Abstract
Cytochrome b 5 (cytb 5) is a membrane protein vital for the regulation of cytochrome P450 (cytP450) metabolism and is capable of electron transfer to many redox partners. Here, using cyt c as a surrogate for cytP450, we report the effect of membrane on the interaction between full-length cytb 5 and cyt c for the first time. As shown through stopped-flow kinetic experiments, electron transfer capable cytb 5 - cyt c complexes were formed in the presence of bicelles and nanodiscs. Experimentally measured NMR parameters were used to map the cytb 5-cyt c binding interface. Our experimental results identify differences in the binding epitope of cytb 5 in the presence and absence of membrane. Notably, in the presence of membrane, cytb 5 only engaged cyt c at its lower and upper clefts while the membrane-free cytb 5 also uses a distal region. Using restraints generated from both cytb 5 and cyt c, a complex structure was generated and a potential electron transfer pathway was identified. These results demonstrate the importance of studying protein-protein complex formation in membrane mimetic systems. Our results also demonstrate the successful preparation of novel peptide-based lipid nanodiscs, which are detergent-free and possesses size flexibility, and their use for NMR structural studies of membrane proteins.
Similar content being viewed by others
Introduction
Microsomal cytochrome b 5 (cytb 5) is a membrane-bound protein that is involved in electron transport to several redox partners including cytochrome P450 (cytP450), several oxygenases and desaturases, and cytochrome b 5 reductase1, 2. Cytochrome b 5 interactions ensure function/activation of cytP450, which plays a vital role in cellular metabolism, including the metabolism of over 70% of drugs in the current market and has been implicated in heart diseases and breast and prostate cancers3,4,5. Because of its membrane-bound native state and the transient nature of cytb 5-cytP450 complex formation, there is relatively little structural insight into this disease relevant complex, especially when compared to the interactions of soluble proteins involved in electron transport.
For decades, it has puzzled investigators how cytb 5 is able to enhance, reduce, or exert no effect on cytP450 metabolism depending on the substrate and the isoform of cytP450 involved6,7,8,9,10. Recent studies with microsomal cytP450 have helped elucidate how cytb 5 causes it apparent contradictory effects6, 11, 12, but few studies have been undertaken in the presence of membrane. There are numerous challenges to obtaining high-resolution structural insights into membrane-bound cytP450 due to its size and the tendency for the full-length protein to aggregate in solution, which make it difficult to use the traditional solution NMR experiments10, 13, 14. Because of these reasons, most reported studies in the literature have focused only on the soluble domain of cytP450, lacking the N-terminal transmembrane domain. On the other hand, previous studies have used cytochrome c (cyt c) as a model for cytP450 in probing the structural interactions between cyt c with other proteins like cytb 5 or CPR15,16,17,18,19. Cyt c has several similarities to cytP450 including: a similar, overlapping binding domain on cytb 5, an overall net positive charge integral for the initial protein-protein complex formation, and membrane has been suggested to promote or enhance activity of both proteins20, 21. Although cyt c is a much smaller protein (104 amino acids compared to 491 amino acids in cytP450 2B4), alignment of these two proteins reveal homologous amino acid sequences including known areas of cytb 5 interaction (Figure S1). Besides being a valid substitute, cyt c also has the advantage of being a well-behaved, NMR-friendly soluble protein. Structural and kinetic details of the electron-transfer complex of cytochrome b 5 – cytochrome c (cyt c) have been the focus of many studies, including NMR spectroscopy22, MD simulations23, and mutagenesis studies16, 18, 24. While cyt c is not a major physiological electron transfer partner of cytb 5, it has been shown to be a productive electron transfer complex25, 26. Previous NMR studies have demonstrated that this complex exists in a 1:1 molar ratio in solution27. In this study, cyt c is used as a model for cyt P450 to examine the role that membrane plays in the formation of an electron transfer cytb 5-cytc complex.
Most studies thus far have focused on the interaction of cyt c with the soluble domain of cytb 5 (without the C-terminal transmembrane (TM) domain) in the absence of membrane15, 16, 28. While several of these studies have shown that the truncated-cytb 5 is capable of slow electron transfer to bacterial soluble cytP450 s29, 30, others have shown truncated-cytb 5 to be incapable of transferring electrons to mammalian membrane-bound cytP45030,31,32. Full-length cytb 5 is necessary to fully understand this protein-protein interaction. Our recent studies have shown that lipid membrane plays an important role in the structural interactions between cytb 5 and cytP45033,34,35,36. Therefore, it is important to investigate the role of membrane on the interactions between cytb 5 and cyt c.
Studying membrane proteins is challenging due to their dynamic nature and difficulties encountered during expression and purification. Additionally, in the case of cytb 5, as with other single transmembrane helix proteins, the TM domain has been shown to increase the tendency to induce protein aggregation in solution. To combat these difficulties, we have used the full-length ~16-kDa rabbit cytb 5 in the presence of membrane mimetics which aid in the monomerization and stabilization of cytb 5 to probe how the inclusion of membrane affects the cytb 5-cyt c complex formation. Solution NMR experiments are used to probe the transient and dynamic protein-protein complex in a near-native membrane environment. NMR is well-suited for this study as it can provide residue specific details of cytb 5-cyt c in the timescale of the complex formation. Two different types of membrane mimetics are used in this study: isotropic bicelles and lipid nanodiscs. Isotropic bicelles have successfully been used to incorporate cytb 5 and perform NMR experiments to study its interaction with cytP45033. These isotropic bicelles have a planar lipid bilayer surrounded with detergents. Using bicelles as a membrane mimetic is preferable to the more traditional micelle as the micelle curvature could distort the structural folding of a membrane protein and the detergent could denature the embedded protein37. A benefit of using isotropic bicelles is that they tumble fast in the NMR time scale to enable the application of solution NMR experiments for structural studies38. The second membrane mimetic used in this study is lipid nanodiscs. Nanodiscs are composed of a lipid bilayer that is typically surrounded by a protein belt. Traditionally, this protein belt is the membrane scaffold protein (MSP)39, 40, but in this study we prepared nanodiscs using a short helical amphipathic peptide scaffold, referred to as 4 F41. This peptide has several advantages for our experiments compared to traditional MSP. Firstly, the nanodiscs can be prepared without the use of detergents, which are known to irreversibly inactivate proteins37. Secondly, a feature of peptide-based nanodisc is that the size is easily controlled through changing the lipid-to-peptide ratio. Lastly, the preparation allows for highly reproducible production of isotropic nanodiscs for solution NMR studies. In peptide-based nanodiscs, cytb 5 is readily monomerized and stable for weeks at room temperature34. As recent studies have identified, cyt c can undergo structural changes in the presence of cardiolipin35, 36, membrane mimetics used in these experiments were cardiolipin-free, such that cyt c would not interact with the membrane environment.
In this study, we investigated the interaction between full-length rabbit cytb 5 and full-length cyt c in different membrane mimetic environments, including lipid-free, isotropic bicelles, and lipid nanodiscs, utilizing solution NMR techniques. Our study provides further evidence that the interaction between these two redox partners is governed by the presence of lipid bilayers. While previous studies of truncated cytb 5 have identified productive binding, our results show an increased, dynamic interaction and changes in the residues most perturbed by complex formation as well as kinetic data demonstrating the formation of a productive complex.
Results
Incorporation of the cytb5-cyt c complex into membrane mimetics
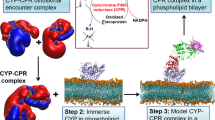
In order to study the interaction between cytb 5 and cyt c, full-length cytb 5 was overexpressed, purified and characterized as reported previously34, 42,43,44. Three different cytb 5 samples were prepared in this study: cytb 5 in buffer (membrane-free cytb 5); cytb 5 in isotropic bicelles; and cytb 5 in lipid nanodiscs. Cytb 5 was added to DMPC/DHPC bicelles. While the isotropic bicelles stabilize the protein, they are not stable enough to be used in size exclusion chromatography. However, the monomerization and incorporation of cytb 5 in lipid nanodiscs can be monitored by Size Exclusion Chromatography (SEC) and Dynamic Light Scattering (DLS). Figure 1(B and C) (green trace) demonstrate that cytb 5 has been incorporated into the lipid nanodiscs in a homologous manner. After purification of the cytb 5 in lipid nanodiscs, cyt c was added to the sample and run through SEC and DLS experiments again (blue trace). These measurements show the complex formation between cytb 5 and cyt c in nanodiscs as indicated by the change in the size of the diameter fit to the DLS data and the elution profile from the SEC (Fig. 1). Cyt c does not interact with the nanodisc, but it only interacts cytb 5 (Figure S2).
Reconstitution of cytb 5 – cyt c in lipid nanodiscs. (A) Schematic of the interaction between cytb 5 (magenta; PDB: 2M33) and cyt c (blue; PDB: 1HRC) in a lipid nanodisc. (B) Dynamic light scattering (DLS) was used to determine the size of the nanodiscs. Radii of empty 4F-DMPC-nanodiscs (black), cytb 5 in nanodiscs (green), and cytb 5 – cyt c in nanodiscs (blue). (C) Size exclusion chromatography (SEC) elution profiles of empty 4F-DMPC-nanodiscs (black) which eluted at 15.1 mL, cytb 5 in nanodiscs (green) which eluted at 12.6 mL, and cytb 5 – cyt c in nanodiscs (blue) which eluted at 12.3 mL.
Formation of productive electron transfer complex between cytb5 and cyt c
Stopped flow kinetic experiments were performed to verify that the cytb 5-cyt c complex was active in all three preparations. The electron transfer rate was monitored between reduced cytb 5 and oxidized cyt c. Ferric cytb 5 was reduced by sodium dithionite and the reduction was monitored by UV-Vis spectrometry (Figure S3). Singular Value Decomposition (SVD) was applied to the difference spectra to deconvolute the principal spectral components (Fig. 2(A,C,E)). For the three samples, two eigenvectors were selected, which variance sum covered 82%, 69%, and 75% of total variance for lipid-free, bicelles, and nanodiscs, respectively. For bicelles, since the cumulative variance covered by the first two eigenvectors was slightly <70%, we used the scree plot to confirm that the first two components (PC1 and PC2) explained most of the variability in the data (Figure S4). The two components and their peak maxima and minima can be identified as either cytb 5 or cyt c based on the difference spectra for each preparation of cytb 5 (lipid-free, bicelles, and nanodiscs). Differences in the peak positions are attributed to the presence of lipids in the solution.
Spectral deconvolution and kinetic modeling reveal membrane environment dependent changes. Time-dependent absorption spectra of the redox couple are depicted for lipid-free (A), bicelles (C), and nanodiscs (E) mimetic membranes. The inset shows the eigenvectors associated with cyt c reduction (PC1) and cytb 5 oxidation (PC1); absorbance maxima are also indicated. The shift of λmax as well as the peak intensity, an indicator of how far along the reaction has progressed, can be attributed to the presence of lipids. Numerical fittings are shown for lipid-free (B), bicelles (D), and nanodiscs (F). (G) Kinetic scheme of electron transfer between cytb 5 and cyt c. In the first reaction, oxidized cytb 5 and reduced cyt c form a complex with association rate k 1 and dissociation rate k 2. Once the first reaction has occurred then the second reaction occurs irreversibly with a rate of k 3 of the electron transfer from oxidized cytb 5 to reduced cyt c giving oxidized cytb 5 and reduced cyt c.
Kinetics traces corresponding to the oxidoreductive reactions (Fig. 2(B,D,F)) were modeled according to the scheme depicted in Fig. 2G. After formation of the complex between ferrous cytb 5 and ferric cyt c (k 1 and k 2), the electron transfer occurs (k 3). We postulate that after the initial electron transfer no further reaction occurs, since it is not possible to distinguish between free and complexed cytochromes based on the absorption spectrum. Two observations were made: 1) the interaction between the redox partners is functional, and 2) the kinetic micro-rates indicate significant differences in the protein binding and electron transfer.
Table 1 reports the micro-rates determined from fitting the time-dependent kinetic traces with the numerical method. For lipid-free systems, the dissociation rate (k 2) was fixed to zero, in order to allow the fitting to converge to a minimum. Bicelles and nanodiscs showed similar association and dissociation rates. Regarding electron transfer, the average micro-rate (k 3) was 6.82 s−1 for lipid-free, 0.92 s−1 for bicelles, and 1.23 s−1 for nanodiscs cytb 5 samples. These results indicate that all three samples form functional complexes with membrane-free cytb 5 having the fastest electron transfer event while nanodiscs reconstituted cytb 5 has slightly faster rate than bicelles reconstituted cytb 5.
NMR experiments probing the interaction between cyt c and 15N-cytb5
Two-dimensional 15N/1H TROSY-HSQC spectra of uniformly-15N-labeled full-length rabbit cytb 5 were recorded to monitor changes induced by the titration of unlabeled cyt c. Both proteins were used in their ferric low spin oxidized forms, and titrations were carried out in a lipid-free solution, DMPC/DHPC bicelles, and DMPC-4F nanodiscs15, 16, 18. N-edited 1H 1D NMR spectra shown in Fig. 3 (top) reveal the changes in the signal intensity of cytb 5 observed from a lipid-free solution, bicelles, or nanodiscs samples as illustrated through spectra acquired over the course of the titration with unlabeled cyt c. In the membrane-free cytb 5 titration (Fig. 3A), there is a significant decrease in signal over the course of the first titration point (1:0, cytb 5: cyt c) to second titration point (1:0.3, cytb 5: cyt c). This decrease in signal could be attributed to a couple of things: (i) as cytb 5 aggregates, signal is lost (or reduced in intensity); (ii) similar to the trend shown in the kinetics data, membrane-free cytb 5 and cyt c are forming a static complex in solution. This larger, static complex could also lead to a decrease in tumbling speed and loss of signal. Membrane bound cytb 5 samples do not exhibit this effect and the NMR signal intensity remains relatively constant throughout the course of the titration with unlabeled cyt c.
2D 1H-15N HSQC-TROSY spectra of 15N-labeled cytb 5 revealing the interaction between cytb 5 and cyt c. The signal intensities over the course of cyt c with the 15N labeled cytb 5 titration experiment are displayed in both 15N- edited 1D spectra (top) and accompanying 2D-TROSY-HSQC spectra (bottom) for the molar ratio of cytb 5: cyt c 1:0 (black) and 1:1 (red). (A) Free cytb 5 has a large loss of signal intensity throughout the course of the experiment, attributed to cytb5 aggregation in solution. (B) bicelle cytb 5 reconstituted in bicelles and (C) nanodisc cytb 5 reconstituted in nanodiscs both maintain signal intensity throughout the course of the experiment. Changes in the signal intensity and line width due to cytb 5-cyt c interactions are shown in Fig. 6 and discussed in the main text. 1H-15N HSQC spectra demonstrating chemical shift perturbations for cytb 5 in buffer (no membrane) (D), bicelles (E), or nanodiscs (F) in the presence of one molar equivalent of cyt c (red). As a reference, the spectrum of cytb 5 is shown without addition of cyt c (black).
The assignment of 1H-15N resonances observed in a HSQC spectrum of cytb 5 has been published previously45 and is shown in Figure S5. The 2D HSQC spectra provide residue specific detail about any changes in cytb 5. Upon titration with cyt c in this experiment, the chemical environment of these backbone amide-NH groups can be affected which can be monitored through changes observed in the 2D HSQC spectra. Two types of information can be gathered from these NMR experiments: chemical shift perturbations (Fig. 3D,E,F) and differential line broadening (Figure S6). It is expected that upon titration of cyt c to cytb 5, complex formation may alter the amide-NH chemical shift values of cytb 5 residues undergoing fast to intermediate time scale exchange between a free and bound forms. Chemical shift perturbations are hallmarks of transient complex formation and may be induced by direct interaction with the binding partner, as well as an overall change in protein conformation. Another possible contribution to CSPs could be due to the ring current effect from the heme groups of these two proteins. However, we expect this effect to be negligible for our system, based on the findings of Shao et al.16. Differential line broadening is indicative of a more stable, tighter complex formation due to the increase in the correlation time of the protein-protein complex. For instance, a rather flexible residue may be rigidified upon binding with cyt c, and its resonance will thus be broadened due to a slower tumbling rate. In addition to correlation time effects, resonance lines may be broadened due to a shift in population levels between different residue conformers46.
After titration of cyt c into cytb 5, the average chemical shift perturbations (CSPs) of cytb 5 measured for lipid-free solution, bicelles, and nanodiscs are 0.016, 0.014, and 0.021 ppm, respectively. These CSPs are not very large which suggests a weak complex formation. The residues with high CSPs are widespread on the surface of the protein, which is a sign of encounter complexes present in the sample that are expected to form with electron transfer proteins47,48,49. While there are unique CSP data for each condition (lipid-free solution, bicelles, or nanodiscs), there is overlap of identified residues and a general region of cytb 5 which seems to be most affected (Fig. 4). All three conditions identify residue E61’s resonance to be highly perturbed and likely to be involved in the interactive interface of the cytb 5–cyt c complex. Most of the residues identified as highly perturbed are located on the upper and lower clefts surrounding the heme group of cytb 5. In the lipid-free sample, Leu99 and Asp104 were identified as highly perturbed. These residues are located in a flexible region of cytb 5 and most likely affected only in this particular sample because cytb 5 is not anchored in a lipid bilayer, unlike in bicelles or nanodiscs.
Cyt c interaction induced chemical shift perturbations of cytb 5. Reported in red are residues with CSPs greater than one standard deviation for cytb 5 in membrane-free solution (A), bicelles (B), and nanodiscs (C). One standard deviation above the mean is represented by the horizontal line and the mean is represented by the dashed line. The significant residues are colored in blue and are mapped onto structures of cytb 5 rotated 180° (PDB: 2M33).
Differential line broadening was seen for all conditions of the cyt c titration into cytb 5. In this protein-protein interaction, differential line broadening is a complementary metric to CSP data to analyze the binding as it covers the timescale of the complex that falls into the time scale of ~10-3 s. The average relative signal intensity observed for the lipid-free solution, bicelles, and nanodiscs samples are 63.9, 79.3, and 65.1% respectively (Fig. 5). These residues are mapped onto cytb 5 structures (PDB: 2 m33) shown in Fig. 6 with lipid-free solution (Fig. 6A), bicelles (Fig. 6B), and nanodiscs (Fig. 6C). In the lipid-free solution (Fig. 5A), there is general line broadening of resonances from cytb 5 in the heme pocket with only one significant residue in the lower cleft identified, His68. The widespread line broadening is concentrated on solvent exposed residues, which can be attributed to the encounter complexes that form in redox partner pairs before the productive binding site is found.
Differential line broadening reveals binding sites on cytb 5. Reported in yellow are residues with depleted signal intensity less than the mean and residues in blue have depleted signal intensity less than one standard deviation below the mean for cytb 5 in no membrane (A), bicelles (B), and nanodiscs (C). The thick horizontal line represents the mean and the dashed horizontal line represents on standard deviation below the mean.
Implicated binding sites on cytb 5 mapped with differential line broadening data. The data from Fig. 5 is mapped onto a solution NMR structure (PDB: 2M33) of cytb 5 for a membrane-free solution (A), bicelles (B), and nanodiscs (C). Residues falling one standard deviation below the mean are colored in blue and residues falling under the mean are colored in yellow.
There is consensus in affected residues from both the chemical shift perturbation data as well as the differential line broadening data. Similar areas are implicated in binding, generally the upper and lower clefts of cytb 5 which surround the heme. Differences in the two types of data can be attributed to that line broadening may be due to direct binding and stabilization of the proteins, whereas CSPs can be induced by global changes. Cytb 5 in the bicelle sample (Fig. 5B) identifies more significant residues than lipid-free cytb 5. These important residues are in the heme pocket, specifically in the lower cleft: Gly56, Gly57, Asn58, Ala59, Thr60, Glu61, Gln62. The lipid nanodisc sample (Fig. 5C) has the similar average relative peak intensity to lipid-free cytb 5 but has eleven important residues: I29, H32, T38, K39, T60, E61, D65, H68, T70, and L84. These residues are mainly clustered on the lower cleft of cytb 5; the residues L51, R52, and R89 in the nanodisc sample could not be assigned.
NMR experiments probing the interaction between 15N-cyt c and cytb5
As with the 15N-labelled cytb5 NMR experiments, two-dimensional 15N-1H TROSY-HSQC spectra of uniformly 15N-labeled equine cyt c were recorded to monitor the cyt c – cytb 5 interaction on the cyt c interface. Full-length cytb 5 in 4F-DMPC nanodiscs was used in this titration experiment based on the previous 15N-labeled cytb 5 NMR experiments. The 1H-15N amide resonances have been reported in the literature50 and the assignment is shown in Figure S2.
CSPs were of similar magnitude to the chemical shifts perturbations of cytb 5, with an average value of 0.027 ppm. For the cyt c CSP calculations, the presented data (Fig. 7B) is calculated from the 1:0.6 (cyt c/cytb 5 molar ratio) rather than the 1:1 ratio as many of the residues in the 1:1 spectra have been broadened beyond detection. The nine residues which have the strongest CSPs (Val3, Val20, Lys22, Leu35, Ala51, Glu62, Leu68, Tyr97, and Lys99) are widespread throughout cyt c.
2D 1H-15N HSQC-TROSY spectra of 15N-labeled cyt c revealing interaction between cyt c and cytb 5. (A) 1H-15N spectra of 15N-labeled cyt c (black) and 15N-labeled cyt c in the presence of one molar equivalent of cytb 5 reconstituted in 4F-DMPC nanodiscs (red). The full 15N-labeled cyt c spectrum with resonance assignment can be found in SI Fig. 5. (B) Chemical shift perturbations of cyt c residues. Reported in red are residues with CSPs greater than one standard deviation. (C) Differential line broadening observed for cyt c residues. The thick horizontal line represents the mean, with the residues colored in yellow, while the dashed horizontal line represents one standard deviation below the mean. The inset in Fig. 8A is the crystal structure of cyt c (PDB: 1HRC) with mapping of experimentally measured differential line broadenings (data reported in 8C) for residues falling under the mean colored in yellow and residues with complete signal intensity depletion in blue.
Substantial differential line broadening was seen for the titration of cytb 5 into cyt c. A complete depletion of signal was observed for 28 residues. The average intensity of the remaining residues was 17%. (Fig. 8C) These affected residues are located around in the unstructured loops surrounding the heme group of cyt c. One explanation for the dramatic decrease in peak intensity is due to a greater increase of size upon complex formation; it should be noted that cyt c alone does not bind to membrane. Viewing the complex from the 10-kDa cyt c, a 15-kDa cytb 5 molecule embedded in a ~120-kDa nanodisc causes a much bigger effect on the tumbling rate than that of cyt c on the cytb 5-ND side.
HADDOCK-generated structures reveal complex between cytb 5 and cyt c. (A) Using the chemical shift perturbations and differential line broadening data from the cytb 5-4F-DMPC-Nanodiscs and cyt c NMR titration experiments, a complex structure was calculated. Cytb 5 is in magenta and cyt c is in blue. (B) Residues implicated in the interface of cytb 5-cyt c from the literature (17) are mapped onto our HADDOCK structure; cytb 5 residues in yellow and cyt c residues in cyan. (C) Residues implicated in the interface of cytb 5-cytP450 from the literature (42) are mapped onto our HADDOCK structure; cytb 5 residues in yellow.
Structural Model of the membrane-bound cytb5-cyt c complex
A structural complex of cytb 5-cyt c was generated using the information driven docking program HADDOCK 2.251, 52 with experimental data obtained in lipid membranes (cyt c and cytb 5 in nanodiscs). NMR based chemical shift perturbation (CSPs) and differential line broadening data were used as proximity restraints to guide the docking process whereby rigid body docking follows the semi-flexible refinement and energy minimization in explicit solvent to allow the free movement of backbone and side chain atoms of the selected amino acids to improve the intermolecular packing at protein interface. Solvent-accessible residues of cyt c identified from CSPs and differential line broadening (Fig. 7) upon complex formation are selected as the active ambiguous restraints including residues Gln16, Cys17, His18, Thr19, Val20, Glu21, Gly29, Thr49, Lys79, Met80, and Ile81. Passive restraints, which are defined as solvent accessible residues around the active restraints, include Lys27, Thr28, Thr78, Phe82, and Ala83. For the cytb 5 side of the complex, active ambiguous restraints (AIRs) were defined from the cytb 5 reconstituted in nanodiscs NMR experiments (Figs 4C and 5C) including: Asp65, His68, and Thr70. The passive residues were defined as Gly67 and Ser69. Some of the residues for cytb 5 that were identified from the NMR experiments were not included in simulations as they were distant from the binding interface of proteins and were observed to penalize the HADDOCK score. Additionally, these residues are involved in significant CSPs because of the encounter complex formation formed prior to productive electron transfer complex. The HADDOCK run was performed as described under “Materials and Methods”.
Careful analysis and comparison of HADDOCK results from various clusters along with experimental data (CSPs and differential line broadening) credits cluster 1 as the most probable model for the complex (Fig. 8). From the 200 lowest energy structures, Cluster 1 is the largest (covers 75%) with better energy and Z score relative to other clusters (Table S1).
In cluster 1, the binding interface of “productive” cytb 5-cyt c complex comprises of polar and charged residues from the helical hairpin of the lower cleft of cytb 5 and the unstructured loops of cyt c. The distance between the two heme groups (Fe-Fe) was estimated as 18.5 Å and is perpendicularly oriented to each other. This is similar to the reported fluorescence quenching studies, where distance between the prosthetic groups was observed to be ~18 Å with the almost perpendicular heme planes17.
Discussion
While it is highly important to determine the high-resolution dynamics-enriched structural interactions between cytP450 and its redox partners such as cytb 5, the large-size and aggregation-prone full-length cytP450 poses numerous challenges to the existing biophysical techniques. To overcome this challenge, cyt c has been used as a substitute for cytP450 as cyt c is a stable and well-behaved protein that has some important features like that of cytP45015, 16, 18, 28. Therefore, we used cyt c in this study, for the first time, to probe the complex formation between cytb 5 and cyt c in the presence of a lipid bilayer membrane. Both bicelles and nanodiscs were used as a model membrane in this study43, 53. Since bicelles are not free of detergents, and detergents are not desirable for studies on a membrane protein or a membrane-bound protein-protein complex, results obtained from bicelles were also compared with that obtained from lipid nanodiscs. A synthetic peptide (denoted as 4 F) capable of solubilizing lipids without the use of any detergents to form nanodiscs54 was used to form a stable lipid bilayer containing nanodisc as characterized by DLS and SEC experiments (Fig. 1). Previous studies reported the physicochemical characterization of 4 F and its use as an apolipoprotein A-I mimetic for atherosclerosis inhibition have been reported54, 55. NMR experiments demonstrate that the nanodiscs are suitable for structural studies by solution NMR spectroscopy. Our NMR experiments also illustrate how cytb 5 is more stable in the presence of lipids and that studies with full-length cytb 5 can be accomplished using membrane mimetics without aggregation related problems16.
Our study reveals that cytb 5 in a lipid-free environment, in isotropic bicelles, and nanodiscs can reduce cyt c. As shown in Fig. 2, stopped-flow experiments revealed that electron transfer from cytb 5 to cyt c occurs faster in the lipid-free cytb 5 sample over the membrane-bound cytb 5 samples. One explanation for this behavior is that the membrane-free cytb 5 can tumble faster than that in bicelles or nanodiscs, and thus able to find cyt c quicker to form a productive complex. This is also revealed by the higher affinity (k 2 ~ 0), indicating the absence of any observable dynamic between the two proteins. The nanodiscs reconstituted cytb 5-cyt c sample appears to support electron transfer function better than the bicelles reconstituted cytb 5-cyt c sample as indicated by a quicker rate of electron transfer in the nanodiscs sample. This is possibility due to the size of the nanodisc versus the size of the bicelle – the nanodiscs are much smaller; another possible reason could be the presence of detergents in bicelles that may cause the observed slow electron transfer rate. Nanodiscs are a highly suitable system to study the interaction between the cytb 5 – cyt c complex as demonstrated by SEC, DLS, and NMR data. The advantageous use of nanodiscs is reflected in the functional complex formation, as well as the high stability of cytb 5, and the well-resolved NMR spectra. In comparison to the bicelle sample, the nanodiscs can both monomerize and stabilize the protein whereas the bicelles can only help to stabilize.
Many of the lipid-free cytb 5 residues implicated in the binding to cyt c are in two clusters: the upper and lower clefts surrounding the active site and the beta sheets at the back of cytb 5. The residues with high chemical shift perturbations in the upper and lower clefts are: Thr38 in the α2 helix, Leu51 in the α3 helix, Glu61 in the α4 helix, and Arg73 in the α5 helix. The other identified residues are: Thr26 in the loop between β2 and β3 strands, Val34 in the β3 strand, Gly46 in the β4 strand, Gly47 in the loop between β4 strand and α3 helix, Ile81 and Gly82 in the β5 strand, and Leu99 and Asp104 located in the flexible linker region between the soluble and transmembrane domains. Differential line broadening data for this sample revealed four residues: His68, one of the axial ligands for the iron in the heme; Asn21, the linker between turn 1 and β2 strand; Tyr35 and Asp36 in β3 strand. While the exact residues implicated are not all the same as reported in the previous study, the implicated residues in this sample are like previous findings16, with residues falling into two different clusters.
In the bicelles-reconstituted cytb 5 sample, the main cluster of residues suggested to be involved in binding to cyt c are found on the upper and lower clefts of the protein. Chemical shift perturbation data indicate the residues in the lower and upper clefts to be: Thr38 in the α2 helix, Val50 and Arg52 in the α3 helix, and Glu61 and Phe63 in the α4 helix. Other residues in more flexible areas of the protein are Leu37 in the linker between β3 strand and α2 helix, Glu54 in turn 3, and Arg89 in turn 4, as well as Asn36 in the β3 strand. The differential line broadening data mostly reveal residues from turn 3 to the α4 helix including Gly56, Gly57, Asp58, Ala59, Thr60, Glu61, and Asn62. The α5 helix residues Ala72, Leu75, and Lys77 were also found to be involved. Three other residues, not located in this area, that were likely to be involved in the interaction are: Ile in the α1 helix, Trp27 in the β3 sheet, and I81 in the β5 sheet. Unlike the Shao et al. 200316 study, these residues mainly fall into one cluster surrounding the active site. The nanodisc reconstituted cytb 5 sample has more overall line broadening effects as the overall 15N-edited proton signal intensity decreased the most in comparison (Fig. 4). Eleven residues are indicated to be highly involved in binding: three in β3 sheet, Ile29, His32, and Tyr35; two in α2 helix, Thr38 and Lys39; three in α4 helix Thr60, Glu61, and Asp65; the axial ligand His68; Thr70 in α5 helix, and Leu84 in a loop. The chemical shift perturbation data from nanodiscs does reveal more residues, with many around the turn3 to α4 helix, similarly to the bicelles reconstituted cytb 5 data: Gly57, Asp58, Glu61, and Phe63. NMR results from both the nanodiscs and bicelles reconstituted cytb 5 also indicate residues from turn 3 to the α4 helix could play a role in the interaction between the protein in the presence of lipids as the membrane-free cytb 5 sample did not identify the residues in this region.
The binding mode of the intermolecular complex (cytb 5-cyt c) was estimated using a data driven docking approach implemented in HADDOCK. The generated structural model (Fig. 8A) presents a binding interface focused on the helical hairpin of the lower cleft of cytb 5 and the unstructured loops cyt c. The predicted electron transfer scheme generated from the cytb 5-cyt c complex structure with HARLEM56 shows a predicted, physiologically-feasible electron transfer path from the heme b of cytb 5 through Lys 27 to Thr 28 to heme c of cyt c. This interface was compared with that calculated in the literature by Deep et al.18. The residues implicated in binding in this study are mapped onto our complex in Fig. 8B in yellow for cytb 5 and cyan for cyt c. These results reveal a difference between our calculated structure and previously reported structures: we propose a front to front interaction as opposed to the reported side to side interaction. Comparing the results from this study on cytb 5-cyt c with that of Ahuja et al.43 on cytb 5-cytP450, we observed that the interacting interface of cytb 5 is similar and contains overlapping residues for interactions with both cytP450 and cyt c. In Fig. 8C, the residues implicated in the cytb 5-cytP450 structure by Ahuja et al.43 are mapped on our model structure, with the cytb 5 residues highlighted in yellow. The cytb 5 residues align very well with our proposed complex structure, illustrating both the importance of utilizing membrane mimetics and the viability of using cyt c as a model. The docking simulations were also performed using the active and passive residues only from the upper cleft of cytb 5. The structures generated post simulation have very low cluster size and high RMSD. Thus, it cannot be considered to be involved in intermolecular interaction. Therefore, NMR detected residues away from binding interface show that enzymes undergo multiple conformational substates.
In summary, our studies illustrate the importance and advantages of studying complex formation of membrane proteins in their native, membrane environment. Both bicelles and nanodiscs stabilize a functional cytb 5 – cyt c complex. To the best of our knowledge, this is the first report on the use of the 4 F peptide based nanodiscs to reconstitute a membrane protein or a protein-protein complex, while previous studies characterized the physiochemical properties and demonstrated the use of 4 F as an apoA-I-mimetic for atherosclerosis inhibition54, 55. With the stability of nanodiscs allowing other characterization methods through SEC and DLS measurements, along with smaller size, nanodiscs make for a better membrane mimetic. The identification of key residues mediating the cytb 5 – cyt c interaction provides important insights into the residues that drive the protein-protein complex formation. The combination of membrane systems and methods utilized in this study are also promising approaches to tackle the structural details of the more physiologically relevant cytb 5 – cytP450 and other membrane-bound electron transfer complexes.
Experimental Procedures
Materials and Reagents
Phosphate buffer components (potassium phosphate monobasic and dibasic) were purchased from Sigma-Aldrich (St. Louis, MO). 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Amino acid sequence of the 4 F peptide used to prepare lipid nanodiscs: DWFKAFYDKV AEKFKEAF. Equine cytochrome c was obtained through a collaborator that was prepared and purified as described previously35. Deuterium oxide and 15N Celtone Base Powder was purchased from Cambridge Isotope Laboratories (Tewksbury, MA). The 5-mm symmetrical D2O-matched Shigemi NMR microtubes were purchased from Shigemi, Inc (Allison Park, PA).
Preparation of cytochrome b5
Full-length uniformly 15N-labeled and unlabeled wild-type rabbit cytb 5 was expressed and purified as described previously10, 43, 57. Briefly, E. coli C41 cells were transformed with a pLW01 plasmid containing the cytb5 gene. The cells were grown in LB medium to an OD of 1 at 600 nm. The culture was diluted 100-fold into 10 mL of 15N-Celtone medium. This culture was grown at 35 °C with shaking at 250 rpm until an OD of 1 at 600 nm was achieved. The cells were pelleted and resuspended in 10 mL of fresh 15N-Celtone medium. The resuspended cell culture was added to the final 1 L of culture minimum medium. Isopropyl β-D-thiogalactopyranoside was added to a final concentration of 10 μM, and incubation was continued for 20 h, at which time the cells were harvested. 15N cytochrome b 5 was purified as described previously58.
Preparation of cytochrome c
Full-length uniformly 15N-labeled cyt c was expressed and purified using a procedure as reported in the literature (34).
Preparation of bicelles
A DMPC/DHPC isotropic bicelle (q = [DMPC]/[DHPC] = 0.25) was prepared by mixing the appropriate amount of DMPC and DHPC in chloroform. The mixture was vortexed and dried under nitrogen to make a thin film, which was further dried under vacuum overnight at room temperature. The film was then hydrated in 10 mM potassium phosphate buffer, pH 7.4 to a concentration of 5% w/v.
Preparation of nanodiscs
DMPC powder was suspended into buffer A (10 mM potassium phosphate, pH 7.4) to make a stock solution at 20 mg/mL. The 4 F peptide was dissolved in buffer A to make a stock solution at 10 mg/mL. The DMPC stock solution was vortexed and sonicated three times for 30 s each to create a suspension, and vortexed thoroughly immediately before use. The stock solution was mixed together at a peptide:lipid ratio of 1:1.5% w/w and incubated at 37 °C o/n with slow agitation. The nanodiscs were purified by size exclusion chromatography (SEC). A Superdex 200 Increase 300/10 GL column was operated on an AKTA purifier (GE Healthcare, Freiburg, Germany).
NMR experiments
NMR experiments were performed at 298 K on a Bruker Avance II 600 MHz spectrometer equipped with a cryoprobe. 2D 15N/1H TROSY HSQC spectra were recorded from 0.1 mM 15N-cytb 5 in buffer A, bicelles containing 15N-cytb 5, or nanodiscs containing 15N-cytb 5. All buffer and nanodisc samples were in buffer A and contained 15N-cytb 5 at a concentration around 0.1 mM. Cytb 5 was incubated with a 5% v/v solution of DMPC/DHPC. Cytb 5 was incubated with 4F-DMPC-Nanodiscs at a ratio of 1:1.
All NMR spectra were recorded with 64 scans and 256 t1 increments. Data was processed using TopSpin 2.0 (Bruker) and analyzed with Sparky (Goddard). The previously reported cytb 5 backbone chemical shift assignments were used in this study45. The weighted amide chemical shift perturbation (Δδavg) was calculated using the following equation:
where Δδ N and Δδ H are the changes in the chemical shifts of amide nitrogen-15 and amide-proton respectively, while F1SW and F2SW represent the spectral width in the first and second dimensions respectively; chemical shift values are given in ppm59, 60.
Stopped Flow Kinetics
All experiments were performed at 25 °C under anaerobic conditions using a Hi-Tech SF61DX2 stopped-flow spectrophotometer (Bradford-on-Avon, UK) housed in an anaerobic Belle Technology glove box (Weymouth, UK). The buffer was purged with nitrogen gas for 30 minutes for deoxygenation prior to being transferred to the glove box. All protein solutions were incubated overnight at 4 °C in the glove box to eliminate oxygen. For measuring the kinetics of cyt c reduction by cytb 5, cytb 5 was pre-reduced anaerobically as described below and then loaded in syringe 3 of the stopped flow, and syringe 2 was loaded with ferric cyt c. Spectra were recorded in the photodiode array mode between the wavelength of 300 and 750 nm after mixing equal volumes of the 12 μM cytb 5 and the 13.2 μM cyt c containing solutions resulting in final concentrations of 6 μM cytb 5 and 6.6 μM cyt c. Each set of samples (free non-membrane bound cytb 5, bicelle cytb 5, nanodisc cytb 5) was measured over different amounts of time between 0.8 and 20 s.
Data analysis and kinetic modeling
Spectra were collected and Singular Value Decomposition (SVD) was applied in the differential spectrum using an algorithm written in Matlab (The Mathworks Inc., v.R2016b)61. Before SVD, Savitzky-Golay smoothing and peak-to-area normalization were performed on all the recorded spectra to increase signal-to-noise ratio. SVD analysis was then performed using the built-in Matlab function, obtaining the corresponding kinetic and spectral eigenvectors. The number of principal components was chosen by visualization of the scree plot (Figure SI6, Supplemental Information), as well as considering the variance covered by the selected eigenvectors. The fraction of variance was computed as following:
where f q represents the fraction of the expression level contributed by the q th eigenvector, \({\sigma }_{q}^{2}\) is the variance of the q th eigenvector, and \({\sigma }_{t}^{2}\) the total variance. In all the performed experiments, two eigenvectors were selected (PC1 and PC2), covering 70–90% of the total variance.
Kinetic Modeling and Simulations
For the three systems (lipid-free, bicelles and nanodiscs), based on the evolution over time and peak maxima, PC1 was assigned to cyt c, and PC2 to cytb 5. The final concentration of ferrous cytochrome c was computed by measuring the absorbance at ~550 nm and using ε = 21.2 mM−1 cm−1. Cytb 5 was considered to be completely oxidized at the end of the experiment. The kinetics model that describe the oxidoreductive reaction is depicted in Fig. 2. The differential equations to model the electron transfer kinetics were parameterized using the NonlinearModelFit function with 1/Y weighting in Mathematica 11.0 (Wolfram Research, Champagne, IL). When fitting parameters, the ParametricNDSolveValue function was used for numerical solutions of the differential equations with MaxSteps → 100,000 and PrecisionGoal → ∞.
Titration of cytb5 by Dithionite under Anaerobic Conditions
Cytb 5 (12 μM) was titrated with a standardized sodium dithionite solution under anaerobic conditions while monitoring the UV-visible spectra with a Cary 4000 spectrometer between 300 and 700 nm. The stock solution of cytb 5 was incubated overnight at 4 °C in an anaerobic Belle Technology glove box (Hi-Tech, Salisbury, UK) to remove oxygen. The titrant (sodium dithionite solution) was prepared in the glove box in oxygen free buffer, and its concentration was calculated using an extinction coefficient of 8.04 mm−1 cm−1 at 315 nm. The titration was carried out in a tonometer, a homemade anaerobic titration apparatus.
Calculation of a membrane-bound cytb5-cyt c complex using NMR data
The HADDOCK (High Ambiguity Driven protein-protein DOCKing) docking software51, 52 was used to calculate the structures of the cytb 5-cyt c complex based on experimental NMR restraints. The HADDOCK algorithm includes three consecutive steps: (1) rigid body docking in which the two molecules are rotated and translated randomly in turn to minimize intermolecular energy; (2) simulated annealing of structures in which annealing in torsion angle space is performed to refine the orientation of the proteins and the side chains and/or backbones of the interface residues; and (3) solvent refinement in which the structures are further refined in an 8.0 Ǻ shell of TIP3P water molecules. The starting structures for this docking were the solution NMR structure of rabbit cytb 5 (PDB: 2M33) and the crystal structure of horse cyt c (PDB: 1 HRC). Ligand topology and parameter files were generated using PRODRG2 server62. Defined ambiguous restraints were generated from NMR experiments performed in this paper, both active and passive residues with solvent accessibility from both proteins. 1000 structures were generated in the rigid body docking step, followed by simulated annealing of the 200 lowest energy structures from the last step, and 200 structures were selected for solvent refinement. The resulting 200 final structures were analyzed and grouped into clusters based on the backbone root mean square deviation values. Molecular structures of the complexes were viewed and graphed using PyMOL.
Data availability
Most of the data generated or analyzed during this study are included in this article (and in the Supporting Information files) and other datasets are available from the corresponding author on reasonable request.
References
Vergeres, G. & Waskell, L. Cytochrome b5, its functions, structure and membrane topology. Biochimie 77, 604–620 (1995).
Schenkman, J. B. & Jansson, I. The many roles of cytochrome b5. Pharmacol. Ther. 97, 139–152 (2003).
Chaudhary, K. R., Batchu, S. N. & Seubert, J. M. Cytochrome P450 enzymes and the heart. IUBMB Life 61, 954–960 (2009).
O’Donnell, A. et al. Hormonal impact of the 17alpha-hydroxylase/C(17,20)-lyase inhibitor abiraterone acetate (CB7630) in patients with prostate cancer. Brit. J. Cancer 90, 2317–2325 (2004).
Orlando, L. et al. Molecularly targeted endocrine therapies for breast cancer. Cancer Treat. Rev. 36(Suppl 3), S67–71 (2010).
Im, S. C. & Waskell, L. The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch. Biochem. Biophys. 507, 144–153 (2011).
Finn, R. D. et al. Defining the in vivo role for cytochrome b5 in cytochrome P450 function through the conditional hepatic deletion of microsomal cytochrome b5. J. Biol. Chem. 283, 31385–31393 (2008).
Zhang, H., Myshkin, E. & Waskell, L. Role of cytochrome b5 in catalysis by cytochrome P450 2B4. Biochem. Biophys. Res. Commun. 338, 499–506 (2005).
McLaughlin, L. A., Ronseaux, S., Finn, R. D., Henderson, C. J. & Roland Wolf, C. Deletion of microsomal cytochrome b5 profoundly affects hepatic and extrahepatic drug metabolism. Mol. Pharmacol. 78, 269–278 (2010).
Durr, U. H., Waskell, L. & Ramamoorthy, A. The cytochromes P450 and b5 and their reductases–promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim. Biophys. Acta 1768, 3235–3259 (2007).
Pearl, N. M. et al. Protonation of the Hydroperoxo Intermediate of Cytochrome P450 2B4 Is Slower in the Presence of Cytochrome P450 Reductase Than in the Presence of Cytochrome b5. Biochemistry 55, 6558–6567 (2016).
Zhang, H., Im, S. C. & Waskell, L. Cytochrome b 5 increases the rate of product formation by cytochrome P450 2B4 and competes with cytochrome P450 reductase for a binding site on cytochrome P450 2B4. J. Biol. Chem. 282, 29766–29776 (2007).
Williams, P. A., Cosme, J., Sridhar, V., Johnson, E. F. & McRee, D. E. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell 5, 121–131 (2000).
Durr, U. H. N., Yamamoto, K., Im, S. C., Waskell, L. & Ramamoorthy, A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b(5). J. Am. Chem. Soc. 129, 6670 (2007).
Ren, Y. et al. Mapping the electron transfer interface between cytochrome b5 and cytochrome c. Biochemistry 43, 3527–3536 (2004).
Shao, W., Im, S. C., Zuiderweg, E. R. & Waskell, L. Mapping the binding interface of the cytochrome b5-cytochrome c complex by nuclear magnetic resonance. Biochemistry 42, 14774–14784 (2003).
Mauk, A. G., Mauk, M. R., Moore, G. R. & Northrup, S. H. Experimental and theoretical analysis of the interaction between cytochrome c and cytochrome b5. J. Bioenerg. Biomembr. 27, 311–330 (1995).
Deep, S., Im, S. C., Zuiderweg, E. R. & Waskell, L. Characterization and calculation of a cytochrome c-cytochrome b5 complex using NMR data. Biochemistry 44, 10654–10668 (2005).
Huang, R., Zhang, M., Rwere, F., Waskell, L. & Ramamoorthy, A. Kinetic and structural characterization of the interaction between the FMN binding domain of cytochrome P450 reductase and cytochrome c. J. Biol. Chem. 290, 4843–4855 (2015).
Bushnell, G. W., Louie, G. V. & Brayer, G. D. High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol. 214, 585–595 (1990).
Vik, S. B., Georgevich, G. & Capaldi, R. A. Diphosphatidylglycerol is required for optimal activity of beef heart cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 78, 1456–1460 (1981).
Hom, K. et al. NMR studies of the association of cytochrome b5 with cytochrome c. Biochemistry 39, 14025–14039 (2000).
Wendoloski, J. J., Matthew, J. B., Weber, P. C. & Salemme, F. R. Molecular dynamics of a cytochrome c-cytochrome b5 electron transfer complex. Science 238, 794–797 (1987).
Qian, C. et al. Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c. Protein Sci. 10, 2451–2459 (2001).
Strittmatter, P. Reversible direct hydrogen transfer from reduced pyridine nucleotides to cytochrome b5 reductase. J. Biol. Chem. 239, 3043–3050 (1964).
Salemme, F. R. An hypothetical structure for an intermolecular electron transfer complex of cytochromes c and b5. J. Mol. Biol. 102, 563–568 (1976).
Eley, C. G. & Moore, G. R. 1H-n.m.r. investigation of the interaction between cytochrome c and cytochrome b5. Biochem. J. 215, 11–21 (1983).
Volkov, A. N., Ferrari, D., Worrall, J. A., Bonvin, A. M. & Ubbink, M. The orientations of cytochrome c in the highly dynamic complex with cytochrome b5 visualized by NMR and docking using HADDOCK. Protein Sci. 14, 799–811 (2005).
Stayton, P. S., Fisher, M. T. & Sligar, S. G. Determination of cytochrome b5 association reactions. Characterization of metmyoglobin and cytochrome P-450cam binding to genetically engineered cytochromeb5. J. Biol. Chem. 263, 13544–13548 (1988).
Chiang, J. Y. Interaction of purified microsomal cytochrome P-450 with cytochrome b5. Arch. Biochem. Biophys. 211, 662–673 (1981).
Bendzko, P., Usanov, S. A., Pfeil, W. & Ruckpaul, K. Role of the hydrophobic tail of cytochrome b5 in the interaction with cytochrome P-450 LM2. Acta Biol. Med. Ger. 41, K1–K8 (1982).
Canova-Davis, E. & Waskell, L. The identification of the heat-stable microsomal protein required for methoxyflurane metabolism as cytochrome b5. J. Biol. Chem. 259, 2541–2546 (1984).
Zhang, M., Huang, R., Im, S. C., Waskell, L. & Ramamoorthy, A. Effects of membrane mimetics on cytochrome P450-cytochrome b5 interactions characterized by NMR spectroscopy. J. Biol. Chem. 290, 12705–12718 (2015).
Zhang, M. et al. Reconstitution of the cytb5-cytP450 complex in nanodiscs for structural studies using NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 55, 4497–4499 (2016).
Kobayashi, H., Nagao, S. & Hirota, S. Characterization of the cytochrome c membrane-binding site using cardiolipin-containing bicelles with NMR. Angew. Chem. Int. Ed. Engl. 55, 14019–14022 (2016).
O’Brien, E. S., Nucci, N. V., Fuglestad, B., Tommos, C. & Wand, A. J. Defining the apoptotic trigger: the interaction of cytochrome c and cardiolipin. J. Biol. Chem. 290, 30879–30887 (2015).
Seddon, A. M., Curnow, P. & Booth, P. J. Membrane proteins, lipids and detergents: not just a soap opera. BBA-Biomembranes 1666, 105–117 (2004).
Durr, U. H., Gildenberg, M. & Ramamoorthy, A. The magic of bicelles lights up membrane protein structure. Chem. Rev. 112, 6054–6074 (2012).
Denisov, I. G. & Sligar, S. G. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 23, 481–486 (2016).
Schuler, M. A., Denisov, I. G. & Sligar, S. G. Nanodiscs as a new tool to examine lipid-protein interactions. Methods Mol. Biol. 945, 415–433 (2013).
Kariyazono, H. et al. Formation of stable nanodiscs by bihelical apolipoprotein A-I mimetic peptide. J. Pept. Sci. 22, 116–122 (2016).
Zhang, M. et al. Insights into the role of substrates on the interaction between cytochrome b5 and cytochrome P450 2B4 by NMR. Sci. Rep. 5, 8392 (2016).
Ahuja, S. et al. A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J. Biol. Chem. 288, 22080–22095 (2013).
Pandey, M. K. et al. Cytochrome-P450-cytochrome-b5 interaction in a membrane environment changes 15N chemical shift anisotropy tensors. J. Phys. Chem. B 117, 13851–13860 (2013).
Vivekanandan, S., Ahuja, S., Im, S. C., Waskell, L. & Ramamoorthy, A. (1)H, (1)(3)C and (1)(5)N resonance assignments for the full-length mammalian cytochrome b(5) in a membrane environment. Biomol. NMR Assign. 8, 409–413 (2014).
Fejzo, J. et al. Dynamic NMR studies of ligand-receptor interactions: design and analysis of a rapidly exchanging complex of FKBP-12/FK506 with a 24 kDa calcineurin fragment. Protein Sci. 5, 1917–1921 (1996).
Bashir, Q., Scanu, S. & Ubbink, M. Dynamics in electron transfer protein complexes. Febs J. 278, 1391–1400 (2011).
Schilder, J., Lohr, F., Schwalbe, H. & Ubbink, M. The cytochrome c peroxidase and cytochrome c encounter complex: The other side of the story. Febs Lett. 588, 1873–1878 (2014).
Guan, J. Y. et al. An ensemble of rapidly interconverting orientations in electrostatic protein-peptide complexes characterized by NMR spectroscopy. Chembiochem. 15, 556–566 (2014).
Liu, W., Rumbley, J., Englander, S. W. & Wand, A. J. Backbone and side-chain heteronuclear resonance assignments and hyperfine NMR shifts in horse cytochrome c. Protein Sci. 12, 2104–8 (2003).
van Zundert, G. C. P. et al. The HADDOCK2.2 webserver: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2015).
Wassenaar et al. WeNMR: Structural Biology on the Grid. J. Grid. Comp. 10, 743–767 (2012).
Soong, R. et al. Proton-evolved local-field solid-state NMR studies of cytochrome b5 embedded in bicelles, revealing both structural and dynamical information. J. Am. Chem. Soc. 132, 5779–5788 (2010).
Datta, G. et al. Effects of increasing hydrophobicity on the physical-chemical and biological properties of a class A amphipathic helical peptide. J. Lipid Res. 42, 1096–1104 (2001).
Datta, G. et al. Bioenergetic programming of macrophages by the apolipoprotein A-I mimetic peptide 4F. Biochem. J. 467, 517–527 (2015).
Kurnikov, I. V. Department of Chemistry, University of Pittsburgh; Pittsburg, PA: 2000. HARLEM molecular modeling package.
Bridges, A. et al. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J. Biol. Chem. 273, 17036–17049 (1998).
Mulrooney, S. B. & Waskell, L. High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b(5). Protein Expres. Purif. 19, 173–178 (2000).
Williamson, R. A., Carr, M. D., Frenkiel, T. A., Feeney, J. & Freedman, R. B. Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry 36, 13882–9 (1997).
Williamson, M. P. Using chemical shift perturbation to characterize ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 (2013).
Maeder, M. & Neuhold, Y. M. Practical Data Analysis in Chemistry, Elsevier (2007).
Schüttelkopf, A. W. & van Aalten, D. M. F. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. Biol. Crystallogr. 60, 1355–1363 (2004).
Acknowledgements
This research was supported by funds from NIH (GM084018 to A.R.). We thank Mr. Kobayashi and Professor Hirota for help with the production of cytochrome c. The FP7 WeNMR (project# 261572) and H2020 West-Life (project# 675858) European e-Infrastructure projects are acknowledged for the use of their web portals, which make use of the EGI infrastructure and DIRAC4EGI service with the dedicated support of CESNET-MetaCloud, INFN-PADOVA, NCG-INGRID-PT, RAL-LCG2, TW-NCHC, IFCA-LCG2, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, South Africa, Malaysia, Taiwan and the US Open Science Grid.
Author information
Authors and Affiliations
Contributions
The study was planned by K.A.G. and A.R. K.A.G., E.P., and M.Z. performed NMR experiments, and the results were interpreted by K.A.G. and A.R. S.I. carried out stopped-flow experiments and the results were characterized by C.B., K.A.G., L.W. and A.R. M.M. and K.A.G. obtained docked structure of the complex using HADDOCK. G.M.A. provided the 4F peptide and S.N. provided cyt c. All reported results were reviewed by K.A.G., E.P., C.B., M.M., L.W. and A.R. K.A.G. and A.R. wrote the paper, and all authors reviewed and approved the final manuscript. A.R. directed the project.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gentry, K.A., Prade, E., Barnaba, C. et al. Kinetic and Structural Characterization of the Effects of Membrane on the Complex of Cytochrome b 5 and Cytochrome c . Sci Rep 7, 7793 (2017). https://doi.org/10.1038/s41598-017-08130-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08130-7
This article is cited by
-
Silybin regulates P450s activity by attenuating endoplasmic reticulum stress in mouse nonalcoholic fatty liver disease
Acta Pharmacologica Sinica (2023)
-
Unraveling cardiolipin-induced conformational change of cytochrome c through H/D exchange mass spectrometry and quartz crystal microbalance
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.