Abstract
Cis-1,2-dichloroethylene (cDCE), which is a common hazardous compound, often accumulates during incomplete reductive dechlorination of higher chlorinated ethenes (CEs) at contaminated sites. Simple monoaromatics, such as toluene and phenol, have been proven to induce biotransformation of cDCE in microbial communities incapable of cDCE degradation in the absence of other carbon sources. The goal of this microcosm-based laboratory study was to discover non-toxic natural monoaromatic secondary plant metabolites (SPMEs) that could enhance cDCE degradation in a similar manner to toluene and phenol. Eight SPMEs were selected on the basis of their monoaromatic molecular structure and widespread occurrence in nature. The suitability of the SPMEs chosen to support bacterial growth and to promote cDCE degradation was evaluated in aerobic microbial cultures enriched from cDCE-contaminated soil in the presence of each SPME tested and cDCE. Significant cDCE depletions were achieved in cultures enriched on acetophenone, phenethyl alcohol, p-hydroxybenzoic acid and trans-cinnamic acid. 16S rRNA gene sequence analysis of each microbial community revealed ubiquitous enrichment of bacteria affiliated with the genera Cupriavidus, Rhodococcus, Burkholderia, Acinetobacter and Pseudomonas. Our results provide further confirmation of the previously stated secondary compound hypothesis that plant metabolites released into the rhizosphere can trigger biodegradation of environmental pollutants, including cDCE.
Similar content being viewed by others
Introduction
Chlorinated ethenes (CEs) are common environmental pollutants due to their intense involvement in anthropogenic activities and improper waste disposal1. Increasing concerns regarding CE toxicity and their adverse effects on human health (https://www.epa.gov/toxics-release-inventory-tri-program) have led to much stricter environmental regulations2 which have limited the use of CEs and tightened the rules on their cleanup in the environment. In this context, microbial bioremediation represents a more promising and cost-effective strategy as compared to traditional physical-chemical methods3. In the subsurface, CEs with higher chlorination levels easily undergo anaerobic biodegradation to less chlorinated CEs but rarely to non-chlorinated compounds3, 4. Despite the possible involvement of many different types of bacteria in this process5, 6, only members of the genus Dehalococcoides have been described as capable of complete anaerobic dechlorination to ethene7,8,9. Consequently, incomplete anaerobic degradation of CEs occurs in many sites, resulting in the accumulation of biodegradation intermediates, such as cis-1,2-dichloroethylene (cDCE) and vinyl chloride (VC), which is more toxic and carcinogenic10, 11 than higher CEs.
Microbial degradation of lower CEs can be achieved under aerobic conditions. The degradation is initiated by di- or monooxygenase enzymes12. Several studies have demonstrated that many microbial species are able to use VC as a sole carbon source and to achieve its complete mineralization5, 12. However, aerobic growth has been far less commonly observed on cDCE, which has only been thoroughly described for Polaromonas sp. strain JS66613,14,15. Little additional evidence of direct aerobic cDCE degradation has been reported for isolates affiliated with Acinetobacter sp., Achromobacter sp., Bacillus sp., Pseudomonas sp.16 or with mixed microbial cultures retrieved from a stream bed sediment17 and from CE-contaminated groundwater18. On the other hand, aerobic microbial degradation of cDCE can occur via aerobic cometabolism (AC), a process driven by oxygenase enzymes which have relaxed substrate specificity19, 20. In this case, cDCE is fortuitously oxidized in the presence of a growth-supporting substrate that stimulates enzymatic activity of the oxygenase21. Growth-supporting substrates, which have been described in relation to cDCE AC processes, are ethene22, methane23, 24, propane25, butane26, propene27, ammonia28 and some less chlorinated hydrocarbons such as VC29, 30. In addition, monoaromatic compounds, such as phenol, toluene and benzene, can efficiently enhance aerobic cometabolic mineralization of cDCE31, 32. Some difficulties with respect to AC implementation arise from the introduction of some of the above-mentioned substrates into the environment mostly due to their toxicity33.
Secondary plant metabolites (SPMEs), which enter the soil via different routes, including root exudation and litter decomposition, constitute an important source of nutrients for soil bacteria and play naturally established roles in plant-microbe communication and chemotactic regulation of soil microbial colonization34, 35. Extensive research has demonstrated that plant-derived compounds also constitute a precious source of promising alternative substrates in order to trigger biodegradative pathways for xenobiotics, including polychlorinated biphenyls (PCBs) and polyaromatic hydrocarbons (PAHs)36,37,38,39. This phenomenon is referred to as the “secondary compound hypothesis”40. In the present study, we sought monoaromatic SPMEs which could be used to assist the degradation of cDCE, possibly through aerobic cometabolism, in a manner similar to that described in relation to phenol and toluene. To our knowledge, this proof-of-concept study is the first to explore the possibility of cDCE degradation induced by monoaromatic SPMEs, which were used to support microbial growth through a laboratory-enrichment cultivation procedure.
Results
Degradation of cDCE by toluene- and phenol-exposed microbial community
At the initial stage of this study, the microbial community in the contaminated soil was shown not to be capable of cDCE degradation when cDCE was provided as the sole carbon source during the first 10 days of cultivation (Fig. 1). Measurement of cDCE concentrations in the culture headspaces performed up to the third month of cultivation revealed the persistence of cDCE in the liquid cultures (with approximately an 11% of variability among all measurements).
Average depletions of cDCE in the water phase of the soil microbial cultures in the presence of only cDCE (filled squares) after the addition of toluene (closed circles) or phenol (closed rectangles). The dashed line indicates the control sodium azide-inhibited phenol-grown cultures. The error bars indicate the standard deviation among three biological replicates prepared per each condition.
On the other hand, when exposed to toluene and phenol, microbial suspensions extracted from soil successfully degraded cDCE over a period of 10 days (Fig. 1). Higher cDCE depletion levels were achieved in the presence of toluene, with a 82.78% ± 3.73 percentage points (pp) removal of the initial amount despite an initial lag phase of approximately three days (Fig. 1); in the presence of phenol, depletion was 57.89% ± 3.43 pp. Much lower cDCE loss (about 11.8% pp) was observed over the same period of time in phenol-supplied control cultures inhibited with sodium azide (Fig. 1), suggesting that cDCE depletion in toluene- and phenol-supplied cultures was attributable to microbial activity.
Degradation of cDCE by SPME-enriched microbial cultures
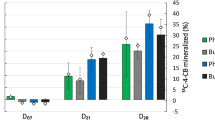
The biodegradation of cDCE was first evaluated for eight microbial cultures where the primary carbon sources were represented by different monoaromatic SPMEs. After approximately a month of cultivation, cDCE depletion over a period of 10 days was monitored for all cultures. The highest cDCE depletion levels were attained in the presence of acetophenone (10.14%), phenethyl alcohol (8%), p-hydroxybenzoic acid (9.34%) and trans-cinnamic acid (7.11%). These four cultures were subjected to further enrichment through serial sub-culturing passages, during which substrate depletion, microbial growth and cDCE degradation were evaluated. At the final enrichment stage, all of the SPMEs tested were totally consumed within the first 18 to 25 hours of incubation, despite a lag phase of approximately 10 hours. The measurements of microbial concentrations at the end of the final sub-culturing stage indicated that microbial growth was more effectively supported by trans-cinnamic acid and phenethyl alcohol (Fig. 2). At the same time, significant cDCE depletion was achieved in the presence of acetophenone (88.59% ± 9.08 pp of the initial amount of cDCE), phenethyl alcohol (79.18% ± 13.53 pp), p-hydroxybenzoic acid (69.05% ± 6.30 pp) and trans-cinnamic acid (46.82% ± 4.10 pp) (Fig. 2). When the concentration of cDCE in the liquid phase was plotted over time (Fig. 3), significant cDCE degradation was found to occur after the first day of cultivation, once the SPME substrates were already consumed, and to slow down considerably after seven days. Negligible cDCE loss (below 5% pp) was observed in sodium azide-inhibited cultures prepared with p-hydroxybenzoic acid as carbon source. Transformation yields of cDCE per SPME, expressed as µmol(cDCE)∙mmol(SPME) −1, were: 16.38 ± 1.55 for acetophenone, 16.32 ± 1.91 for p-hydroxybenzoic acid, 10.53 ± 1.31 for phenethyl alcohol and 9.80 ± 0.76 for trans-cinnamic acid.
Microbial concentration, estimated as microbial protein content, and depletion of cDCE (% out of the initial amount) after 10 incubation days for the cultures enriched on 50 mg∙L−1 acetophenone (AP), trans-cinnamic acid (CA), p-hydroxybenzoic acid (HB) and phenethyl alcohol (PH). The error bars indicate the standard deviation calculated on the basis of three biological replicates.
Average depletion of cDCE over time in the aqueous phase for the cultures enriched on acetophenone (AP); trans-cinnamic acid (CA); p-hydroxybenzoic acid (HB); and phenethyl alcohol (PH) (three biological replicates were used per each condition). Dashed line indicates the over-time cDCE concentration in sodium azide-inhibited cultures grown on acetophenone and p-hydroxybenzoic acid (two biological replicates were used per control condition). The error bars indicate the standard deviation among biological replicates.
Structural diversity of SPME-enriched microbial populations
The highest levels of richness, as assessed by the number of sequence variants observed, or phylogenetic diversity, as assessed by the Shannon, Simpson and reciprocal Simpson diversity indexes, were clearly observed for the initial soil microbial community (Fig. 4). Remarkable differences were observed between the SPME-enriched microbial populations, where the highest diversity indexes were found in acetophenone-enriched cultures, followed by p-hydroxybenzoic acid and phenethyl alcohol-amended cultures, while trans-cinnamic-enriched populations were characterized by the lowest diversity indexes (Fig. 4). In order to highlight the structural differences between the SPME-enriched microbial populations, a non-metric multidimensional scaling (NMDS) analysis was carried out. This showed that the cultures enriched on p-hydroxybenzoic acid and trans-cinnamic acid formed distinct clusters, while cultures enriched on acetophenone and phenethyl alcohol were more related (Fig. 5).
Number of observed sequence variants and diversity indexes of the microbial communities from the original soil or microbial populations enriched on each SPME (acetophenone AP; trans-cinnamic acid CA; p-hydroxybenzoic acid HB; phenethyl alcohol PH; each dot represents one biological replica, mean values are shown as red bars). Abbreviations: Observed – number of observed sequence variants, Shannon – Shannon-Wiener index, Simpson – Simpson’s index of diversity (1–D), InvSimpson – Inverse Simpson’s index (1/D).
Phylogenetic analysis of the most abundant taxa
The relative abundance profiles of the thirty most abundant sequence variants in the treatments were evaluated. A ubiquitous enrichment of taxa within Proteobacteria and Actinobacteria occurred in the enrichment cultures (Table 1). The most commonly detected sequence variants amongst Betaproteobacteria clustered with Burkholderiaceae, with Cupriavidus being the most abundant taxon in cultures enriched on trans-cinnamic acid and phenethyl alcohol and quite abundant in cultures enriched on the other SMPEs. Members of the families Comamonadaceae (Simplicispira, unclassified Comamonadaceae) and Oxalobacteraceae (Massilia) were also profusely detected. Gammaproteobacteria were mainly represented by Xanthomonadaceae (genera Dokdonella, Pseudoxanthomonas, Rhodanobacter) and the genera Pseudomonas, Acinetobacter and Legionella. Alphaproteobacteria were more abundant in acetophenone-enriched cultures, which particularly included Devosia and Sphingopyxis. Acetophenone-enriched populations were characterized by a number of additional phylogenetic classes, such as Verrucomicrobiae, Alphaproteobacteria, Flavobacteria, Sphingobacteria, Actinobacteria (Rhodococcus) and Planctomycetia, as well as an enrichment of unclassified bacteria. A significant enrichment of Deltaproteobacteria (mainly Bdellovibrio) was observed in the presence of p-hydroxybenzoic acid and also in the other enriched microbial populations (Table 1
). Interestingly, none of the enriched taxa were detected with frequencies of more than 0.001% in the original soil.
Discussion
This proof-of-concept study demonstrates the possibility of cDCE aerobic degradation promoted by the presence of SPMEs used as carbon and energy sources during the enrichment of specific soil microbial populations. The initial microbial community extracted from CE-contaminated soil was shown to be incapable of cDCE biodegradation when exposed to cDCE without the addition of any other carbon and energy source.
Successful cDCE degradation was first achieved upon exposure of the soil microbial community to toluene or phenol (Fig. 1). A number of studies have demonstrated that cDCE biodegradation is attainable by using aerobic cometabolism with aromatic hydrocarbons such as toluene and phenol41, 42.
Promising bioremediation potential has often been ascribed to microorganisms associated with plant roots43,44,45,46. Some studies have even shown that, in polluted environments, the composition of root exudates can drive a selection of rhizosphere microbial populations with a greater capacity to degrade pollutants47, 48. Given the structural analogy between toluene, phenol and some monoaromatic SPMEs, we hypothesized that these SPMEs might promote the degradation of cDCE in a manner similar to that for toluene and phenol. Microbial growth on some plant metabolites has already been demonstrated with regard to the cDCE-assimilating bacterium Polaromonas sp. JS666 which is capable of growing on catechol, gentisate and salicylate14. Moreover, other plant compounds, including certain types of terpenes, have been used as growth-supporting substrates to enhance microbial degradation of other CEs49. The choice of the initially screened SPMEs depended on their monoaromatic structure and on the fact that they are widespread natural compounds produced by different plant species (details of the natural distribution of the SPMEs mentioned in this study are available from the U.S. Department of Agriculture at https://phytochem.nal.usda.gov). This study provides evidence that selected monoaromatic SPMEs can support microbial growth and cDCE depletion. In order to obtain enrichment cultures, the SPME concentrations used in this study were based on previous studies, in which same-class SPMEs were used during research on the cometabolic degradation of other xenobiotic compounds, including PCBs and trichloroethylene49, 50. At the final stage of enrichment, the presence and type of substrate were found to greatly influence cDCE degradation activity. According to the monitoring data (Fig. 3), cDCE depletion was limited during the first day of cultivation in the presence of acetophenone and zero in the presence of p-hydroxybenzoic acid. This might be due to substrate inhibition phenomena, which are frequently observed during cometabolism51. By contrast, cDCE depletion was detectable from the first day of incubation for cultures enriched on phenethyl alcohol and trans-cinnamic acid. In all enrichment cultures, cDCE degradation activity was notable until day 7 of cultivation and decreased considerably thereafter. The overall transformation of cDCE was more effectively driven by acetophenone-, p-hydroxybenzoic acid- and phenethyl alcohol-enriched microbial populations (Fig. 2) compared to that enriched on trans-cinnamic acid. In particular, the highest yields of cDCE removal per SPME used were observed for acetophenone- and p-hydroxybenzoic acid-enriched cultures. It is noteworthy that increased cDCE biodegradation activity took place in acetophenone- and p-hydroxybenzoic acid-enriched cultures, in which lower final microbial concentrations (Fig. 2) were observed along with higher alpha diversity indices for the enriched microbial populations (Fig. 4). In addition, a clear match was found between the increase in alpha diversity and biodegradative activity, expressed as cDCE transformation yields, which is in line with the finding that increased diversity is generally an indication of greater community resilience and is associated with a higher degree of bioremediation efficiency52. As highlighted by NMDS analysis, the enrichment of diverse microbial populations was strictly dependent on the type of SPME used. The two-dimensional NMDS plot in Fig. 5 shows how enrichment on p-hydroxybenzoic acid and trans-cinnamic acid shapes the microbial community in a very different way to that on acetophenone and phenethyl alcohol.
16S rRNA gene-based phylogenetic analysis revealed ubiquitous enrichment of several sequence variants affiliated with microbial taxa, which have been reported to act as degraders of many classes of environmental pollutants. These include members of the following genera: Cupriavidus, previously found to be involved in the degradation of phenolic and chloroaromatic compounds as well as toxic aromatic petroleum hydrocarbons53; Rhodococcus, whose members are notorious for their biodegradation capabilities, also involving chlorinated aliphatic hydrocarbons such as trichloroethylene and VC54, 55; Sphingopyxis, with S. chilensis being reported to be capable of growing on certain chlorophenolic compounds56; Sediminibacterium, whose member S. salmoneum was detected in a butane-enriched mixed microbial culture capable of aerobic cometabolic degradation of trichloroethylene57; Dokdonella, detected in the aqueous phase of a model wetland supplied with cDCE- and trans-DCE-contaminated water58; and Pseudomonas, whose members are known to contain di- and monooxygenases which can initiate cDCE oxidation upon induction of cometabolic growth on monoaromatic substrates59, 60. In this study, different pseudomonads were detected, including those closely related to P. fulva, P. flavescens, P. benzenivorans and P. argentinensis enriched in acetophenone, phenethyl alcohol and p-hydroxybenzoic acid cultures; and those associated with P.corrugata, P. kilonensis and P. jessenii, enriched in trans-cinnamic acid cultures. Furthermore, enrichment of several unclassified taxa of the family Comamonadaceae occurred in all enriched populations. All the most abundant taxa enriched on SPMEs were mostly below the detection limit in the initial microbial community, which provides further evidence that these taxa were involved in cDCE degradation, as the community initially present in the contaminated soil was not capable of cDCE degradation.
Growth on p-hydroxybenzoic acid was the only condition to raise the abundance levels of Bdellovibrio spp. (Table 1). A previous study has reported the enrichment of members of the family Bdellovibrionaceae in microbial communities, which occurred at a contaminated aquifer after toluene-based biostimulation experiments were performed on the in situ cometabolization of trichloroethylene61. However, Bdellovibrio spp. are known to parasitize other Gram-negative bacteria62, which may also explain their abundance in SPME-enriched populations. Other microbial sequence variants were exclusively observed in some of the SPME-enriched cultures; this was the case for Acinetobacter johnsonii detected in acetophenone cultures as well as Acinetobacter calcoaceticus and Acinetobacter pittii in trans-cinnamic acid cultures (Table 1). To date, a limited number of Acinetobacter spp. have been shown to participate in the degradation of pollutants such as diesel fuel hydrocarbons via alkane hydroxylase enzymes63, 64. 16S rRNA gene data analysis demonstrated that the type of SPME remarkably influenced the structure and composition of enriched microbial populations, which was a reflection of diverse cDCE degradative activities. However, the enrichment of microbial genera, such as Cupriavidus, Burkholderia, Pseudomonas, and Rhodococcus (Table 1), throughout the SPME-enriched cultures suggests that microorganisms affiliated with those genera might play a role in the degradation of cDCE.
In conclusion, this study provides evidence of enhanced cDCE degradation by microbial communities selected in the presence of SPMEs. Our results suggest that natural attenuation of cDCE in soil might occur as a result of the activity of plant-associated soil microorganisms. The study of SPME-stimulated microbial degradation of CEs could open up new avenues for the exploitation of SPMEs in biotechnology in areas such as the development of purpose-built phytoremediation techniques. In basic research terms, additional data are required on the fate of the original substrates – both SPMEs and cDCE – including the identity of microbial populations which assimilate them as well as the pathways used for their degradation. This could be done with the aid of parallel stable isotope probing experiments as shown elsewhere65. Furthermore, the isolation of cDCE-degrading pure microbial cultures could shed light on cDCE degradation mechanisms, including the molecular basis of SPME action, the genes/enzymes involved and their regulation.
Materials and Methods
Chemicals, culture medium and cultivation conditions
Liquid cDCE (97%) and all the SPMEs (purity higher than 97% for caffeic acid and vanillic acid, while higher than 99% for all the other SPMEs) used were obtained from Sigma-Aldrich (USA). Toluene was purchased from Penta (Czech Republic), and phenol (99.5%) was obtained from Fluka (USA). All other chemicals were of analytical grade. Liquid microbial cultures were prepared in an inorganic chloride-free minimal salt medium (MSM) with the following composition per liter of deionized water: Na2HPO4∙12H2O 11.0 g, KH2PO4 2.7 g, (NH4)2SO4 1 g, MgSO4∙7H2O 0.2 g, FeSO4∙7H2O 0.02 g and Ca(NO3)2 0.03 g, pH 7. No vitamins or complex growth factors were added to the MSM. All the microbial cultures were aerobically established in 119 mL-serum vials (Sigma-Aldrich), sealed with teflon-lined butyl rubber septa (Sigma-Aldrich) and shaken on a rotary shaker at 130 rpm at 20 °C. The liquid phase to headspace (air) ratio in each vial was 1:3. Unless otherwise specified, all the experiments described were set up in biological triplicates.
Contaminated site and soil microbial culture generation
A complex microbial community was retrieved from the shallow fraction (0 to 1.2 meter depth) of a long-term CE-contaminated soil, sampled with a pedological needle on the right bank of the river Ploučnice in SAP Mimoň (Czech Republic, 50°37′55.353″N, 14°43′16.401″E). The site was contaminated by the leakage of tetrachloroethylene used during improper management of animal waste. Before the preparation of microbial cultures, the contaminated soil was aerated overnight in a fume hood to facilitate the evaporation of highly volatile CEs. At the preliminary stage of the present study, the capacity of the initial soil microbial community to degrade cDCE was investigated. Microbial suspensions were prepared as 10% (w/v) soil dilutions in MSM in order to minimize initial cDCE concentration levels. Each suspension was spiked with ∼0.35 µmol of cDCE, corresponding to a concentration of ~7 µmol∙L−1 in the aqueous phase. Depletion of cDCE was regularly evaluated over a time frame of 10 days and only once in two weeks up to a period of three months.
Cultivation on toluene and phenol
A 10% (w/v) soil suspension was prepared in MSM without replication, incubated for 24 hours and shaken at 130 rpm at 20 °C. This suspension was then used as an inoculum (25% v/v in fresh MSM) for the preparation of microbial cultures which were exposed to 20 µmol∙L−1 of toluene or phenol with 0.5 mg∙L−1 yeast extract. These cultures were spiked with ∼0.35 µmol of cDCE per vial initially, corresponding to a concentration of ~7 µmol∙L−1 in the aqueous phase. The cDCE depletion was monitored over time.
Cultivation on SPMEs
Microbial cultures were separately established in the same way as cultures exposed to toluene or phenol but were amended with one of the following eight monoaromatic SPMEs as sole carbon sources: salicylic acid, acetophenone, phenethyl alcohol, p-hydroxybenzoic acid, trans-cinnamic acid, ferulic acid, vanillic acid and caffeic acid. An initial concentration of 50 mg∙L−1 of each SPME (equivalent to molar concentration values between 0.3 and 0.4 mM) was supplied to each culture along with a spike of ∼0.25 µmol of cDCE, corresponding to an initial concentration of ~5 µmol∙L−1 of cDCE in the aqueous phase. No biological replicates were used in this initial screening experiment. The cultures described above were kept for a period of 33 days and were newly propagated into fresh medium containing SPME and cDCE each time cDCE was completely depleted. Four microbial cultures exhibiting higher cDCE depletion rates over time were selected for further enrichment on SPMEs and cDCE-degradation experiments.
SPME-based liquid enrichment cultures
Each culture selected was used to inoculate new enrichment cultures in three replicates (25% v/v into fresh MSM), which were propagated every 10 days. A total of seven propagation steps were performed. At the beginning of each culturing step, each vial was provided with 50 mg∙L−1 of each SPME and was spiked with ∼0.25 µmol cDCE (corresponding to an initial concentration of ~5 µmol∙L−1 in the aqueous phase). SPME and cDCE depletions were monitored over time. Microbial growth was monitored through protein content in microbial cell pellets obtained from 4 mL of culture. The pellets were further resuspended in 0.5 mL of deionized water adjusted with 0.1 M NaOH and boiled for 1 minute. The protein concentration (mg∙L−1) was measured according to the Folin-Ciocalteau reagent-based method66, the test colorimetric reaction was conducted in the presence of 0.1 M NaOH and quantification was based on bovine serum albumin standard solutions.
Depletion of cDCE in the selected SPME-enriched cultures
The cDCE transformation yield was calculated as the ratio between total cDCE depleted (µmol) and total SPME consumed (mmol) in each vial during the 10 days of cultivation.
Analytical methods used to quantify SPMEs and cDCE
The concentration of SPMEs in the MSM was determined by means of High Performance Liquid Chromatography (HPLC) on a HP1100 device (Agilent) equipped with a Luna column (C18-2, 150 × 2 mm, 5 µm, Phenomenex) connected to a diode array detector. Analytical conditions were as follows: a mobile phase flow of 0.4 mL∙min−1; isocratic elution; injection volume of 10 µl. The mobile phase was constituted from 0.1% H3PO4 acidified analytical water and methanol in a 3:1 ratio for the detection of p-hydroxybenzoic acid, 1:1 for trans-cinnamic acid and 3:2 for acetophenone and phenethyl alcohol. The cDCE was measured by sampling 200 μL of the headspace with a gas-tight syringe (Hamilton) which was then injected into a GC-µECD Agilent 6890 N instrument. The concentration of cDCE (µmol∙L−1) in the aqueous phase was deduced according to Henry’s law while assuming equilibrium between the liquid and gas phases. The dimensionless Henry coefficient for cDCE at 20 °C was 0.127, as indicated by U.S. E.P.A. On-line Tools for Site Assessment Calculation (https://www3.epa.gov/ceampubl/learn2model/part-two/onsite/esthenry.html). Standard solutions of cDCE were prepared in MSM with the same liquid-to-headspace gas ratio as experimental vials. The GC instrument was equipped with an electron capture detector and a capillary column (DB-XLB, J&B, 30 m, 0.25 mm, film 0.25 µm, mobile phase: nitrogen). The analytical conditions were as follows: nitrogen flow, 165 ml ∙ min−1; speed, 59 cm ∙ s−1; injection chamber temperature, 135 °C; column temperature, 135 °C; detector temperature, 250 °C; split ratio, 30:1; time analysis, 2 min. The microbial cultures, which were previously grown to reach microbial concentrations comparable to those obtained in the active cultures but inhibited with 1 g∙L−1 sodium azide, were used to evaluate possible losses of cDCE through non-biological processes.
DNA isolation and 16S rRNA gene sequencing
Total metagenomic DNA was isolated from the triplicate samples of the initial contaminated soil and from the SPME-enriched microbial cultures (three biological replicates per each treatment) obtained at the end of the enrichment procedure. DNA extraction was performed using the FastDNA Spin Kit for Soil (MP Bio, USA) according to the manufacturer’s instructions. Primers 515 forward 5′-GTGYCAGCMGCNGCGG-3′ and 926 reverse 5′-CCGYCAATTYMTTTRAGTTT-3′ (adapted from Walters, et al.67 with modification of the forward primer) were used to target hypervariable regions V4–V5 of the 16S rRNA genes amplified by PCR in a final volume of 15 µL with: KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Boston, MA, USA) containing 0.02 U/µL of KAPA HiFi HotStart DNA Polymerase, 2.5 mM MgCl2 and 0.3 mM of each dNTP; 0.3 µM of each primer (Generi Biotech, Czech Republic); and template DNA (~20 ng). The cycling program was set as follows: 5 min at 95 °C, 20 cycles of 20 s at 98 °C, 15 s at 56 °C, 15 s at 72 °C and a final extension of 5 min at 72 °C. A volume of 0.5 µL of the PCR product was used as the template for another round of PCR, which was performed under the same conditions except that the final reaction volume was 25 µL, with 1 µM of each primer, and the cycle number was decreased to 5-10. The forward and reverse primers used for the second PCR were modified with sequencing adapters and internal barcodes of variable length (5–8 bp) using the TaggiMatrix spreadsheet courtesy of Travis C. Glenn at the University of Georgia (http://www.baddna.org). The resultant PCR products were purified using AMPure XP Beads (Agencourt, Beckman Coulter, USA). Further amplicon-sample library preparation and sequencing analysis on an Illumina MiSeq instrument were performed at the Core Facility for Nucleic Acid Analysis at the University of Alaska Fairbanks.
Sequencing data analysis
A pipeline for the analysis of amplicon sequences was adapted from Callahan, et al.68 using DADA2. The slight modifications made to the DADA2 SOP resulted from the analysis of a mock community, which consisted of 12 bacterial strains and was amplified along with the sample sequences. Briefly, octuplicate sequences for each biological replicate (fastaq files) were filtered according to their quality (parameters truncLen = c(260,260), maxN = 0, maxEE = 2, truncQ = 2), while the primer sequences were trimmed off to the final degenerated position. Upon dereplication, a DADA2-based removal of sequencing errors was performed, followed by the merging of the denoised forward and reverse reads and the removal of chimeric sequences. Based on the results of mock community analysis, sequences differing by one base were clustered together, with the most abundant sequence being regarded as correct; octuplicate sequences were merged, while keeping only those sequences that occurred in at least three of the octuplicates. Finally, taxonomy was assigned to the valid sequences using rdp_train_set_1469 to create a table of sequence variants.
Further sequence analyses were carried out in R project70 using the phyloseq package71, with diversity indexes being calculated on the basis of rarefied data (14,000 valid reads); ordination analysis was carried out on CSS-normalized data72, while abundance barcharts were constructed from merged biological replicates. The structural diversity of the community was described by the number of sequence variants observed, Simpson’s Index of Diversity (1–D), Simpson’s reciprocal index (1/D) and the Shannon index73, 74. The Bray-Curtis distance-based unconstrained ordination method for NMDS was used to evaluate the ecological distance between the four enriched microbial cultures.
Sequence deposition
MiSeq reads were deposited in the NCBI Short Read Archive under SRA study number SRP090758.
References
Olaniran, A. O., Pillay, D. & Pillay, B. Chloroethenes contaminants in the environment: Still a cause for concern. Afr. J. Biotechnol. 3, 675–682 (2004).
Agency for Toxic Substances and Disease Registry. Toxicological Profile for 1,2-Dichloroethene, https://www.atsdr.cdc.gov/toxprofiles/tp.asp?id=464&tid=82 (1996).
Tiehm, A. & Schmidt, K. R. Sequential anaerobic/aerobic biodegradation of chloroethenes–aspects of field application. Curr. Opin. Biotechnol. 22, 415–421 (2011).
Smidt, H. & de Vos, W. M. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58, 43–73 (2004).
Field, J. A. & Sierra-Alvarez, R. Biodegradability of chlorinated solvents and related chlorinated aliphatic compounds. Rev. Environ. Sci. Biotechnol. 3, 185–254 (2004).
Bhatt, P., Kumar, M. S., Mudliar, S. & Chakrabarti, T. Biodegradation of chlorinated compounds—a review. Crit. Rev. Environ. Sci. Technol. 37, 165–198 (2007).
Maymo-Gatell, X., Chien, Y.-t, Gossett, J. M. & Zinder, S. H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276, 1568–1571 (1997).
Cupples, A. M., Spormann, A. M. & McCarty, P. L. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl. Environ. Microbiol. 69, 953–959 (2003).
Löffler, F. E. et al. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum. Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635 (2013).
Schmidt, K. & Tiehm, A. Natural attenuation of chloroethenes: identification of sequential reductive/oxidative biodegradation by microcosm studies. Water Sci. Technol. 58, 1137–1145 (2008).
Kielhorn, J., Melber, C., Wahnschaffe, U., Aitio, A. & Mangelsdorf, I. Vinyl chloride: still a cause for concern. Environ. Health Perspect. 108, 579 (2000).
Mattes, T. E., Alexander, A. K. & Coleman, N. V. Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol. Rev. 34, 445–475 (2010).
Coleman, N. V., Mattes, T. E., Gossett, J. M. & Spain, J. C. Biodegradation of cis-dichloroethene as the sole carbon source by a β-proteobacterium. Appl. Environ. Microbiol. 68, 2726–2730 (2002).
Mattes, T. E. et al. The genome of Polaromonas sp. strain JS666: insights into the evolution of a hydrocarbon-and xenobiotic-degrading bacterium, and features of relevance to biotechnology. Appl. Environ. Microbiol. 74, 6405–6416 (2008).
Jennings, L. K. et al. Proteomic and transcriptomic analyses reveal genes upregulated by cis-dichloroethene in Polaromonas sp. strain JS666. Appl. Environ. Microbiol. 75, 3733–3744 (2009).
Olaniran, A. O., Pillay, D. & Pillay, B. Aerobic biodegradation of dichloroethenes by indigenous bacteria isolated from contaminated sites in Africa. Chemosphere 73, 24–29 (2008).
Bradley, P. M. & Chapelle, F. H. Aerobic microbial mineralization of dichloroethene as sole carbon substrate. Environ. Sci. Technol. 34, 221–223 (2000).
Schmidt, K. R., Augenstein, T., Heidinger, M., Ertl, S. & Tiehm, A. Aerobic biodegradation of cis-1,2-dichloroethene as sole carbon source: Stable carbon isotope fractionation and growth characteristics. Chemosphere 78, 527–532 (2010).
Rojo, F. Specificity at the end of the tunnel: understanding substrate length discrimination by the AlkB alkane hydroxylase. J. Bacteriol. 187, 19–22 (2005).
Behrendorff, J. B., Huang, W. & Gillam, E. M. Directed evolution of cytochrome P450 enzymes for biocatalysis: exploiting the catalytic versatility of enzymes with relaxed substrate specificity. Biochem. J. 467, 1–15 (2015).
Arp, D. J., Yeager, C. M. & Hyman, M. R. Molecular and cellular fundamentals of aerobic cometabolism of trichloroethylene. Biodegradation 12, 81–103 (2001).
Le, N. B. & Coleman, N. V. Biodegradation of vinyl chloride, cis-dichloroethene and 1,2-dichloroethane in the alkene/alkane-oxidising Mycobacterium strain NBB4. Biodegradation 22, 1095–1108, doi:10.1007/s10532-011-9466-0 (2011).
Fogel, M. M., Taddeo, A. R. & Fogel, S. Biodegradation of chlorinated ethenes by a methane-utilizing mixed culture. Appl. Environ. Microbiol. 51, 720–724 (1986).
Freedman, D. L., Danko, A. & Verce, M. Substrate interactions during aerobic biodegradation of methane, ethene, vinyl chloride and 1,2-dichloroethenes. Water Sci. Technol. 43, 333–340 (2001).
Frascari, D., Kim, Y., Dolan, M. E. & Semprini, L. A kinetic study of aerobic propane uptake and cometabolic degradation of chloroform, cis-dichloroethylene and trichloroetylene in microcosms with groundwater/aquifer solids. Water Air Soil Pollut. 3, 285–298 (2003).
Kim, Y., Arp, D. J. & Semprini, L. Chlorinated solvent cometabolism by butane-grown mixed culture. J. Environ. Engineer. 126, 934–942 (2000).
Ensign, S., Hyman, M. & Arp, D. Cometabolic degradation of chlorinated alkenes by alkene monooxygenase in a propylene-grown Xanthobacter strain. Appl. Environ. Microbiol. 58, 3038–3046 (1992).
Vannelli, T., Logan, M., Arciero, D. M. & Hooper, A. B. Degradation of halogenated aliphatic compounds by the ammonia-oxidizing bacterium Nitrosomonas europaea. Appl. Environ. Microbiol. 56, 1169–1171 (1990).
Verce, M. F., Gunsch, C. K., Danko, A. S. & Freedman, D. L. Cometabolism of cis-1,2-dichloroethene by aerobic cultures grown on vinyl chloride as the primary substrate. Environ. Sci. Technol. 36, 2171–2177 (2002).
Tiehm, A., Schmidt, K. R., Pfeifer, B., Heidinger, M. & Ertl, S. Growth kinetics and stable carbon isotope fractionation during aerobic degradation of cis-1,2-dichloroethene and vinyl chloride. Water Res. 42, 2431–2438 (2008).
Hopkins, G. D. & McCarty, P. L. Field evaluation of in situ aerobic cometabolism of trichloroethylene and three dichloroethylene isomers using phenol and toluene as the primary substrates. Environ. Sci. Technol. 29, 1628–1637 (1995).
Elango, V., Kurtz, H. D. & Freedman, D. L. Aerobic cometabolism of trichloroethene and cis-dichloroethene with benzene and chlorinated benzenes as growth substrates. Chemosphere 84, 247–253 (2011).
Semprini, L. Strategies for the aerobic co-metabolism of chlorinated solvents. Curr. Opin. Biotechnol. 8, 296–308 (1997).
Bacilio-Jiménez, M. et al. Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 249, 271–277 (2003).
Bais, H. P., Broeckling, C. D. & Vivanco, J. M. in Secondary metabolites in soil ecology 241–252 (Springer, 2008).
Shukla, K. P. et al. Nature and role of root exudates: efficacy in bioremediation. Afr. J. Biotechnol. 10, 9717–9724 (2011).
Jha, P., Panwar, J. & Jha, P. Secondary plant metabolites and root exudates: guiding tools for polychlorinated biphenyl biodegradation. Int. J. Environ. Sci. Technol. 12, 789–802 (2015).
Singer, A. C., Thompson, I. P. & Bailey, M. J. The tritrophic trinity: a source of pollutant-degrading enzymes and its implications for phytoremediation. Curr. Opin. Microbiol. 7, 239–244 (2004).
Singer, A. C., Crowley, D. E. & Thompson, I. P. Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol. 21, 123–130 (2003).
Musilová, L., Rídl, J., Polívková, M., Macek, T. & Uhlík, O. Effects of Secondary Plant Metabolites on Microbial Populations: Changes in Community Structure and Metabolic Activity in Contaminated Environments. Int. J. Molec. Sci. 17, 1205 (2016).
Shim, H., Ryoo, D., Barbieri, P. & Wood, T. Aerobic degradation of mixtures of tetrachloroethylene, trichloroethylene, dichloroethylenes, and vinyl chloride by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1. Appl. Microbiol. Biotechnol. 56, 265–269 (2001).
Futamata, H., Harayama, S. & Watanabe, K. Diversity in kinetics of trichloroethylene-degrading activities exhibited by phenol-degrading bacteria. Appl. Microbiol. Biotechnol. 55, 248–253 (2001).
Rui, L., Cao, L., Chen, W., Reardon, K. F. & Wood, T. K. Active site engineering of the epoxide hydrolase from Agrobacterium radiobacter AD1 to enhance aerobic mineralization of cis-1,2-dichloroethylene in cells expressing an evolved toluene ortho-monooxygenase. J. Biol. Chem. 279, 46810–46817 (2004).
Kuiper, I., Lagendijk, E. L., Bloemberg, G. V. & Lugtenberg, B. J. Rhizoremediation: a beneficial plant-microbe interaction. Molec. Plant-Microbe Interact. 17, 6–15 (2004).
Pham, T. T., Pino Rodriguez, N. J., Hijri, M. & Sylvestre, M. Optimizing Polychlorinated Biphenyl Degradation by Flavonoid-Induced Cells of the Rhizobacterium Rhodococcus erythropolis U23A. PLoS One 10, e0126033 (2015).
Toussaint, J.-P., Pham, T., Barriault, D. & Sylvestre, M. Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl. Microbiol. Biotechnol. 95, 1589–1603 (2012).
Siciliano, S. D. et al. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl. Environ. Microbiol. 67, 2469–2475 (2001).
Powell, C. L. & Agrawal, A. Biodegradation of trichloroethene by methane oxidizers naturally associated with wetland plant roots. Wetlands 31, 45–52 (2011).
Suttinun, O., Lederman, P. B. & Luepromchai, E. Application of terpene-induced cell for enhancing biodegradation of TCE contaminated soil. Songklanakarin J. Sci. Technol. 26, 131–142 (2004).
Gilbert, E. S. & Crowley, D. E. Plant compounds that induce polychlorinated biphenyl biodegradation by Arthrobacter sp. strain B1B. Appl. Environ. Microbiol. 63, 1933–1938 (1997).
Alvarez-Cohen, L. & Speitel, G. E. Jr. Kinetics of aerobic cometabolism of chlorinated solvents. Biodegradation 12, 105–126 (2001).
Romantschuk, M. et al. Means to improve the effect of in situ bioremediation of contaminated soil: an overview of novel approaches. Environ. Pollut. 107, 179–185 (2000).
Bacosa, H., Suto, K. & Inoue, C. Preferential degradation of aromatic hydrocarbons in kerosene by a microbial consortium. Int. Biodeter. Biodeg. 64, 702–710 (2010).
Frascari, D. et al. A kinetic study of chlorinated solvent cometabolic biodegradation by propane-grown Rhodococcus sp. PB1. Biochem. Engineer. J. 42, 139–147 (2008).
Larkin, M. J., Kulakov, L. A. & Allen, C. C. Biodegradation and Rhodococcus–masters of catabolic versatility. Curr. Opin. Biotechnol. 16, 282–290 (2005).
Aranda, C., Godoy, F., Becerra, J., Barra, R. & Martínez, M. Aerobic secondary utilization of a non-growth and inhibitory substrate 2,4,6-trichlorophenol by Sphingopyxis chilensis S37 and Sphingopyxis-like strain S32. Biodegradation 14, 265–274 (2003).
Frascari, D. et al. Development of an attached-growth process for the on-site bioremediation of an aquifer polluted by chlorinated solvents. Biodegradation 25, 337–350 (2014).
Imfeld, G. et al. Characterization of microbial communities in the aqueous phase of a constructed model wetland treating 1,2-dichloroethene-contaminated groundwater. FEMS Microbiol. Ecol. 72, 74–88 (2010).
Clingenpeel, S. R., Moan, J. L., McGrath, D. M., Hungate, B. A. & Watwood, M. E. Stable carbon isotope fractionation in chlorinated ethene degradation by bacteria expressing three toluene oxygenases. Front. Microbiol. 3, 63 (2012).
Lang, E., Burghartz, M., Spring, S., Swiderski, J. & Spröer, C. Pseudomonas benzenivorans sp. nov. and Pseudomonas saponiphila sp. nov., represented by xenobiotics degrading type strains. Curr. Microbiol. 60, 85–91 (2010).
Ha, C. et al. Natural gradient drift tests for assessing the feasibility of in situ aerobic cometabolism of trichloroethylene and evaluating the microbial community change. Water Air Soil Pollut. 219, 353–364 (2011).
Strauch, E., Beck, S. & Appel, B. in Predatory Prokaryotes: Biology, Ecology and Evolution (ed Edouard Jurkevitch) 131–152 (Springer Berlin Heidelberg, 2007).
Fondi, M. et al. The genome sequence of the hydrocarbon-degrading Acinetobacter venetianus VE-C3. Res. Microbiol. 164, 439–449 (2013).
Pepi, M., Minacci, A., Di Cello, F., Baldi, F. & Fani, R. Long-term analysis of diesel fuel consumption in a co-culture of Acinetobacter venetianus, Pseudomonas putida and Alcaligenes faecalis. Antonie Van Leeuwenhoek 83, 3–9 (2003).
Leewis, M. C., Uhlík, O. & Leigh, M. B. Synergistic Processing of Biphenyl and Benzoate: Carbon Flow Through the Bacterial Community in Polychlorinated-Biphenyl-Contaminated Soil. Sci. Reports 6, 22145 (2016).
Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951).
Walters, W. et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 1, e00009–00015 (2016).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014).
R Development Core Team. R: A language and environment for statistical computing (2013).
McMurdie, P. J. & Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 8, e61217 (2013).
Paulson, J. N., Stine, O. C., Bravo, H. C. & Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202 (2013).
Bibi, F. & Ali, Z. Measurement of diversity indices of avian communities at Taunsa Barrage Wildlife Sanctuary, Pakistan. J. Anim. Plant Sci. 23, 469–474 (2013).
Hill, T. C., Walsh, K. A., Harris, J. A. & Moffett, B. F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43, 1–11 (2003).
Acknowledgements
We wish to thank the Czech Science Foundation for funding this research (project no. 14-32432S), Michael O’Shea for proof-reading the manuscript and two independent reviewers for their valuable comments.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: S.F. and O.U. Performed the experiments: S.F. Analyzed the data: S.F., M.S. and O.U. Contributed reagents/materials: O.U., I.D. and T.M. Wrote the paper: S.F., M.S. and O.U.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fraraccio, S., Strejcek, M., Dolinova, I. et al. Secondary compound hypothesis revisited: Selected plant secondary metabolites promote bacterial degradation of cis-1,2-dichloroethylene (cDCE). Sci Rep 7, 8406 (2017). https://doi.org/10.1038/s41598-017-07760-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07760-1
This article is cited by
-
Compost, plants and endophytes versus metal contamination: choice of a restoration strategy steers the microbiome in polymetallic mine waste
Environmental Microbiome (2023)
-
Prokaryotes of renowned Karlovy Vary (Carlsbad) thermal springs: phylogenetic and cultivation analysis
Environmental Microbiome (2022)
-
Soil microbial communities following 20 years of fertilization and crop rotation practices in the Czech Republic
Environmental Microbiome (2022)
-
Genomic analysis of Acinetobacter pittii CEP14 reveals its extensive biodegradation capabilities, including cometabolic degradation of cis-1,2-dichloroethene
Antonie van Leeuwenhoek (2022)
-
Chemically degraded soil rehabilitation process using medicinal and aromatic plants: review
Environmental Science and Pollution Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.