Abstract
Despite therapeutic advances, glioblastoma represents a lethal brain tumor. Recently, research to identify prognostic markers for glioblastoma has intensified. Our previous study demonstrated that median progression-free survival (PFS) and overall survival (OS) of patients with high cysteine-rich protein 61 (CCN1) expression was significantly shorter than that of patients with low CCN1 expression. To understand the molecular mechanisms that regulate CCN1 expression, we examined 147 tumour samples from 80 patients with glioblastoma and 67 patients with lower grade glioma. Next-generation and Sanger sequencing showed that PIK3R1Met326Ile was more frequent in the CCN1 high expression group (10/37 cases, 27.0%) than the CCN1 low expression group (3/38 cases, 7.9%) in glioblastoma. This mutation was also detected in corresponding blood samples. In multivariate analysis, high CCN1 expression and PIK3R1Met326Ile in glioblastoma patients were prognostic factors for OS [HR = 2.488 (1.298–4.769), p = 0.006] and [HR = 2.089 (1.020–4.277), p = 0.0439], respectively. Thus, the PIK3R1Met326Ile germline appears to be correlated with CCN1 expression and poor prognosis in glioblastoma.
Similar content being viewed by others
Introduction
Glioblastoma is the most common type of primary malignant brain tumor in adults, with a median survival time of 14 to 16 months, even after the most aggressive treatment, including maximum resection, radiation and chemotherapy1,2,3. Recently, the poor prognosis of glioblastoma patients has been linked to intratumoral genetic heterogeneity4, and many researchers have investigated the genetic and epigenetic alterations of glioblastoma5, 6.
Concurrently, several prognostic molecular biomarkers for glioblastoma have been reported. The cellular DNA repair protein, O6-methylguanine-DNA methyltransferase (MGMT), inhibits the cross-linking of double-stranded DNA by removing alkylation lesions, determines the effect of temozolomide, and is independently associated with overall survival (OS) of malignant glioma patients7, 8. Somatic mutations in the metabolic enzyme isocitrate dehydrogenase (IDH) have also been found in glioma, and glioblastoma patients with wild-type IDH had a poorer prognosis than those with mutant IDH9, 10. In previous research, high expression of cysteine-rich protein 61 (CCN1; also known as CYR61) correlated with a poorer prognosis in glioblastoma patients11.
CCN1, a 42-kDa, secreted, heparin-binding protein, is one of the prototypical members of the CCN family of matricellular proteins12. It was first identified as a growth factor-inducible immediate-early gene, which promoted proliferative responses to growth factors through its interaction with cell surface integrins13, 14. In many cancers, CCN1 may play an important role in tumor progression, and expression levels of CCN1 are related to patient prognosis15, 16. The mechanism of CCN1 expression and genetic alterations in malignant tumors have not been well-characterized.
Somatic mutations in PIK3R1 are associated with several cancers types, in particular, endometrial carcinoma (33.8%), metastatic prostate adenocarcinoma (11.5%), and colorectal adenocarcinoma (9.7%)17. According to The Cancer Genome Atlas (TCGA) database, PIK3R1 is the 11th most commonly mutated gene across 4,429 tumors covering 20 diseases18. With respect to glioblastoma, PIK3R1 mutations represent one of the most common genetic aberrations, and the phosphoinositide 3-kinase (PI3K) pathway is one of the most frequently targeted signaling pathways for therapeutic strategies. However, in TCGA, somatic mutations were analyzed, but germline mutations were not6.
Some germline mutations are known to participate in different types of familial glioma. Germline mutations in p53 may give rise to Li-Fraumeni syndrome, germline mutations in the mismatch DNA repair genes MLH1 and PMS2 are implicated in Turcot syndrome, and individuals with germline mutations in the NF1 or NF2 gene are susceptible to neurofibromatosis, which progresses to glioma19. However, the relationship between germline mutations and tumorigenesis or prognosis has not been fully elucidated.
In this study, the focus was to identify genetic alterations that correlate with CCN1 expression. The results demonstrated that a germline mutation in PIK3R1 (p85α, the regulatory subunit of PI3K) occurred at a higher rate in patients with high CCN1 expression.
Results
The expression level of CCN1 in immunohistochemical staining correlated with cleaved CCN1 expression in western blotting
To identify genetic alterations that correlate with CCN1 expression, we first examined CCN1 levels in glioblastoma cases. A total of 147 tumour samples from 80 patients with glioblastoma and 67 patients with lower grade glioma were analysed and we classified cases based on high or low CCN1 expression level (as described in Methods). Representative high and low CCN1 immunohistochemical stainings are shown in Fig. 1A. Subsequently, the expression level of cleaved CCN1 (cCCN1), which is the truncated form of CCN1 and associated with biological activity in cancer20, 21, was analyzed. Western blotting showed that the NH2-terminal fragment of CCN1 protein was more highly expressed in the group with high CCN1 compared with low CCN1 in immunohistochemical staining (Fig. 1B and C, p < 0.05).
The relationship between CCN1 in immunohistochemical staining and cleaved CCN1 (cCCN1) in western blotting. (A) Representative immunohistochemical staining of CCN1 expression in glioblastoma multiforme patients. The photomicrographs show Left; CCN1 high, Right; CCN1 low. Scale bar = 50 μm. (B,C) Expression level of the NH2-terminal fragment of CCN1 protein (cCCN1, 28 kDa) determined by western blotting compared with immunohistochemical staining of CCN1 performed on formalin-fixed paraffin-embedded tissues from 16 corresponding tumors. Correlation was assessed using the Mann-Whitney U test with p < 0.05 considered as significant. Data are shown as the mean ± SEM. Full-length blots are presented in Supplementary Figure S2.
Genetic alterations in high and low CCN1 glioblastoma
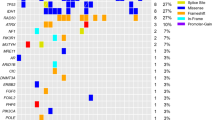
The genetic alterations related to CCN1 expression were investigated using a HaloPlex Cancer Research Panel, which targets Catalogue of Somatic Mutations in Cancer (COSMIC) mutations within 47 genes known to be associated with cancer. Using immunohistochemical staining, 7 samples in the CCN1 high expression group, 7 samples in the CCN1 low expression group, and 1 normal brain tissue were evaluated (Fig. 2). All 14 glioblastoma samples were wild-type IDH. The total number of nonsynonymous amino acid alterations was 47 and 38 in the CCN1 high and low expression groups, respectively (F = 0.450, t = 0.739, p = 0.474, Student’s t-test). In contrast, the number of specific PIK3R1 mutations (chromosome 5, 67588148 G- > A, Met326Ile) tended to be higher in the CCN1 high expression group (4/7 cases, 57.1%) compared with the CCN1 low expression group (1/7 cases, 14.3%) (p = 0.266, Fisher’s exact test) (Table 1). Sanger sequencing of corresponding blood samples demonstrated that the PIK3R1Met326Ile mutation was detected in patients with PIK3R1Met326Ile-mutant glioma specimens (Table 1). Copy-number variation analysis showed no significant difference in gain (14 and 28 genes in the CCN1 high and low expression group, respectively, F = 17.455, t = 1.173, p = 0.263, Student’s t-test) or loss (86 and 81 genes in the CCN1 high and low expression group, respectively, F = 0.05, t = 0.374, p = 0.715, Student’s t-test).
Mutations in high and low CCN1 expression glioblastoma. Left; Specific genes harboring nonsynonymous and synonymous mutations are shown. Middle; The frequency of nonsynonymous mutations in each of the listed genes in both CCN1 groups. Right; The frequency of genetic alterations in each of the listed genes in The Cancer Genome Atlas (TCGA) dataset (2013).
The frequency of PIK3R1Met326Ile was higher in the CCN1 high expression group
Deep sequencing data revealed that the frequency of the PIK3R1Met326Ile mutation tended to be higher in the CCN1 high expression group on immunohistochemical staining. To examine the relevance of the PIK3R1Met326Ile mutation and CCN1 expression, PIK3R1Met326Ile mutations were analyzed in additional glioblastoma specimens and lower grade glioma specimens (Tables 2 and 3, and Supplementary Table S1). We investigated 80 glioblastoma specimens and 67 lower grade glioma specimens, including 39 anaplastic astrocytoma, 8 diffuse astrocytoma, 14 anaplastic oligodendroglioma, and 6 oligodendroglioma samples. Five paediatric glioblastoma patients and 7 paediatric lower grade glioma were excluded from the cohorts, because recent studies showed that children and adult glioma represent molecularly distinct entities with differing biological backgrounds22,23,24,25. The overall frequencies of the PIK3R1Met326Ile mutation in adult glioblastoma patients and adult lower grade glioma patients were 17.3% (13/75 cases) and 16.7% (10/60 cases), respectively, and conforms to Hardy-Weinberg proportions. The frequency of PIK3R1Met326Ile in glioblastoma was significantly higher in the CCN1 high expression group (10/37 cases, 27.0%) than the CCN1 low expression group (3/38 cases, 7.9%) (p = 0.036, Fisher’s exact test) (Fig. 3A and B). In all except 1 case of glioblastoma, PIK3R1Met326Ile mutations were heterozygotic (Fig. 3C). In lower grade glioma, the CCN1 high expression group tended to have a high frequency of PIK3R1Met326Ile, but it was not statistically significant (Table 3 and Supplementary Table S1; astrocytoma, p = 0.2488; oligodendroglioma, p = 0.3158, Fisher’s exact test).
PIK3R1Met326Ile mutations in glioblastoma patients. (A) Electropherogram of patients with heterozygous mutation of PIK3R1Met326Ile. (B) The frequency of PIK3R1Met326Ile mutations was significantly higher in the CCN1 high expression group compared with the CCN1 low expression group. Statistical significance was calculated by Fisher’s exact test. (C) Electropherogram of patients with homozygous mutation of PIK3R1Met326Ile.
Glioblastoma patients with PIK3R1Met326Ile had poor PFS and OS
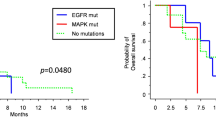
To investigate the relationship between clinical characteristics and prognosis, PFS and OS curves relative to PIK3R1Met326Ile mutations were obtained using the Kaplan-Meier method. The survival curves were compared using a log-rank test based on age, sex, surgical status, CCN1 expression and PIK3R1 mutational status. The results showed that gross total resection, high CCN1 expression and PIK3R1Met326Ile (Fig. 4A) were significantly associated with PFS (p = 0.0138, 0.0013 and 0.0407, respectively). Furthermore, gross total resection, high CCN1 expression and PIK3R1Met326Ile (Fig. 4B) were significantly associated with OS (p = 0.0163, 0.0008 and 0.0125, respectively) (Table 4).
Kaplan-Meier survival curves for patients with and without PIK3R1Met326Ile mutations. (A) Progression-free survival of patients with PIK3R1Met326Ile was significantly shorter than that of patients without PIK3R1Met326Ile (p = 0.0407). (B) Overall survival of patients with PIK3R1Met326Ile was significantly shorter than that of patients without PIK3R1Met326Ile (p = 0.0125).
In multivariate analysis, high CCN1 was a prognostic factor for PFS [hazard ratio (HR) = 2.109 (1.105–4.025), p = 0.0236] and OS [HR = 2.488 (1.298–4.769), p = 0.0060], while the PIK3R1Met326Ile mutation was a prognostic factor for OS [HR = 2.089 (1.020–4.277), p = 0.0439] (Table 5).
In contrast, among lower grade patients, only age was a prognostic factor and neither CCN1 nor PIK3R1Met326Ile was a prognostic factor (Supplementary Tables S2 and S3).
Glioblastoma patients with high CCN1 expression showed high Akt phosphorylation (p-Akt)
To investigate the relationship between CCN1 and PI3K signalling in glioblastoma cases, we evaluated p-Akt levels and p85 protein expression using western blotting (n = 25) and immunohistochemistry (n = 70), respectively. Western blotting demonstrated that the CCN1 high expression group showed high p-Akt at Thr308 compared with the CCN1 low expression group (Fig. 5A and B; p < 0.05). We also evaluated p85 protein expression in 70 glioblastoma patients using the immunohistochemical scoring system as previously described26. However, there was no difference in p85 expression between the CCN1 high and low expression groups (Supplementary Fig. S1A,C, and D). This result corresponds to data from cBioPortal27, 28 (Supplementary Fig. S1B).
PI3K/AKT/mTOR pathway is upregulated in CCN1 high expression group. (A) Immunoblot analysis of levels of p-Akt and Akt total protein in CCN1 high and low expression cases. (B) Quantification of data from panel A. Statistical significance was calculated by the Student’s t-test. *p < 0.05. Data are shown as the mean ± SEM. Full-length blots are presented in Supplementary Figure S2.
Overexpression of either CCN1 or PIK3R1Met326Ile promoted glioma cell invasion in vitro.
Finally, to address the biological relevance of CCN1 and PIK3R1Met326Ile expression, we evaluated the role of CCN1 and the PIK3R1Met326Ile mutation on cellular activities using WST-1 and invasion assays. Plasmids expressing CCN1, wild-type PIK3R1, mutant PIK3R1, or a combination of these plasmids were transfected into U87MG glioma cells, which carry low CCN1 expression29, no PIK3R1 somatic mutations30 and PIK3R1Met326Ile germline mutation (Fig. 6A). Western blot analysis confirmed that overexpression vectors induced detectable levels of CCN1, PIK3R1 and PIK3R1 mutant (Fig. 6B). WST-1 assay showed that overexpression of CCN1, PIK3R1 or PIK3R1 mutant had no effect on cell proliferation (Fig. 6C). In contrast, invasion assays showed that CCN1 and PIK3R1 mutant both significantly increased cell invasion compared to the control (Fig. 6D and E; p < 0.05). Furthermore, the combination of CCN1 together with wild-type PIK3R1, and CCN1 with PIK3R1 mutant showed enhanced cell motility; however, there was no difference between these groups.
The role of PIK3R1Met326Ile on cell process. (A) Sequencing of PIK3R1 in U87MG. (B) Western blotting of CCN1 and PIK3R1 protein in U87MG cells transfected with the indicated plasmids. Full-length blots are presented in Supplementary Figure S2. (C) WST-1 assays for cell proliferation in U87MG cells transfected with the indicated plasmids (n = 5). (D,E) Migration assays in U87MG cells transfected with the indicated plasmids (n = 3). Quantification of migrated cells are shown on the graph in D. Representative images of each cell transfection group are shown in E. Statistical significance was calculated by ANOVA with Dunnett’s post hoc test. *p < 0.05. Data in C and D are shown as the mean ± SEM. Scale bar = 100 μm.
Discussion
In this study, high expression of CCN1 was related to cCCN1 expression and highly germline mutation of PIK3R1Met326Ile, and PIK3R1Met326Ile was found to be a prognostic factor in glioblastoma patients.
CCN1 is widely expressed in several tumor types and has been linked to a poor prognosis in cancer patients, including those with glioma15, 16, 31, 32. Previously, CCN1 expression was revealed to be an independent prognostic factor for glioblastoma patients11. Full-length CCN1 (flCCN1) is a 42-kDa protein, which is converted into cCCN1 by secreted plasmin in the extracellular matrix, resulting in two peptide fragments: the 28-kDa fragment corresponds to the NH2-terminal of CCN1 and the 21-kDa fragment corresponds to the COOH-terminal20, 33. Because the NH2-terminal fragment (28-kDa) promotes cell migration in breast cancer20, this fragment was analyzed by western blotting, and cCCN1 in western blotting was significantly correlated with CCN1 in immunohistochemical staining.
Activation of the PI3K pathway, which comprises PIK3R1, is evident in many cancers, including glioblastoma. Reports suggest that activation of PI3K signaling with overexpression of Akt or loss of PTEN leads to gliomagenesis34,35,36, while Bai et al. showed that PI3K signaling was one of the three important pathways in IDH1-mutant glioma progression37. To date, somatic mutations of PIK3R1 in glioma have been well-researched. Quayle et al. found that several types of somatic mutation in the iSH2 domain of PIK3R1 promoted gliomagenesis30. However, germline mutations in PIK3R1 were not fully elucidated. A single nucleotide polymorphism in codon 326 of PIK3R1, comprising Met substituted by Ile and located near to the NH2-terminal SH2 domain coding region, is known as a germline mutation. Only a few reports have been published on PIK3R1Met326Ile mutations in cancer. Li et al. showed that individuals carrying one or two copies of the 326Ile variant had a 47% increased risk of colon cancer compared with Met/Met38. Hasheminasab et al. reported that cancer patients with the PIK3R1Met326Ile variation were predisposed to skin rashes induced by epidermal growth factor receptor (EGFR) inhibitors, but tended to have better OS39. The PIK3R1Met326Ile mutation was found to be a novel genetic biomarker in gastric cancer, and patients with this germline mutation displayed higher sensitivity to GSK2636771, a PI3Kβ-selective inhibitor40. Kim J. H. et al. reported that in metastatic renal cell carcinoma, there was a tendency for poorer responses to vascular endothelial growth factor receptor (VEGFR) tyrosine kinase inhibitors in patients harboring PIK3R1Met326Ile germline mutations compared with wild-type PIK3R1 41.
Functional studies of the PIK3R1Met326Ile mutation are scarce. Almind et al. reported that the PIK3R1Met326Ile mutation was associated with a lower level of cellular p85α protein in diabetes mellitus42, but it had no impact on PI3K activity43. Cheung also found that the PIK3R1Met326Ile mutation had no effect on Ba/F3 cell survival or Akt phosphorylation44. However, in gastric cancer, PIK3R1Met326Ile was associated with increased p85α expression40. In this study, we found that U87MG cells transfected with plasmids expressing either CCN1 or PIK3R1Met326Ile showed significantly increased invasion ability compared with control cells. Some researchers have already reported that CCN1 is involved in glioma invasion45, 46. Also, somatic mutation of PIK3R1 is known to be related with the increasing of cell proliferations and invasion in cancer, however, the role of germline mutation of PIK3R1 have not been fully revealed18.
Some germline mutations are known to participate in certain familial gliomas; however, the relationship between germline mutations and tumorigenesis or prognosis remains unclear. As described above, PIK3R1Met326Ile germline mutations are related to chemosensitivity in several cancers, including EGFR inhibitors, a PI3Kβ-isoform selective inhibitor, and tyrosine-kinase inhibitors39, 40. Recently, clinical trials have been conducted to identify therapeutic targets for cancers based on specific genetic alterations24, 47, and our results may be highly relevant to these trials.
Limitations of this study is the data were from a single center. Therefore, further investigations using large cohorts, such as TCGA data, are necessary to demonstrate that the key findings are applicable to other centers. Our results also showed the significance of PIK3R1Met326Ile in glioblastoma but not in lower grade glioma. One reason may be that lower grade glioma comprises many types of glioma. Our study included 41 astrocytoma and 19 oligodendroglioma cases, but the patients were diagnosed based on histology rather than molecular confirmations. This might affect the result, and further studies should include lower grade glioma cases diagnosed by molecular patterns.
In conclusion, high expression of CCN1 by immunohistochemical staining was significantly correlated with cCCN1 on western blotting, and the PIK3R1Met326Ile germline mutation existed at a high rate in the CCN1 high expression group. In addition, the PIK3R1Met326Ile germline mutation was significantly correlated with the prognosis of glioblastoma patients. These results may contribute toward the development of novel therapeutic strategies based on molecular alterations.
Material and Methods
Patients
One hundred forty-seven glioma specimens were excised from 80 glioblastoma patients and 67 lower grade glioma patients treated at the Okayama University Hospital between 2006 and 2017. The study (No. 1603–070) was approved by the ethical committee of the Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan and carried out in accordance with the approved guidelines. All patients provided informed written consent.
Immunohistochemistry
For immunohistochemistry, all specimens were fixed in 10% formaldehyde solution, embedded in paraffin blocks, and then sliced into 4.5-µm sections. Paraffin-embedded sections were deparaffinized, epitope-retrieved with an autoclave, and blocked with 5% normal goat serum in phosphate-buffered saline (PBS). Slices were incubated with primary antibodies (anti-CYR61, 1:100, Novus Biologicals, Littleton, CO, USA; anti-PIK3R1, 1:100, Sigma-Aldrich, St Louis, MO, USA). The Dako Cytomation Envision + System-HRP Kit was then used in accordance with the manufacturer’s protocol (DakoCytomation, Carpentaria, CA, USA). After washing in PBS, the sections were counterstained with hematoxylin. Immunohistochemistry samples were observed with a BZ-8100 fluorescence microscope (Keyence, Osaka, Japan). CCN1 protein is expressed in the nucleus, cytoplasm and membrane, and also extracellular areas. The amount of CCN1 immunoreactivity was measured according to the immunoreactive intensity and the estimated extent of the immunopositive area, expressed as a percentage of the total (0–14%, low; 15–100%, high). The cut-off value of CCN1 was previously reported to be 15% of the immunopositive area11.
Western blotting
For immunoblot analysis, cells were lysed in RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) containing phenylmethanesulfonyl fluoride (PMSF, Cell Signaling Technology), and then sonicated. Samples were loaded for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and the separated proteins were blotted onto PVDF membranes. After blocking in 5% skimmed milk, the membranes were incubated overnight with primary antibodies at 4 °C. The membranes were washed with Tris-buffered saline-Tween 20 (TBST), incubated with secondary antibodies at room temperature for 1 h, and rinsed with TBST. HRP signals were visualised using an ECL prime western blotting detection system (GE Healthcare, Little Chalfont, UK) and detected by VersaDoc (Bio-Rad, Hercules, CA, USA). For the immunoblot analysis, the primary antibodies were anti-CYR61 (1:300), p-Akt (1:1000, Cell Signaling Technology), Akt (1:1000, Cell Signaling Technology) and β-actin (1:5000, Sigma-Aldrich), with anti-mouse and anti-rabbit IgG HRP-linked secondary antibodies (both 1:5000, Cell Signaling Technology).
DNA extraction
The QIAamp DNA Micro kit (QIAGEN, Santa Clarita, CA, USA) and QIAamp DNA FFPE Tissue kit (QIAGEN) were used according to the manufacture’s protocol to extract genomic DNA from freshly frozen tumor tissue, FFPE tissue and blood.
Next-generation sequencing
Each genomic DNA sample from tissue specimens was subjected to target amplification and library preparation for next-generation sequencing analysis using a Haloplex Cancer Panel according to the manufacturer’s protocol (Agilent Technologies, Santa Clara, CA, USA). The target enrichment library pool was sequenced using Miseq (Illumina, San Diego, CA, USA). Sequence data were aligned, analyzed, and visualized using SureCall 2.0 software (Agilent Technologies).
Sanger sequencing
Oligo primers were designed for amplifying a target lesion of PIK3R1 (forward; 5′-ATTGCATGGAATTGTGAACTAATGC-3′, reverse; 5′-TGTTCTTAGGCAGTGCCACTTCA-3′). The Pfu DNA polymerase (Agilent technologies) and optimized thermal conditions were used for polymerase chain reaction (PCR). The PCR products were analyzed using the Big Dye Terminator kit (Applied Biosystems, CA, USA) and an ABI 3130xl capillary sequencer (Applied Biosystems).
Cell culture
The U87MG cell line was purchased from DS Pharma Biomedical (Osaka, Japan) in September 2016 and authenticated by Promega (Madison, WI, USA) via STR profiling in December 2016. U87MG cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% fatal bovine serum, 100 U of penicillin, and 0.1 mg mL−1 of streptomycin.
Plasmids
The PIK3R1 plasmids were purchased from GeneCopoeia (Rockville, MD, USA). Site-directed mutagenesis was performed using the GENEART Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA, USA) to produce the PIK3R1Met326Ile plasmid. The CCN1 plasmid was provided by Dr. H. Phillip Koeffler, Cedars-Sinai Medical Center, UCLA School of Medicine (LA, USA). Cells were transfected with plasmids using Lipofectamine® 3000 (Invitrogen) according to the manufacturer’s instructions.
Water soluble tetrazolium-1 (WST-1) assay
For analyses of glioma cell proliferation, the WST-1 assay was performed according to the manufacturer’s protocol (Roche, Mannheim, Germany).
Invasion assay
Invasion assays were performed using a BioCoat Matrigel invasion chamber (BD Bioscience, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. In brief, 2 × 105 cells were seeded in DMEM with 0.1% FBS in the upper chamber. The lower chamber was filled with DMEM with 10% FBS. After 24 h incubation, the filters of inserts were fixed with methanol and stained with Giemsa solution. The number of invading cells on the lower surface of the filter was counted.
Statistical analysis
Protein expression was assessed with the Mann-Whitney U test. The frequency of each mutation was analysed using a paired t-test or Fisher’s exact test. Kaplan-Meier curves were compared using the log-rank test. The results of WST-1 assay and invasion assay were calculated by ANOVA with Dunnett’s post hoc test. Statistical analyses were performed using the SPSS statistical software (version 20; SPSS, Chicago, IL, USA). P-values < 0.05 were considered to denote statistically significant differences.
References
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352, 987–996 (2005).
Chinot, O. L. et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med 370, 709–722 (2014).
Gilbert, M. R. et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med 370, 699–708 (2014).
Bedard, P. L., Hansen, A. R., Ratain, M. J. & Siu, L. L. Tumour heterogeneity in the clinic. Nature 501, 355–364 (2013).
Verhaak, R. G. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Brell, M. et al. Prognostic significance of O6-methylguanine-DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res 11, 5167–5174 (2005).
Hegi, M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352, 997–1003 (2005).
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360, 765–773 (2009).
Ishida, J. et al. Evaluation of extracellular matrix protein CCN1 as a prognostic factor for glioblastoma. Brain Tumor Pathol 32, 245–252 (2015).
Walsh, C. T. et al. Thrombin receptor and RhoA mediate cell proliferation through integrins and cysteine-rich protein 61. FASEB J 22, 4011–4021 (2008).
O’Brien, T. P., Yang, G. P., Sanders, L. & Lau, L. F. Expression of cyr61, a growth factor-inducible immediate-early gene. Mol Cell Biol 10, 3569–3577 (1990).
Babic, A. M., Kireeva, M. L., Kolesnikova, T. V. & Lau, L. F. CYR61, a product of a growth factor-inducible immediate early gene, promotes angiogenesis and tumor growth. Proc Natl Acad Sci USA 95, 6355–6360 (1998).
Xie, D. et al. Breast cancer. Cyr61 is overexpressed, estrogen-inducible, and associated with more advanced disease. J Biol Chem 276, 14187–14194 (2001).
Xie, J. J. et al. Expression of cysteine-rich 61 is correlated with poor prognosis in patients with esophageal squamous cell carcinoma. Eur J Surg Oncol 37, 669–674 (2011).
Costa, C. & Engelman, J. A. The double life of p85. Cancer Cell 26, 445–447 (2014).
Cheung, L. W. et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell 26, 479–494 (2014).
Robertson, L. B. et al. Survey of familial glioma and role of germline p16INK4A/p14ARF and p53 mutation. Fam Cancer 9, 413–421 (2010).
Pendurthi, U. R., Tran, T. T., Post, M. & Rao, L. V. Proteolysis of CCN1 by plasmin: functional implications. Cancer Res 65, 9705–9711 (2005).
Guillon-Munos, A. et al. Kallikrein-related peptidase 12 hydrolyzes matricellular proteins of the CCN family and modifies interactions of CCN1 and CCN5 with growth factors. J Biol Chem 286, 25505–25518 (2011).
Paugh, B. S. et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol 28, 3061–3068 (2010).
Jones, C. et al. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro Oncol (2016).
Project, I. C. G. C. P. T. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med 22, 1314–1320 (2016).
Kameda, M. et al. Congenital Glioblastoma with Distinct Clinical and Molecular Characteristics: Case Reports and a Literature Review. World Neurosurg (2017).
Cizkova, M. et al. PIK3R1 underexpression is an independent prognostic marker in breast cancer. BMC Cancer 13, 545 (2013).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012).
Kurozumi, K. et al. Oncolytic HSV-1 infection of tumors induces angiogenesis and upregulates CYR61. Mol Ther 16, 1382–1391 (2008).
Quayle, S. N. et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS One 7, e49466 (2012).
Xie, D. et al. Levels of expression of CYR61 and CTGF are prognostic for tumor progression and survival of individuals with gliomas. Clin Cancer Res 10, 2072–2081 (2004).
Sabile, A. A. et al. Cyr61 expression in osteosarcoma indicates poor prognosis and promotes intratibial growth and lung metastasis in mice. J Bone Miner Res 27, 58–67 (2012).
Moon, H. G. et al. CCN1 secretion and cleavage regulate the lung epithelial cell functions after cigarette smoke. Am J Physiol Lung Cell Mol Physiol 307, L326–337 (2014).
Holland, E. C. et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 25, 55–57 (2000).
Zheng, H. et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455, 1129–1133 (2008).
Sonoda, Y. et al. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res 61, 6674–6678 (2001).
Bai, H. et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet 48, 59–66 (2016).
Li, L., Plummer, S. J., Thompson, C. L., Tucker, T. C. & Casey, G. Association between phosphatidylinositol 3-kinase regulatory subunit p85alpha Met326Ile genetic polymorphism and colon cancer risk. Clin Cancer Res 14, 633–637 (2008).
Hasheminasab, S. M. et al. High-throughput screening identified inherited genetic variations in the EGFR pathway contributing to skin toxicity of EGFR inhibitors. Pharmacogenomics 16, 1605–1619 (2015).
Kim, C. et al. Whole-exome sequencing of gastric cancer identifies germline PIK3R1 variant as a novel genetic biomarker for a PI3K beta-isoform selective inhibitor, GSK2636771. In: Proceedings of the American Association for Cancer Research; 2015 Apr 18–22; Philadelphia, PA: AACR; 2015. Abstract nr 4694.
Kim, J. H. et al. Novel biomarkers for VEGFR inhibitors in metastatic renal cell carcinoma: BIM expression, and germline polymorphisms of BIM and PIK3R1. In: Proceedings of the American Association for Cancer Research; 2016 Apr 16–20; New Orleans, LA: AACR; 2016. Abstract nr 430.
Almind, K. et al. Characterization of the Met326Ile variant of phosphatidylinositol 3-kinase p85alpha. Proc Natl Acad Sci USA 99, 2124–2128 (2002).
Baynes, K. C. et al. Natural variants of human p85 alpha phosphoinositide 3-kinase in severe insulin resistance: a novel variant with impaired insulin-stimulated lipid kinase activity. Diabetologia 43, 321–331 (2000).
Cheung, L. W. et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov 1, 170–185 (2011).
Young, N., Pearl, D. K. & Van Brocklyn, J. R. Sphingosine-1-phosphate regulates glioblastoma cell invasiveness through the urokinase plasminogen activator system and CCN1/Cyr61. Mol Cancer Res 7, 23–32 (2009).
Haseley, A. et al. Extracellular matrix protein CCN1 limits oncolytic efficacy in glioma. Cancer Res 72, 1353–1362 (2012).
Worst, B. C. et al. Next-generation personalised medicine for high-risk paediatric cancer patients - The INFORM pilot study. Eur J Cancer 65, 91–101 (2016).
Acknowledgements
The authors would like to thank M. Arao and Y. Ukai in the Department of Neurological Surgery and A. Ueda in the Department of Physiology for their technical assistance. This study was supported by grants-in-aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology to K.K. (No. 23592125; No. 26462182) and T.I. (No. 25462261).
Author information
Authors and Affiliations
Contributions
Y.O., K.K., J.I., T.O., T.S., Y.T., Y.H., A.U., Y.M., H.M., T.I. and D.I. conceived the study; Y.O. and K.K. wrote the manuscript; and Y.O., J.I., T.S., Y.T., Y.H., A.U., Y.M., S.T. and T.M. performed experiments and bioinformatics analyses. All authors reviewed and contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Otani, Y., Ishida, J., Kurozumi, K. et al. PIK3R1Met326Ile germline mutation correlates with cysteine-rich protein 61 expression and poor prognosis in glioblastoma. Sci Rep 7, 7391 (2017). https://doi.org/10.1038/s41598-017-07745-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07745-0
This article is cited by
-
Differentiated glioblastoma cells accelerate tumor progression by shaping the tumor microenvironment via CCN1-mediated macrophage infiltration
Acta Neuropathologica Communications (2021)
-
The CCN axis in cancer development and progression
Journal of Cell Communication and Signaling (2021)
-
New insights for precision treatment of glioblastoma from analysis of single-cell lncRNA expression
Journal of Cancer Research and Clinical Oncology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.