients likely have translational significance, given that a small molecule inhibitor of HDAC11 would target conventional T cells and other cellular components of host alloresponses, in addition to the Treg population.
Pharmacologic targeting of HDAC11 promotes Treg suppression and long-term allograft acceptance
The effects of HDAC11 gene deletion of allograft survival (Fig. 4) led us to assess the effects of an HDAC11i (JB3-22) on Treg function in vitro, and allograft survival in vivo. Our studies were informed by our own (Supplementary Fig. S7) and previous findings that HDAC11 can physically associate, in both the cytoplasm and nucleus, with HDAC614, and that pharmacologic inhibitors of HDAC6 boost Treg function and, in conjunction with a 14-day course of low-dose RPM, promote long-term cardiac allograft survival in murine models (BALB/c- > C57BL/6)11. Accordingly, we began by comparing the effects of HDAC11i on WT, HDAC6−/− and HDAC11−/− Tregs in vitro (Fig. 5A). Both HDAC6−/− and HDAC11−/− Tregs had increased suppressive function as compared to WT Treg cells, despite addition of DMSO-containing compound diluent, whereas in the absence of Tregs (0:1 ratio) B6 conventional T cells proliferated to a comparable extent (Fig. 5A, upper panel). Addition of the HDAC11i (0.1 µM) significantly increased Treg suppressive function of WT Tregs compared to DMSO-treated Treg cells, and also increased the suppressive function of HDAC6−/− Tregs compared to DMSO-treated Tregs, but had no added benefit when HDAC11−/− Tregs were used (Fig. 5A, lower panel). These data indicate that an HDAC11i was able to increase Treg function in HDAC6−/− Tregs but, consistent with specificity of the compound for HDAC11, had no effect using HDAC11−/− Treg cells. We then tested the HDAC11i (10 mg/kg/d, 14 d, i.p.) plus low-dose RPM (0.2 mg/kg/d, 14 d) in the BALB/c- > C57BL/6 cardiac allograft model. Allografts in WT recipients were acutely rejected, whereas allograft recipients receiving HDAC11i therapy showed long-term (>100 d) cardiac allograft survival (p < 0.01) (Fig. 5B). We conclude that HDAC11 targeting for 2 weeks post-transplant can promote Treg suppressive activity and lead to long-term allograft survival.
Effects of pharmacologic inhibition of HDAC11 on Treg function and allograft survival. (A) Testing of the in vitro effects of the HDAC11i, JB3-22, on WT, Hdac6−/− and HDAC11−/− Treg suppressive function. Upper 3 rows show baseline function of WT vs. HDAC6−/− and HDAC11−/− Tregs in the presence of DMSO carrier alone; proliferation of CFSE-labeled conventional T cells is shown in each panel. The lower 3 rows show the effects of HDAC11i (0.1 µM) on Treg function using the same cell types as tested above. The point is that HDAC11i further increases the suppressive function of WT and HDAC6−/− Tregs, but does not have any additional effects on the suppressive function of HDAC11−/− Treg cells. Data are representative of 3 separate assays. (B) Effects of HDAC11i (JB3-22, (10 mg/kg/d, 14 d, i.p.) plus low-dose RPM (0.2 mg/kg/d, 14 d) in the BALB/c- > C57BL/6 cardiac allograft model. Control mice receiving low dose RPM rejected their allografts within 2–3 weeks, but cardiac allografts were maintained long-term (>100 days, p < 0.01) in recipients treated for 2 weeks with low dose RPM plus HDAC11i therapy; 6 allografts/group.
Discussion
Since its discovery, cloning and report of its limited tissue distribution in 2002, HDAC11 garnered relatively little attention compared to most other HDACs and its functions were largely unknown, until two reports in 2009. First, HDAC11 was shown to contribute to control of DNA replication involving sequential recruitment of Cdc6, Cdt1 and several Mcm proteins29. HDAC11 deacetylates Cdt1, regulating its ubiquitination and proteasomal degradation, and thereby limits DNA replication. Second, HDAC11 was shown to regulate chromatin accessibility within the Il10 promoter region, such that knockdown of HDAC11 enhanced antigen-presenting cells (APC) production of IL-10 in response to LPS stimulation15. In a follow-up study by the same group, HDAC11 was shown to inhibit the development of myeloid derived suppressor cells (MDSC)30.
Moving beyond the role of HDAC11 in myelopoiesis, the current study assessed the role of HDAC11 in Foxp3+ Treg cells. This work was undertaken as part of our ongoing studies of the roles of individual HDAC isoforms in Treg cells, and whether such isoforms may be usefully targeted for therapeutic effect. Deletion of certain isoforms such as HDAC611, 31 and HDAC99, 11, 12 increase Treg function in vitro and in vivo, though only HDAC6 has been targetable pharmacologically32, 33. Deletion of other HDAC enzymes, such as HDAC313 and HDAC534 impaired Treg function, and in the case of HDAC3 led to lethal autoimmunity. Hence, to date, HDAC6 targeting appears to be the best option for pharmacologic enhancement of Treg function, with potential utility in the management of patients undergoing organ transplantation or in patients suffering from autoimmune diseases. The current study presents data showing that genetic and pharmacologic targeting of HDAC11, like that of HDAC6, has therapeutic potential.
Deletion of HDAC11 promotes Foxp3 protein acetylation, which increases its DNA binding9, 35. However, additional mechanisms likely also contribute to the increased Treg suppressive function in HDAC11-targeted mice. These include promoting an open chromatin structure at the Foxp3 promoter and all 3 CNS regions within Foxp3 intronic regions; facilitating the actions of transcription factors such as NFκB and AP-1 that regulate Foxp3 expression; and promoting expression of Foxp3 and several other genes important to Treg function, including TGF-β, CTLA-4 and GITR, though not IL-10. The net effect of these effects of HDAC11 deletion was an increase in Treg suppressive function, that in vitro, was largely attributable to increased TGF-β production. Treatment of WT Tregs with an HDAC11 inhibitor also increased Treg suppressive function.
Translation of these findings to in vivo models was assessed using transplant studies. Remarkably, in conjunction with a 2-week sub-therapeutic course of RPM, deletion of HDAC11, or concomitant use of an HDAC11i in WT recipients, resulted in long-term cardiac allograft survival, as well as protection against the development of transplant arteriosclerosis, a hallmark of chronic allograft dysfunction and a common complication of clinical organ transplantation28. Adoptive transfer of human Tregs into chimeric humanized mice was previously shown to protect arterial grafts from developing transplant arteriosclerosis36, especially when combined with RPM therapy37. The current studies show that comparable protection in a more biologically relevant model can be achieved by pharmacologic therapy. The pathogenesis of transplant arteriosclerosis is complex and multi-faceted, though roles for T and B cells are implicated, in addition to the well-established involvement of macrophages and smooth muscle cells that undergo activation and infiltration into the expanded intima38, 39. While additional research will be required to assess the points at which Foxp3+ Tregs modulate such arterial injury and gradual occlusion, beyond their effects on other T cells, as well as monocytes, macrophages, dendritic cells, natural killer cells, neutrophils and B cells, Foxp3+ Tregs are capable of suppressing various deleterious functions of non-immune cells, including that of endothelial and vascular smooth muscle cells, in disease models40,41,42.
Clinical trials involving the adoptive transfer of large numbers of Foxp3+ Tregs are currently at the Phase I stage in various centers worldwide. While no data are yet publicly available, these studies entail considerable expense and expertise to isolate and expand the requisite Foxp3+ Treg populations, and are envisaged in most cases as a single injection of cells in the early post-transplant period. Treg biology is more complex in humans than in mice, and even their isolation and characterization is not straightforward43. Thus, unlike in rodents in which Foxp3 has been engineered to be linked with various fluorescent proteins to allow efficient cell isolation, the surface signatures of human Tregs identifiable by flow cytometry are not diagnostic, and conventional T cells can transiently express FOXP3 upon cell activation. As a result, isolated preparations of CD4 + CD25 + CD127lo Tregs used for clinical cell therapy involve purities as low as 76% FOXP3+ cells44, and there are important concerns as to the safety of infusion of such mixed cell populations in transplant recipients. An important confounding variable is these trials is use of concomitant immunosuppression with calcineurin inhibitors that are a mainstay of modern immunosuppression, but which impair Treg function even when used at low concentrations45. The current studies suggest that the epigenetic regulation of Foxp3 expression and function using a combination of an HDAC11i and RPM may offer considerable advantages over Treg cell therapy, including ease of use, titratable dosing and the option of intermittent or sustained administration.
Lastly, while HDAC11 is overexpressed in various carcinomas compared to normal tissues, its depletion causes inhibition of tumor cell metabolism and their death, and comparable effects are seen when tumor cells are transfected with catalytically inactivated HDAC11 constructs46. However, HDAC11 depletion in normal cells does not affect their metabolism or promote their deatth46. These findings, along with the data from the current study, including the good health of globally HDAC11-deficient mice, suggest that HDAC11 targeting would not necessarily increase risk of tumor formation or normal cell injury. Such considerations support our ongoing efforts to develop potent and selective HDAC11i compounds that are suitable for clinical therapy.
Methods
Mice
We used male WT BALB/c, WT C57BL/6, and MHC class II variant H2-Ab1bm12 (Bm12/B6) mice from The Jackson Laboratory, plus Foxp3YFP-Cre mice47, and HDAC11fl/fl and globally HDAC11-deleted mice15. The latter 3 strains were backcrossed on the C57BL/6 background at least 8 times, and all mice were used at 6–8 weeks of age unless specified. Animal studies were approved and undertaken in accordance with the regulations and guidelines of the Institutional Animal Care and Use Committee of The Children’s Hospital of Philadelphia.
Antibodies and flow cytometry
Single cell suspensions were stained with mAbs directed against CD4 (BD Bioscience, Pacific blue, clone RM4-5, #558107), CD8 (PE-Cy7, eBioscience, clone 53-6.7, #25-0081), Foxp3 (PE-Cy5, eBioscience, clone FJK-16s, #15-5773), and CD25 (APC, eBioscience, clone PC61.5, #17-0251), and acquired with a Cyan flow cytometer. We also purchased unconjugated CD3 (clone 145-2C11, #553057) and CD28 (clone 37.51, #553294) mAbs from BD Bioscience. Anti-TGF-β mAb that neutralizes isoforms 1, 2 and 3 was purchased from R&D Systems (clone MAB135).
Treg suppression assays
For in vitro studies, 5 × 104 cell-sorted CD4+ YFP+ Tregs, or bead-isolated CD4+ CD25+ Tregs from CD4cre mice, were added to 96-well plates and serially diluted in medium (RPMI-1640 and 10% FBS, plus pen/strep). Thereafter, equal numbers of CFSE-labeled CD4+ CD25- T cells and γ-irradiated antigen-presenting cells were added, plus CD3 mAb (1 μg/ml), and cultured for 3 days. Thereafter, cells were stained with CD4 mAb (Pacific blue) and CFSE and CD4 positive T cell proliferation was acquired and data analyzed with FlowJo.
Cardiac transplantation and histopathology
Heterotopic abdominal cardiac transplantation was performed using BALB/c or Bm12/B6 donors and C57BL/6 recipients38. Recipients of BALB/c cardiac allografts were treated with RPM (0.2 mg/kg/d, i.p.), for 14 days beginning on the day of engraftment, and grafts were collected for histology at day 7 or day 100 post-transplant. In the case of class II-disparate Bm12- > B6 cardiac allografts (using WT or conditional HDAC11−/− recipients), hearts were collected at day 30 post-transplant and all intramyocardial arteries within elastin-stained paraffin sections were scored for the extent of intimal proliferation as <5% occlusion (0), >5–20% (1), >20–40% (2), >40–60% (3), >60–80% (4), or >80–100% (5)39.
Luciferase assays
293 T cells were maintained (37 °C, 5% CO2) in RPMI-1640 plus 10% heat-inactivated fetal bovine serum (FBS), penicillin, and streptomycin. Cells at 80% to 90% confluence were transfected with murine Foxp3 promoter luciferase reporter plus expression vectors for HDAC11, p65, Foxp3, p300, or empty vector utilizing Lipofectamine 2000 according to manufacturer’s recommendations. Combinations of expression vectors were utilized wherever specified and empty vector was co-transfected to ensure equal amount of DNA is used per condition. After 48 h, cells were treated with 10 ng/ml phorbol myristate acetate (PMA) and 1 μM ionomycin (Sigma) for 6 h, and luciferase activities of whole-cell lysates were analyzed with a dual-luciferase reporter assay kit (Promega). HDAC11i (JB-3–22) was added at a concentration of 1 μM for 6 h at 48 h post-transfection.
ChIP assays
ChIP assays were performed using Pierce Magnetic ChIP kit (Cat# 26157). Briefly, TE or Treg cells were fixed with 1% formaldehyde and fragmented by sonication. Chromatin was immunoprecipitated with antibodies directed against Foxp3 (eBioscience # 14-4774-82), HDAC11 (Invitrogen, #PA5-11249) and anti-acetyl-histone H3 (Millipore#06-599). The resultant DNA was purified and used for real-time PCR using SYBR green master mix (Applied Biosystems) and primers for IL-2 promoter, CNS1, CNS2, and CNS3 primer sets (reported previously) and TGFβ1 promoter (forward: GCTCTGAGCCGCACTCG and reverse: CTCCTCGGCTGCTCCTTT).
Microarray and real-time qPCR
RNA was isolated using RNeasy kits (Qiagen), and RNA integrity and quantity analyzed by NanoDrop ND-1000 and Nanochip 2100 Bioanalyzer (Agilent Technologies). Microarray experiments were performed using whole-mouse-genome oligoarrays (Mouse430a, Affymetrix), and array data analyzed using MAYDAY 2.12 software48. Array data were subjected to robust multiarray average (RMA) normalization and analyzed SAM (significance analysis of microarrays). Only data with a false discovery rate adjusted p-value of p < 0.05 and at least 1.5-fold differential expression were included in the analysis. Data underwent z-score transformation for display. Enrichment scores of the up- or downregulated genes were calculated using DAVID Functional Annotation Clustering49. Our microarray data was deposited at the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) and has the accession number GSE95316. Expression of individual genes was verified by qPCR. RNA was reverse transcribed to cDNA (Applied Biosystems), and qPCR performed using Taqman primer and probe sets; data were normalized to endogenous 18 s rRNA and relative expression determined by the formula 2−ΔCT.
Immunoprecipitation and Western blotting
Proteins were isolated using RIPA solution plus protease inhibitor (Sigma). Pull-down antibody was incubated with pre-cleared samples for 2 h at 4 °C, then overnight with protein G agarose. Western blots were performed using anti-Flag and anti-Foxp3 Abs (Cell Signaling).
Immunofluorescent microscopy
Cytospins of resting Treg cells or Treg cells activated overnight with CD3/CD28 mAbs were fixed, permeabilized with 0.2% Triton-100, blocked with normal goat serum for 2 h, incubated overnight with Abs to Foxp3 and HDAC11, washed, stained with secondary antibodies, and analyzed with a fluorescent microscope.
Statistics
Data were analyzed using GraphPad Prism 5.0d software. Data were presented as mean ± SD unless specified otherwise. Measurements between two groups were done with a 2-tailed Student’s t-test if data were normally distributed, or Mann-Whitney U unpaired test when the populations were not normally distributed. Groups of three or more were analyzed by 1-way ANOVA with corresponding Tukey’s multiple comparison test if normally distributed, or the Kruskal-Wallis with Dunn’s multiple comparison test if not. Graft survival was evaluated with Kaplan-Meier followed by Log-rank test. P < 0.05 was considered significant. Graphs show mean ± SD, unless otherwise stated in the figure legend.
Study approval
Animal studies were approved by the Institutional Animal Care and Use Committee of The Children’s Hospital of Philadelphia (protocol 2013-000561).
References
Leipe, D. D. & Landsman, D. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res 25, 3693–3697 (1997).
Gregoretti, I. V., Lee, Y. M. & Goodson, H. V. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 338, 17–31 (2004).
Glozak, M. A., Sengupta, N., Zhang, X. & Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 363, 15–23 (2005).
Sadoul, K., Boyault, C., Pabion, M. & Khochbin, S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie 90, 306–312 (2008).
Singh, B. N. et al. Nonhistone protein acetylation as cancer therapy targets. Expert Rev Anticancer Ther 10, 935–954 (2010).
Seto, E. & Yoshida, M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology 6, a018713 (2014).
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009).
Joshi, P. et al. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol 9, 672 (2013).
Tao, R. et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med 13, 1299–1307 (2007).
Wang, L., de Zoeten, E. F., Greene, M. I. & Hancock, W. W. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov 8, 969–981 (2009).
Beier, U. H. et al. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci Signal 5, ra45 (2012).
de Zoeten, E. F., Wang, L., Sai, H., Dillmann, W. H. & Hancock, W. W. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology 138, 583–594 (2010).
Wang, L. et al. FOXP3+ regulatory T cell development and function require histone/protein deacetylase 3. J Clin Invest 125, 1111–1123 (2015).
Gao, L., Cueto, M. A., Asselbergs, F. & Atadja, P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem 277, 25748–25755 (2002).
Villagra, A. et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol 10, 92–100 (2009).
Cheng, F. et al. Divergent roles of histone deacetylase 6 (HDAC6) and histone deacetylase 11 (HDAC11) on the transcriptional regulation of IL10 in antigen presenting cells. Mol Immunol 60, 44–53 (2014).
Toropainen, S., Vaisanen, S., Heikkinen, S. & Carlberg, C. The down-regulation of the human MYC gene by the nuclear hormone 1alpha,25-dihydroxyvitamin D3 is associated with cycling of corepressors and histone deacetylases. J Mol Biol 400, 284–294 (2010).
Zheng, Y. et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445, 936–940 (2007).
Liu, Y. et al. Two histone/protein acetyltransferases, CBP and p300, are indispensable for Foxp3+ T-regulatory cell development and function. Mol Cell Biol 34, 3993–4007 (2014).
Zheng, Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010).
Wong, P. G. et al. Chromatin unfolding by Cdt1 regulates MCM loading via opposing functions of HBO1 and HDAC11-geminin. Cell Cycle 9, 4351–4363 (2010).
Das, M., Singh, S., Pradhan, S. & Narayan, G. MCM paradox: Abundance of eukaryotic replicative helicases and genomic integrity. Molecular biology international 2014, 574850 (2014).
Mantel, P. Y. et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol 176, 3593–3602 (2006).
Xu, L. et al. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity 33, 313–325 (2010).
Ogawa, C. et al. TGF-beta-mediated Foxp3 gene expression is cooperatively regulated by Stat5, Creb, and AP-1 through CNS2. J Immunol 192, 475–483 (2014).
Wang, L. et al. B7-H3 promotes acute and chronic allograft rejection. Eur J Immunol 35, 428–438 (2005).
Gao, W. et al. Targeting of the chemokine receptor CCR1 suppresses development of acute and chronic cardiac allograft rejection. J. Clin. Invest. 105, 35–44 (2000).
Costello, J. P., Mohanakumar, T. & Nath, D. S. Mechanisms of chronic cardiac allograft rejection. Texas Heart Institute journal 40, 395–399 (2013).
Glozak, M. A. & Seto, E. Acetylation/deacetylation modulates the stability of DNA replication licensing factor Cdt1. J Biol Chem 284, 11446–11453 (2009).
Sahakian, E. et al. Histone deacetylase 11: A novel epigenetic regulator of myeloid derived suppressor cell expansion and function. Mol Immunol 63, 579–585 (2015).
de Zoeten, E. F. et al. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol 31, 2066–2078 (2011).
Kalin, J. H., Butler, K. V., Akimova, T., Hancock, W. W. & Kozikowski, A. P. Second-generation histone deacetylase 6 inhibitors enhance the immunosuppressive effects of Foxp3+ T-regulatory cells. J Med Chem 55, 639–651 (2012).
Segretti, M. C. et al. Thiol-Based Potent and Selective HDAC6 Inhibitors Promote Tubulin Acetylation and T-Regulatory Cell Suppressive Function. ACS Med Chem Lett 6, 1156–1161 (2015).
Xiao, H. et al. HDAC5 controls the functions of Foxp3( + ) T-regulatory and CD8(+) T cells. Int J Cancer 138, 2477–2486 (2016).
Liu, Y. et al. Inhibition of p300 impairs Foxp3+ T regulatory cell function and promotes antitumor immunity. Nat Med 19, 1173–1177 (2013).
Nadig, S. N. et al. In vivo prevention of transplant arteriosclerosis by ex vivo-expanded human regulatory T cells. Nat Med (2010).
Hester, J., Schiopu, A., Nadig, S. N. & Wood, K. J. Low-dose rapamycin treatment increases the ability of human regulatory T cells to inhibit transplant arteriosclerosis in vivo. Am J Transplant 12, 2008–2016 (2012).
Hancock, W. W., Buelow, R., Sayegh, M. H. & Turka, L. A. Antibody-induced transplant arteriosclerosis is prevented by graft expression of anti-oxidant and anti-apoptotic genes. Nat Med 4, 1392–1396 (1998).
Russell, M. E. et al. Chronic cardiac rejection in the LEW to F344 rat model. Blockade of CD28-B7 costimulation by CTLA4Ig modulates T cell and macrophage activation and attenuates arteriosclerosis. J Clin Invest 97, 833–838 (1996).
Tamosiuniene, R. et al. Regulatory T cells limit vascular endothelial injury and prevent pulmonary hypertension. Circ Res 109, 867–879 (2011).
Maganto-Garcia, E. et al. Foxp3+ -inducible regulatory T cells suppress endothelial activation and leukocyte recruitment. J Immunol 187, 3521–3529 (2011).
Dinh, T. N. et al. Cytokine therapy with interleukin-2/anti-interleukin-2 monoclonal antibody complexes expands CD4 + CD25 + Foxp3 + regulatory T cells and attenuates development and progression of atherosclerosis. Circulation 126, 1256–1266 (2012).
Akimova, T., Levine, M. H., Beier, U. H. & Hancock, W. W. Standardization, evaluation and area-under-curve analysis of human and murine Treg suppressive function. Methods in Molecular Biology 1371, 43–78 (2016).
Bluestone, J. A. et al. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci Transl Med 7, 315ra189 (2015).
Akimova, T. et al. Differing effects of rapamycin or calcineurin inhibitor on T-regulatory cells in pediatric liver and kidney transplant recipients. Am J Transplant 12, 3449–3461 (2012).
Deubzer, H. E. et al. HDAC11 is a novel drug target in carcinomas. Int J Cancer 132, 2200–2208 (2013).
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Battke, F., Symons, S. & Nieselt, K. Mayday–integrative analytics for expression data. BMC Bioinformatics 11, 121 (2010).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009).
Acknowledgements
Supported by grants from the National Institutes of Health to W.W.H. (AI073489 and AI095276) and to U.H.B. (AI095353).
Author information
Authors and Affiliations
Contributions
J.H. performed surgery and analyzed data, L.W. performed surgery and molecular and flow cytometry assays, analyzed data and edited the manuscript, S.D. and A.S. contributed biochemical studies, R.H. bred mice and performed histologic studies, U.H.B. analyzed data and edited the manuscript, J.B. and A.P.K. developed HDAC11i, E.M.S. and E.S. developed Hdac11-targeted mice and analyzed data, and W.W.H. supervised the project, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Accession code: GSE95316 for microarray data deposited at GEO (www.ncbi.nlm.nih.gov/geo).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, J., Wang, L., Dahiya, S. et al. Histone/protein deacetylase 11 targeting promotes Foxp3+ Treg function. Sci Rep 7, 8626 (2017). https://doi.org/10.1038/s41598-017-09211-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-09211-3
This article is cited by
-
Gut microbiota as a target in the bone health of livestock and poultry: roles of short-chain fatty acids
Animal Diseases (2023)
-
Targeting the epigenome to reinvigorate T cells for cancer immunotherapy
Military Medical Research (2023)
-
Salt and Gut Microbiota in Heart Failure
Current Hypertension Reports (2023)
-
Epigenetic modulation of antitumor immunity for improved cancer immunotherapy
Molecular Cancer (2021)
-
Design, synthesis and biological evaluation of a series of CNS penetrant HDAC inhibitors structurally derived from amyloid-β probes
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.