Abstract
Myopia and astigmatism, two common refractive errors frequently co-exist, are affecting vision at all working distances in the affected populations worldwide. Eyeballs having these refractive errors are known to exhibit abnormal eye shape at the anterior and posterior eye segments, but whether the outer coats of these abnormal eyeballs, cornea anteriorly and sclera posteriorly, are regulated by region-specific molecular mechanism remains unclear. Here we presented the changes in mRNA expression levels of three genes (MMP2, TIMP2, and TGFB2), all known to participate in extracellular matrix organization, at five regions of the cornea and sclera in chickens developing high myopia and astigmatism induced by form deprivation. We found that, compared to normal chicks, the highly myopic-astigmatic chicks had significantly higher expression of all three genes in the superior sclera (Mann-Whitney tests, all p ≤ 0.05), as well as higher TIMP2 expression in the central cornea and nasal sclera (Mann-Whitney tests, both p ≤ 0.05). Strikingly, the superior scleral region stood out as showing the strongest and most widespread correlations between mRNA expression and biometry parameters including axial and astigmatic components (r = + 0.52~ + 0.85, all p < 0.05). These results imply that local molecular mechanism may manipulate the eye shape remodeling across the globe during refractive-error development.
Similar content being viewed by others
Introduction
Myopia, or short-sightedness, is a highly prevalent visual condition associated with significant risks of morbidity. Despite its worldwide public health impact, the etiology of myopia remains poorly understood. Epidemiological studies on geographically diverse populations showed a higher prevalence of myopia in Chinese ethnicity1; of particular concern is the increasing trend of highly myopic population2. Although the vision of high myopes may be corrected by ophthalmic aids, their larger and longer eyeballs are associated with increased risks of sight-threatening eye diseases (e.g., glaucoma and cataract) and posterior segment anomalies (e.g., retinal detachment, staphyloma and tilted optic disc)3, 4.
The external ocular tissues responsible for maintaining the eye’s biomechanical properties – the cornea anteriorly, and the sclera posteriorly – exhibit abnormal structural properties in highly myopic eyes3, 5,6,7. High myopia is frequently associated with an irregular posterior eye shape in humans5, 8,9,10; and in animal models (monkeys11; chickens12). Importantly, astigmatism, another refractive error due mainly to non-uniform corneal curvature, also frequently co-exists with high myopia (humans13, monkeys14, and chicks15), suggesting abnormal refractive error development may involve structural remodeling of both the anterior and posterior eye segments. While it remains unclear how eye shape is regulated across the globe, there is ample evidence that eye shape remodeling during myopia development involves significant alterations of the components of extracellular matrix (ECM) in humans and animal models16,17,18,19,20,21,22. However, converging evidence indicates that the structural changes in ametropic eyes may be modulated by mechanisms localized within the eye itself23: First, optic-nerve blockade does not prevent the myopia induced by form deprivation or spherical defocus24,25,26,27,28,29. Second, restricting visual manipulations to particular regions of the visual field induces refractive-error and structural changes only in areas corresponding to the deprived retinal regions12, 30,31,32,33,34,35. Whether this local, region-specific mechanism plays a role in the development of myopic-astigmatic error is unclear. One working hypothesis is that the astigmatism associated with high myopia may be a byproduct of asynchronous and/or region-specific structural remodeling during myopia development. We tested this hypothesis by comparing the mRNA expression of 3 target genes (matrix metalloproteinase-2, MMP2; tissue inhibitor of metalloproteinase-2, TIMP2; transforming growth factor-beta 2, TGFB2), all known to participate in scleral structural remodeling (summarized in Supplementary Table S1). Transforming growth factor β (TGF-β) is a multi-functional factor that regulates cells growth and differentiation. TGF-β has been associated with myopia development. With functions in the proliferation regulation of scleral fibroblast cells and ECM production, TGFB2 plays an important role in biological activities36. However, the expression patterns of TGFB2 are controversial. For example, TGFB2 protein is upregulated in the posterior sclera of myopic chicks37,38,39, whereas TGFB2 mRNA expression decreased after form deprivation in tree shrew sclera39. Matrix-metalloproteinases (MMPs) are some of the enzymes that degrade ECM molecules. MMP2 is capable of degrading a range of components of the scleral ECM including collagens and proteoglycans40. During myopia development, upregulated MMP2 expression and increased activity are reported in myopia models, both mammalian16, 41 and avian42, 43. MMP2 can subsequently be regulated by tissue inhibitor of metalloproteinases (TIMPs) such as TIMP2 40. Balance between MMP2 and TIMP2 is critical to normal scleral matrix turnover and subsequently, regulation of eye development. Alteration of TIMP levels has been reported during slowed or increased eye growth in different animal models and human studies18, 44, 45. Recent study showed that downregulation of MMP2 expression levels and increased TIMP2 levels accompanied the pirenzepine-induced suppression of myopia in guinea pig44.
The experiments in this study were carried out using the chick form-deprivation paradigm, since large amounts of high myopia and astigmatism are reliably induced using this model, and general (non-region-specific) structural and molecular changes have been widely documented15, 22, 23, 35.
Results
Effects of form deprivation on refractive and axial components
In the treated group, the form-deprived eyes developed significantly higher myopia (mean ± SEM: −21.56 ± 2.78D vs. –0.66 ± 0.21D, p < 0.001), deeper anterior chamber depth (mean ± SEM: 1.65 ± 0.05 mm vs. 1.45 ± 0.02 mm, p < 0.01), and longer vitreous chamber depth (mean ± SEM: 6.12 ± 0.10 mm vs. 5.28 ± 0.07 mm, p < 0.001) than the fellow untreated eyes. As expected, the inter-ocular differences (treated/right eye– fellow/left eye) of these parameters were all significantly larger (independent t-tests, all p < 0.01) in the treated group than the normal group (mean±SEM: spherical equivalent = −22.22 ± 7.93D vs. −0.25 ± 0.93D; anterior chamber depth = 0.20 ± 0.05 mm vs. 0.04 ± 0.01 mm; vitreous chamber depth = 0.84 ± 0.26 mm vs. 0.07 ± 0.04 mm). Figure 1 illustrates the distributions of the inter-ocular differences in spherical-equivalent refractive errors versus anterior chamber depths (left) and vitreous chamber depths (right) for the normal (open symbols) and treated birds (filled symbols) at the end of the 7-day treatment period. In addition to the myopic errors, the treated chicks also developed significantly higher inter-ocular differences in refractive astigmatism, (mean ± SEM: 3.17 ± 0.32D vs. 0.47 ± 0.14D, p < 0.001) corneal astigmatism (mean ± SEM: 2.74 ± 0.24D vs. 0.43 ± 0.11D, p < 0.001), and J45 astigmatic components (mean ± SEM: refractive: −1.09 ± 0.19D vs. −0.00 ± 0.09D; corneal: −1.14 ± 0.19D vs. 0.06 ± 0.08D; both p < 0.001) when compared to normal birds. The polar plots in Fig. 2 illustrate the distributions of inter-ocular differences in refractive (left) and corneal astigmatism (right) for normal (open) and treated (filled) chicks: The magnitude and axis of the astigmatism for each bird are represented by the distance and angle from the origin respectively. As shown, the refractive astigmatism and corneal astigmatism in the treated birds were much higher than those in normal birds and the axes were oriented slightly obliquely from 90 axis. In addition, correlation analyses showed that refractive, corneal, and ocular axial components were interrelated (Table 1).
Effects of form deprivation on refractive and axial parameters. Spherical-equivalent refractive errors are plotted against the anterior chamber (left) and vitreous chamber depths (right) at the end of the one-week treatment period of monocular form deprivation. All data are expressed as inter-ocular differences (treated/right eye – fellow/left eye). The data for individual birds are represented by open (normal) or filled (treated) symbols. The mean ± SEM values for each group are represented with grey symbols with error bars, note that the error bars for normal group are masked by the filled symbols.
Effects of form deprivation on astigmatism. Refractive (left) and corneal (right) astigmatism at the end of the treatment period for individual normal (open circles) and treated birds (filled diamonds). The magnitude and axis of astigmatism are represented by the distance and angle from the origin, respectively. Average values calculated by power vector analyses are represented by grey symbols with a cross.
Effects of treatment on gene expression
When the inter-ocular differences in gene expressions (ΔΔCT) of all five regions from each bird were averaged and compared between groups, the scleral MMP2 and TIMP2 expressions in treated group were significantly higher (i.e., lower ΔΔCT) than those in normal groups (Mann-Whitney tests, both p < 0.05). No such differences were found in all three genes from the cornea (Mann-Whitney tests, all p > 0.05).
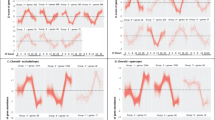
Analyses of the treatment effects on each gene at individual regions showed that, compared to normal group, the treated group had significantly higher gene expressions in all three genes at the superior sclera (Mann-Whitney tests: MMP2, p < 0.01; TIMP2, p < 0.001; TGFB2, p < 0.05), and in TIMP2 gene expression in the central cornea and nasal sclera (both p < 0.05). Figure 3 plots the significant fold-change differences in mRNA expressions for individual data and group means.
Differences in mRNA expressions at specific regions. Messenger RNA expression differences between the treated group (treated minus fellow untreated; filled symbols) and normal group (right minus left; open symbols) for regions of sclera or cornea that displayed significant between-group differences. Symbols represent individual data and bars represent mean ± SEM for each group. P values represent results from Mann-Whitney test.
Correlation between mRNA expression and ocular component dimensions
To understand if regional mRNA expression level was correlated with refractive error and/or axial ocular component dimensions, we pooled all the data from both treated and normal groups for Pearson’s correlation analyses. Data from both groups were chosen to cover a wide range of changes in refractive errors and ocular components. Fig. 4 shows the statistically significant correlation coefficients between individual biometric components and genes expressions at different scleral regions. The strengths of the correlation are represented using different colors, with warmer colors representing stronger correlations. The superior scleral region stood out as showing the strongest and most widespread correlations between mRNA expression and biometry parameters. Significant correlations were also found at the temporal, nasal and central sclera, but not at the inferior region.
Multiple significant correlations at superior sclera. Pearson’s correlation coefficients (r) between the mRNA expressions (ΔΔCT) and multiple ocular parameters at individual scleral regions (boxes). Only statistically significant coefficients are shown, the warmer the color the higher the correlations (see color code). VC, vitreous chamber depth; RT, retinal thickness; AC, anterior chamber depth; ST, scleral thickness; SE, spherical-equivalent refractive error; R.J45, refractive J45 astigmatic component; CC, average corneal curvature; C.J45, corneal J45 astigmatic component.
In contrast to the sclera, the corneal samples only showed significant correlations between gene expression level and biometry parameters along the horizontal meridian and inferior region (Fig. 5). Here the central cornea showed the highest number of significant correlations. However, even in this region, the correlations were not as high or as numerous as those found in the superior sclera (cf. Fig. 4).
Correlations in cornea. Pearson’s correlation coefficients (r) between the mRNA expressions (ΔΔCT) with multiple ocular parameters at four corneal regions. Only statistically significant correlations are shown; green boxes highlight higher correlations than blue boxes (similar color code as in Fig. 4). VC, vitreous chamber depth; RT, retinal thickness; AC, anterior chamber depth; ST, scleral thickness; SE, spherical-equivalent refractive error; CC, average corneal curvature.
Correlations of mRNA expressions between genes
Figure 6 shows the significant correlations between pairs of genes, in scleral (Fig. 6A) and corneal (Fig. 6B) regions. Only statistically significant Pearson’s coefficients are presented, with warmer colors representing stronger correlations. In both sclera and cornea, significant correlations between MMP2 and TIMP2 were found for all five regions. On the other hand, whereas all pairwise correlations were statistically significant across the five scleral regions, significant correlations between all three genes were found in all except the central cornea. Interestingly, the highest correlations between gene expressions were found at the inferior region of cornea (r = 0.97 for MMP2-TIMP2) and superior region of sclera (r = 0.93 for MMP2-TIMP2).
Correlations between pairs of genes. Pearson correlation coefficients of mRNA expression levels (ΔΔCT) between pairs of genes at specific scleral (A) and corneal (B) regions. Only statistically significant coefficients are shown; the warmer the color the higher the correlations ( , 0.50~0.59;
, 0.50~0.59;  , 0.60~0.69;
, 0.60~0.69;  , 0.70~0.79;
, 0.70~0.79;  , 0.80~0.89;
, 0.80~0.89;  , 0.90~0.99). All coefficients are positive values.
, 0.90~0.99). All coefficients are positive values.
Discussion
Deprivation of form vision has consistently been shown to induce axial myopia in a wide variety of animal models, making this a very common visual manipulation for the study of myopia development. Recent studies from rhesus monkeys14 and chicks15, 35 showed that form deprivation promoted not only posterior axial elongation but also significant magnitudes of ocular astigmatism, suggesting a mechanistic link between the changes in posterior and anterior eye growth. In the current study, eight of the 39 form-deprived birds exhibited axial elongation and highly myopic-astigmatic errors, the percentage of birds (20.5%) exhibiting myopia <−10D and astigmatism >2D was similar to the percentage (27.2%) from a separate group of birds receiving identical treatment paradigm in a previous study15, excluding the possibility that the selected chicks in this study were outliers due to the form deprivation paradigm. As shown in Table 1, significant correlations were found not only between anterior chamber depth and posterior ocular components (vitreous chamber depth and retinal thickness), but also between corneal parameters and posterior ocular dimensions (e.g., corneal J45 astigmatic component with vitreous chamber depth and retinal thickness). These results support the hypothesis that eye shape remodeling in the highly myopic eyeballs involves changes in both the anterior and posterior eye segments.
An important finding in this study is that the mRNA expression levels of MMP2 and TIMP2 genes were higher in form-deprived eyes than in normal eyes in sclera as a whole (average data), as well as in all three genes at the superior scleral region specifically (Fig. 3). As summarized in Supplementary Table S1, the three genes have shown differential expression patterns in sclera during the development of ametropia in previous studies. It should be noted that, unlike several studies which performed analyses on individual scleral layers (i.e., fibrous and/or cartilaginous) in chicks, we measured the mRNA expression level of the full-thickness scleral tissue. There is evidence that the molecular changes are opposite in the two scleral layers of chicks but the changes in fibrous sclera in chicks resemble those in tree shrew43, 46. Nevertheless, in agreement with our results, MMP2 mRNA level in sclera have previously been found to increase in the treated eyes of form-deprived chicks43, in both treated and fellow eyes of chicks after 4 hours of monocular −7D lens wear45, and in the treated eyes of tree shrews after either 11 days of form deprivation47 or 4 days of −5D lens wear18. Compared to MMP2, expression levels of TIMP2 and TGFB2 in myopic sclera have shown inconsistent results across the different studies in the literature (Supplementary Table S1): TIMP2 levels were not altered at multiple early time points after wearing +7D or −7D lenses45, but were found to decrease significantly after 10 days of form deprivation in chicks43. On the other hand, although the cartilaginous scleral TGFB2 mRNA levels were found to increase after wearing +7D lens for 24 hrs in chicks45, the scleral TGFB2 levels decreased after up to five days of form deprivation17, 39, 48 or −5D lens wear17, 48 in tree shrews (see Supplementary Table S1 for details). In short, the higher MMP2 expression in myopic sclera was consistently found in the current and previous studies, regardless of the differences in experimental protocol and animal model. On the other hand, the patterns of change in TIMP2 and TGFB2 varied across studies. Nevertheless, the fact that differential mRNA levels were found at scleral/corneal regions in myopic chicks (Fig. 3) and regional variations in scleral proteoglycan synthesis were found in humans49 and myopic chicks50 underscores the importance of understanding the role of local mechanisms in controlling corneal and scleral remodeling.
A second important and novel finding was the regional specificity of the correlations between gene expression levels and refractive/structural parameters, these being most pronounced in the superior sclera. For example, the two key indicators for myopia development, the vitreous chamber depth and spherical-equivalent refractive error, showed moderate correlations with the mRNA levels of MMP2 and TIMP2 genes in the superior scleral (Fig. 4). These region-specific differential gene expressions are in line with our recent findings that full-field form deprivation actually induced a bigger eye expansion superiorly than inferiorly near the posterior pole (up to 20° eccentricity), i.e., an asymmetric posterior eye shape35. These results suggest the potential involvement of molecular changes concentrated at the superior scleral region in modulating the structural changes that occur in eyes developing myopia and astigmatism (see Table 1): a thinner retina, a deeper anterior chamber, a steeper corneal curvature, and a higher J45 astigmatism. It is possible that this combination of structural and molecular changes led to the unique astigmatic axis repeatedly found in form-deprived chicks, i.e., an axis typically oriented near the 90° meridian15. In contrast to the significant correlations between the two J45 astigmatic components and mRNA expressions at the superior sclera, only weak correlations were found between the astigmatic components and mRNA expressions at the nasal and inferior corneal regions, suggesting a less important and/or passive role of differential corneal gene expressions in manipulating the characteristics of astigmatism, at least at the time point we tested in this study. In light of the significant changes in mRNA expression concentrated at the superior sclera and the potential roles of the three genes in regulating eye shape remodeling, further studies are strongly in need for understanding the role of local, region-specific mechanisms during myopia development.
Active scleral remodeling has been shown to occur in a range of myopia models22, 51. Evidences from time-course experiments at the structural52 and molecular levels19, 53 indicate a more rapid, dramatic change during the early phase of myopic eye growth. In addition, recent studies in tree shrews53 and chicks45 have also shown that mRNA expression levels related to collagen fibril reorganization are time-dependent. In the current study, significantly higher expression of MMP2, TIMP2, and TGFB2 mRNAs were found in the superior sclera of highly myopic-astigmatic eyes after 1 week of form deprivation in chicks, suggesting that local scleral structural remodeling was still in progress at this time point. However, given the potential differences in the mechanisms underlying form-deprivation and lens-induced myopia23, whether similar results would be found in highly myopic-astigmatic eyes induced through a closed loop condition (e.g., induced by wearing negative lens or sphero-cylindrical lens) awaits further studies.
Overall, our results demonstrated the structural and regional molecular changes in highly myopic-astigmatic eyes. It should be noted that the biometry and molecular changes were expressed as inter-ocular differences. The limitation of this approach is the potential interocular effects on the fellow untreated eyes of treated birds. However, given that no significant differences were found in the refractive parameters (refractive and corneal), axial structural parameters, or regional CT values between the fellow untreated (left) eyes of the treated birds and the left eyes of the normal birds, these indicate that form deprivation did not produce significant effects on the fellow untreated eyes. The second limitation of this study is the use of full-thickness scleral tissue for molecular analysis. Although on average the form-deprived sclera showed higher expressions of MMP2 and TIMP2 than normal sclera, it remains unresolved if these changes were contributed by one or both scleral layers in chicks at this time point. Lastly, because the primary goal of this study was to test the hypothesis of whether genes known to participate in scleral structural remodeling show region-specific expressions in eyeball with abnormal anterior and posterior eye shapes, we only included chicks that developed high myopia with significant astigmatism. Further works are needed to confirm if similar region-specific expressions also occur in eyes with different refractive errors.
In conclusion, highly myopic and astigmatic eyes induced by monocular form deprivation not only exhibited elongated eyeball in both anterior and posterior segments, but also showed higher mRNA expressions of genes at the posterior sclera. The high correlations between biometry parameters and expression levels at specific scleral region indicate that local mechanism may manipulate the eye shape remodeling. Further studies are needed to confirm if these mechanisms found mostly in sclera are unique only to eyes with high myopia and astigmatism.
Materials and Methods
Form-deprivation treatment
Forty-seven White Leghorn (Gallus Gallus domesticus) chicks were used (treated, n = 39; normal, n = 8). Chicks were raised in the Centralized Animal Facilities of The Hong Kong Polytechnic University under a 12-hr light/12-hr dark cycle with free access to food and water. The average illuminance was ~100 lux at the chick’s eye level and the room temperature was 25 °C. All experiments were conducted in accordance with the Association for Research in Vision and Ophthalmology (ARVO) statement for the use of animals in Ophthalmic and Vision Research. The experimental protocols were approved by the Department of Health, Government of the Hong Kong Special Administrative Region (10–28 in DH/HA&P/8/2/4 Pt.3) and Animal Subject Experiment Subcommittee of The Hong Kong Polytechnic University (ASESC#0916).
To induce high myopia with astigmatism, chicks were reared with translucent occluders (diffusers) placed in front of their right eyes from post-hatch day 5 to day 12 (P5 - P12). At the end of the one-week treatment period, a modified Hartinger refractometer54 was used to identify birds that had developed high magnitudes of myopia (<−10D) and refractive astigmatism (>2D). Of the 39 treated birds, eight fulfilled these refractive-error criteria and additional biometric measures were taken to determine the corneal parameters and ocular axial dimensions by means of a custom-made corneal topographer55 and a high resolution (20 mHz) A-scan ultrasonography system, respectively52. Refractometry and A-scan ultrasonography were carried out while birds were anaesthetized (isoflurane inhalation, 1.0% to 1.5% in oxygen) whereas corneal topography were measured in awake chicks55. The age-matched control group (referred to as ‘normal group’, n = 8) received no treatment, but underwent the same biometry measurements. Following the above measurements, chicks were sacrificed by carbon dioxide asphyxiation.
Tissue preparation
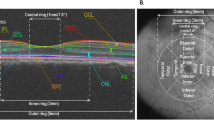
Immediately after the bird was sacrificed, eyes were enucleated carefully. To minimize potential confounding effects on RNA integrity due to the sequence of tissue preparation, the treated (or right eye) and fellow untreated eyes (or left eye) were enucleated in random sequence in different birds. The muscles and connective tissues remaining on the enucleated eye were removed, eyes were bisected with a razor blade along the equator of the eyeball and the vitreous body was removed. The remaining tissue was placed in ice-chilled 1% phosphate-buffered saline. The retina, lens and uvea were removed and discarded, and punches from the cornea and sclera were collected, using 1.5 mm- and 5 mm-diameter disposable trephines respectively, from 5 regions. These regions were chosen based on our previous findings that form deprivation induced both asymmetric posterior eye shape and significant amounts of astigmatism with axis oriented near 90° axis35, 54. As illustrated in Fig. 7, a mark made at the 12 o’clock conjunctiva was used as a reference to collect corneal tissues near the limbus (Fig. 7A), whereas the pecten was used as a landmark when collecting scleral tissues (Fig. 7B). The tissue punches were stored in 1.5 ml centrifuge tubes, snap-frozen in liquid nitrogen and stored at −80 °C until RNA was extracted.
Corneal and scleral tissues used for mRNA analyses. Messenger RNA expressions of three genes were measured at different regions in chick’s cornea (A) and sclera (B). Arrows indicate the ink mark and pecten used to identify the orientations of cornea and sclera respectively. C, central; T, temporal; N, nasal; S, superior; I, inferior.
RNA quality check, RNA purification, and cDNA synthesis
The RNA quality of samples (0.5 μg) collected with and without using RNAlater was checked in a preliminary study using 1% denaturing agarose gels in formaldehyde containing running buffer (1xMOPS and formaldehyde). Both central corneal and scleral punches at the posterior pole were collected separately from a normal and a treated eye (separately from two birds) using 5 mm-diameter disposable trephines. The isolated tissues were further divided into 2 parts: one half was treated with RNAlater and the other half was not. As shown in the electrophoresis patterns in Supplementary Fig. 1, both bands of ribosomal RNAs (18S and 28S) were clearly visible and RNA qualities were comparable between the samples treated with or without RNAlater. Based on this result, all tissue samples in this study were collected without using RNAlater.
Total RNA from both tissue punches was purified with the RNeasy Fibrous Tissue Mini Kit (Qiagen, Utraco Greentech, Singapore) according to the manufacturer’s instructions. Tissues were dispersed and homogenized at a speed of 1600 rpm by a freezer mill (Mikro-Dismembrator, B. Braun Biotech, Melsungen, Germany). All samples were treated with DNase I according to the supplier’s instructions. The purity of RNA was determined by Nanodrop ND 1000 (NanoDrop Technologies), the optical density ratio (OD260/OD280) was between 1.8 and 2.1. All RNA samples were diluted to a concentration of 10 ng/μl in distilled water. Each RNA sample (50 ng) was reverse-transcribed to cDNA by using high capacity RNA to cDNA master mix (Life technologies, Tubas, Singapore) according to the manufacturer’s instructions. The cDNA reverse transcription reactions were prepared by mixing 10 μl of 2xRT buffer, 1μl of 20xRT enzyme mix, 5μl of RNA sample, and 4μl of nuclease-free water. Reverse transcription was carried out at 37 °C for 60 min, and halted by heating the sample to 95 °C for 5 min. Paired samples from right and left eyes were reverse-transcribed together to minimize batch effects.
RT-PCR (reverse transcription –polymerase chain reaction) and sequencing
Primers for conventional RT-PCR were designed using Primer 3 Plus software (http://www.bioinformatics.nl/primer3plus; Life technologies, Hong Kong, China). Primers were designed to amplify a product size of 200–600 bp with GC content between 40% and 55%, and Tm values between 55 °C and 60 °C. To avoid the amplification of genomic DNA, the primers were designed to cross at least one exon junction for the specific amplification of cDNA. All primers were located within the coding region of the target genes. The detailed information of primer is shown in Supplementary Table S2. PCR was performed by using Hotstart Taq PCR master mix (QIAGEN, Utraco Greentech, Singapore) with primers specific for chick MMP2, TIMP2, and TGFB2. Both chick glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and 18S ribosomal RNA (RN18S) were used as the reference gene for this step. In some samples the template was omitted as a negative control.
PCR was run under the following conditions: The initial denaturation was started at 95 °C for 5 min and followed by 32 cycles for GAPDH and RN18S genes, and 35 cycles for MMP2, TIMP2 and TGFB2 genes. Each cycle consisted of denaturation at 94 °C for 30 sec, annealing at 60 °C for 30 sec and extension at 72 °C for 30 sec, and a final extension step was performed at 72°C for 10 min.
PCR products were verified by DNA sequencing (Dragon technology Ltd., Hong Kong). Data were sent back and analyzed by a special Chromas software (provided by the Dragon technology Ltd., Hong Kong) and BLAST available from NCBI. All genes had 99% matching identities from BLAST.
Real-time RT-PCR
Messenger RNA sequences for chicken MMP2, TIMP2, TGFB2 and RN18S (accession numbers: MMP2, U07775; TIMP2, AF004664; TGFB2, X59080; and RN18S, AF173612) were obtained from the European Molecular Biology Laboratory sequence database (EMBL, Heidelberg, Germany), identical to those used in a previous chicks study45. The Taqman® qPCR gene expression assays used in this study were designed and the primers sequences for MMP2, TIMP2, TGFB2 and RN18S are shown in Supplementary Table S3. Quantitative real-time PCR was performed in 96-well plates on an ABI 7500HT Real–Time PCR System (Applied Biosystems, New York, US). A total reaction volume of 20μl contained 10μl of 2 × TaqMan Universal PCR Master Mix (without uracil-N–glycosylase), 7μl of sterile water, 2μl of cDNA template, and 1μl of 20x Gene Expression Assay mix (including the primers and the marked probes). The thermal cycling conditions were: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 sec and 60 °C for 1 min. Samples were run in triplicate. One negative (i.e., no-template) control sample was included in each plate. In order to confirm the size of amplicons, MMP2, TIMP2, TGFB2 and RN18S qPCR products were separated by electrophoresis on a 3% agarose gel with a 100 bp DNA ladder (Thermo Fisher Scientific, USA), and visualized using 1x Gel Red (Biotium, Hayward, CA, USA). The result shown in Supplementary Fig. 2 indicated that the primers (Supplementary Table S3) in TaqMan assays were specific and only one qPCR product was identified in the agarose gel.
The stability of GAPDH and RN18S at different regions was tested by performing TaqMan assay qPCR for both corneal and scleral tissues collected from 2 birds (4 eyes). Analyses of CT values indicated that while RN18S did not show significant variability in either cornea (SD = 0.88) or sclera (SD = 0.80), the variability of GAPDH in sclera (SD = 3.26) was much higher than those in cornea (SD = 1.11). These results are in agreement with a previous study showing larger variability in GAPDH mRNA levels in chick’s fibrous sclera but 18 SRNA did not show significant difference between lens-treated and control eyes45. As a consequence, we chose RN18S as our endogenous control gene in subsequent analyses.
Real time RT-PCR Data Analysis
RT-PCR quantification was performed using CT values. “CT” is defined as the PCR cycle at which the fluorescence signal crosses a threshold line, which occurs during the exponential phase of the amplification curve. The smaller the CT value, the higher the expression level. Because comparing the refractive parameters (refractive and corneal), axial parameters, and CT values between the fellow untreated (left) eyes of the treated birds and the left eyes of the normal birds showed no significant differences (independent t-tests, all p > 0.05), we calculated the inter-ocular difference in gene expression (ΔΔCT) between the fellow eyes (i.e., treated and untreated fellow eyes in treated group, and right and left eyes in normal group) to compare the effects of visual manipulation on gene expressions between the treated and normal groups. Thus, at each of the five regions for each bird, the ΔΔCT in gene expression between the treated (or right eye in normal group) and fellow eyes (or left eye in normal group) was calculated56, 57 with the equation:
The geometric mean of the triplicate samples for each bird was used for statistical analyses. To illustrate the expression levels in figure (Fig. 3), fold-change values above 1 (corresponding to gene expression that was greater in the treated eye sample compared to the fellow eye sample) was further expressed as 2−ΔΔCT, whereas fold-changes below 1 were expressed as −1/2−ΔΔCT.
Statistical analyses
Refractive errors and corneal curvature (CC) were represented as spherical-equivalent (SE for refraction and CC for average corneal curvature), JO (=−0.5*C*cos2α) and J45 (=−0.5*C*sin2α) astigmatic components using power vectors, where C is the magnitude of the cylindrical power and α is the axis of the minus cylinder correcting lens58. To correlate with inter-ocular difference in gene expressions (ΔΔCT), effects of form deprivation on refractive errors and ocular dimensions were expressed as inter-ocular differences (treated/right eye minus fellow untreated/left eye). Paired t-tests were used to compare parameters between treated and fellow eyes. Independent t-tests or Mann-Whitney tests were used to test the difference in refractive and ocular dimensions between groups, depending on whether the within-group distribution was normally distributed. Spearman’s correlations were used to examine monotonic relationships between variables. Correction method such as Bonferroni’s method was not applied for multiple comparisons because the 3 genes and the biometric parameters could have inter-dependent effects (see results above59). All statistical tests were performed using Minitab 15.1.30.0 (Minitab Inc., USA) or SPSS Statistics 23.0 (IBM, USA) with a significance level set at α < 0.05.
References
Pan, C. W., Ramamurthy, D. & Saw, S. M. Worldwide prevalence and risk factors for myopia. Ophthalmic Physiol Opt 32, 3–16, doi:10.1111/j.1475-1313.2011.00884.x (2012).
Holden, B. A. et al. Global prevalence of myopia and high myopia and temporal trends from 2000 through 2050. Ophthalmology 123, 1036–1042, doi:10.1016/j.ophtha.2016.01.006 (2016).
Curtin, B. J. Ocular findings and complications in The Myopias: Basic Science And Clinical Management. Ch. 13, 279–280 (Harper & Row, 1985).
Morgan, I. G., He, M. & Rose, K. A. Epidemic of pathologic myopia: what can laboratory studies and epidemiology tell us? Retina, doi:10.1097/IAE.0000000000001272 (2016).
Curtin, B. J. & Teng, C. C. Scleral changes in pathological myopia. Trans Am Acad Ophthalmol Otolaryngol 62, 777–790 (1958).
McBrien, N. A., Jobling, A. I. & Gentle, A. Biomechanics of the sclera in myopia: extracellular and cellular factors. Optom Vis Sci 86, E23–E30 (2009).
Shen, M. et al. Biomechanical properties of the cornea in high myopia. Vision Research 48, 2167–2171 (2008).
Atchison, D. A. et al. Eye shape in emmetropia and myopia. Invest Ophthalmol & Vis Sci 45, 3380–3386 (2004).
Logan, N. S., Gilmartin, B., Wildsoet, C. F. & Dunne, M. C. Posterior retinal contour in adult human anisomyopia. Invest Ophthalmol Vis Sci 45, 2152–2162 (2004).
Stone, R. A. & Flitcroft, D. I. Ocular shape and myopia. Ann Acad Med Singapore 33, 7–15 (2004).
Wiesel, T. N. & Raviola, E. Myopia and eye enlargement after neonatal lid fusion in monkeys. Nature 266, 66–68 (1977).
Wallman, J. & Adams, J. I. Developmental aspects of experimental myopia in chicks: susceptibility, recovery and relation to emmetropization. Vision Res 27, 1139–1163 (1987).
Farbrother, J. E., Welsby, J. W. & Guggenheim, J. A. Astigmatic axis is related to the level of spherical ametropia. Optom Vis Sci 81, 18–26 (2004).
Kee, C.-s, Hung, L.-F., Qiao-Grider, Y., Ramamirtham, R. & Smith, E. L. III Astigmatism in monkeys with experimentally induced myopia or hyperopia. Optom Vis Sci 82, 248–260 (2005).
Kee, C.-s & Deng, L. Astigmatism associated with experimentally induced myopia or hyperopia in chickens. Invest Ophthalmol Vis Sci 49, 858–867, doi:10.1167/iovs.06-1370 (2008).
Guggenheim, J. A. & McBrien, N. A. Form-deprivation myopia induces activation of scleral matrix metalloproteinase-2 in tree shrew. Invest Ophthalmol & Vis Sci 37, 1380–1395 (1996).
Gao, H., Frost, M. R., Siegwart, J. T. Jr. & Norton, T. T. Patterns of mRNA and protein expression during minus-lens compensation and recovery in tree shrew sclera. Mol Vis 17, 903–919, doi:101 [pii] (2011).
Siegwart, J. T. & Norton, T. T. Selective regulation of MMP and TIMP mRNA levels in tree shrew sclera during minus lens compensation and recovery. Invest Ophthalmol & Vis Sci 46, 3484–3492, doi:10.1167/iovs.05-0194 (2005).
Gentle, A., Truong, H. T. & McBrien, N. A. Glycosaminoglycan synthesis in the separate layers of the chick sclera during myopic eye growth: comparison with mammals. Curr Eye Res 23, 179–184 (2001).
McBrien, N. A., Metlapally, R. & Jobling, A. I. & Gentle, A. Expression of collagen-binding integrin receptors in the mammalian sclera and their regulation during the development of myopia. Invest Ophthalmol Vis Sci 47, 4674–4682, doi:10.1167/iovs.05-1150 (2006).
Rada, J. A., Thoft, R. A. & Hassell, J. R. Increased aggrecan (cartilage proteoglycan) production in the sclera of myopic chicks. Dev Biol 147, 303–312 (1991).
Rada, J. A., Shelton, S. & Norton, T. T. The sclera and myopia. Exp Eye Res 82, 185–200 (2006).
Wallman, J. & Winawer, J. Homeostasis of eye growth and the question of myopia. Neuron 43, 447–468 (2004).
Norton, T. T., Essinger, J. A. & McBrien, N. A. Lid-suture myopia in tree shrews with retinal ganglion cell blockade. Vis Neurosci 11, 143–153, doi:S0952523800011184 [pii] (1994).
Troilo, D., Gottlieb, M. D. & Wallman, J. Visual deprivation causes myopia in chicks with optic nerve section. Curr Eye Res 6, 993–999 (1987).
Wildsoet, C. & Pettigrew, J. D. Experimental myopia and anomalous eye growth patterns unaffected by optic nerve section in chickens: Evidence for local control of eye growth. Clin Vis Sci 3, 99–107 (1988).
Troilo, D. & Wallman, J. The regulation of eye growth and refractive state: an experimental study of emmetropization. Vision Res 31, 1237–1250, doi:0042-6989(91)90048-A [pii] (1991).
Wildsoet, C. & Wallman, J. Choroidal and scleral mechanisms of compensation for spectacle lenses in chicks. Vision Res 35, 1175–1194 (1995).
Wildsoet, C. Neural pathways subserving negative lens-induced emmetropization in chicks–insights from selective lesions of the optic nerve and ciliary nerve. Curr Eye Res 27, 371–385 (2003).
McFadden, S. Partial occlusion produces local form deprivation myopia in the guinea pig eye. Invest. Ophthalmol. Vis. Sci. 43, 189- (2002).
Diether, S. & Schaeffel, F. Local changes in eye growth induced by imposed local refractive error despite active accommodation. Vision Res 37, 659–668 (1997).
Hodos, W. & Kuenzel, W. J. Retinal-image degradation produces ocular enlargement in chicks. Invest Ophthalmol Vis Sci 25, 652–659 (1984).
Wallman, J., Gottlieb, M. D., Rajaram, V. & Fugate-Wentzek, L. A. Local retinal regions control local eye growth and myopia. Science 237, 73–77 (1987).
Smith III, E. L. et al. Hemiretinal form deprivation: evidence for local control of eye growth and refractive development in infant monkeys. Invest Ophthalmol Vis Sci 50, 5057–5069, doi:10.1167/iovs.08-3232 (2009).
Chu, C. H., Deng, L. & Kee, C. S. Effects of hemiretinal form deprivation on central refractive development and posterior eye shape in chicks. Vision Res 55, 24–31 (2012).
Laiho, M., Saksela, O., Andreasen, P. A. & Keski-Oja, J. Enhanced production and extracellular deposition of the endothelial-type plasminogen activator inhibitor in cultured human lung fibroblasts by transforming growth factor-beta. J Cell Biol 103, 2403–2410 (1986).
Seko, Y., Shimokawa, H. & Tokoro, T. Expression of bFGF and TGF-beta 2 in experimental myopia in chicks. Invest Ophthalmol Vis Sci 36, 1183–1187 (1995).
Seko, Y., Tanaka, Y. & Tokoro, T. Influence of bFGF as a potent growth stimulator and TGF-beta as a growth regulator on scleral chondrocytes and scleral fibroblasts in vitro. Ophthalmic Res 27, 144–152 (1995).
Jobling, A. I., Nguyen, M., Gentle, A. & McBrien, N. A. Isoform-specific changes in scleral transforming growth factor-beta expression and the regulation of collagen synthesis during myopia progression. J Biol Chem 279, 18121–18126, doi:10.1074/jbc.M400381200M400381200 [pii] (2004).
Sternlicht, M. D. & Werb, Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol 17, 463–516, doi:10.1146/annurev.cellbio.17.1.463 (2001).
Chen, M., Qian, Y., Dai, J. & Chu, R. The sonic hedgehog signaling pathway induces myopic development by activating matrix metalloproteinase (MMP)-2 in Guinea pigs. PloS one 9, e96952, doi:10.1371/journal.pone.0096952 (2014).
Rada, J. A. & Brenza, H. L. Increased latent gelatinase activity in the sclera of visually deprived chicks. Invest Ophthalmol Vis Sci 36, 1555–1565 (1995).
Rada, J. A., Perry, C. A., Slover, M. L. & Achen, V. R. Gelatinase A and TIMP-2 expression in the fibrous sclera of myopic and recovering chick eyes. Invest Ophthalmol Vis Sci 40, 3091–3099 (1999).
Qian, L. et al. Pirenzepine Inhibits Myopia in Guinea Pig Model by Regulating the Balance of MMP-2 and TIMP-2 Expression and Increased Tyrosine Hydroxylase Levels. Cell Biochem Biophys 71, 1373–1378, doi:10.1007/s12013-014-0359-9 (2015).
Schippert, R., Brand, C., Schaeffel, F. & Feldkaemper, M. P. Changes in scleral MMP-2, TIMP-2 and TGF beta-2 mRNA expression after imposed myopic and hyperopic defocus in chickens. Exp Eye Res 82, 710–719 (2006).
McBrien, N. A. & Gentle, A. Role of the sclera in the development and pathological complications of myopia. Prog Retin Eye Res 22, 307–338, doi:S1350946202000630 [pii] (2003).
Siegwart, J. T. Jr. & Norton, T. T. Steady state mRNA levels in tree shrew sclera with form-deprivation myopia and during recovery. Invest Ophthalmol Vis Sci 42, 1153–1159 (2001).
Guo, L., Frost, M. R., He, L., Siegwart, J. T. Jr. & Norton, T. T. Gene expression signatures in tree shrew sclera in response to three myopiagenic conditions. Invest Ophthalmol Vis Sci 54, 6806–6819, doi:10.1167/iovs.13-12551 (2013).
Rada, J. A., Achen, V. R., Penugonda, S., Schmidt, R. W. & Mount, B. A. Proteoglycan composition in the human sclera during growth and aging. Invest Ophthalmol Vis Sci 41, 1639–1648 (2000).
Rada, J. A., Fini, M. E. & Hassell, J. R. Regionalized growth patterns of young chicken corneas. Invest Ophthalmol Vis Sci 37, 2060–2067 (1996).
McBrien, N. A., Cornell, L. M. & Gentle, A. Structural and ultrastructural changes to the sclera in a mammalian model of high myopia. Invest Ophthalmol Vis Sci 42, 2179–2187 (2001).
Kee, C.-s, Marzani, D. & Wallman, J. Differences in time course and visual requirements of ocular responses to lenses and diffusers. Invest Ophthalmol Vis Sci 42, 575–583 (2001).
Siegwart, J. T. Jr. & Norton, T. T. The time course of changes in mRNA levels in tree shrew sclera during induced myopia and recovery. Invest Ophthalmol Vis Sci 43, 2067–2075 (2002).
Kee, C. S., Deng, L. & Ho, W. C. Time course of changes in astigmatic components in chicks with myopia Induced by form-deprivation or lens-wear. Invest Ophthalmol Vis Sci 49, 3716- (2008).
Chu, C. H., Zhou, Y., Zheng, Y. & Kee, C. S. Bi-directional corneal accommodation in alert chicks with experimentally-induced astigmatism. Vision Res 98, 26–34, doi:10.1016/j.visres.2014.03.002 (2014).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols 3, 1101–1108 (2008).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408, doi:10.1006/meth.2001.1262S1046-2023(01)91262-9 [pii] (2001).
Thibos, L. N., Wheeler, W. & Horner, D. Power vectors: an application of fourier analysis to the description and statistical analysis of refractive error. Optom & Vision Sci. 74, 367–375 (1997).
Armstrong, R. A. When to use the Bonferroni correction. Ophthalmic Physiol Opt 34, 502–508, doi:10.1111/opo.12131 (2014).
Acknowledgements
This study was supported by Hong Kong government grants RGC-GRF (#5612/09 M, #151011/14 M) and Centre for Myopia Research, School of Optometry, The Hong Kong Polytechnic University. We thank Dr. Jeremy Guggenheim for giving constructive comments and helpful suggestions for an earlier manuscript of this article. We also thank Dr. Wang Jianchao for her technical assistance.
Author information
Authors and Affiliations
Contributions
L.X., S.P.Y., J.S.R. and C.K. designed the study; L.X. wrote the first draft of manuscript; L.X., S.W.S. and C.K. prepared all figures and tables; L.X. and C.K. performed all analyses; C.K. and S.W.S. edited the manuscript; S.P.Y. and J.S.R. supplied material and provided expertise; all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xi, L.Yy., Yip, S.P., Shan, S.W. et al. Region-specific differential corneal and scleral mRNA expressions of MMP2, TIMP2, and TGFB2 in highly myopic-astigmatic chicks. Sci Rep 7, 11423 (2017). https://doi.org/10.1038/s41598-017-08765-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08765-6
This article is cited by
-
TGF-β2 levels in the aqueous humor are elevated in the second eye of high myopia within two weeks after sequential cataract surgery
Scientific Reports (2022)
-
Corneal proteome and differentially expressed corneal proteins in highly myopic chicks using a label-free SWATH-MS quantification approach
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.