Abstract
Non-typhoidal Salmonella are associated with gastrointestinal disease worldwide and invasive disease in Africa. We constructed novel bivalent vaccines through the recombinant expression of heterologous O-antigens from Salmonella Choleraesuis in Salmonella Typhimurium. A recombinant Asd+ plasmid pCZ1 with the cloned Salmonella Choleraesuis O-antigen gene cluster was introduced into three constructed Salmonella Typhimurium Δasd mutants: SLT11 (ΔrfbP), SLT12 (ΔrmlB-rfbP) and SLT16 (ΔrfbP ∆pagL::TT araCPBAD rfbP). Immunoblotting demonstrated that SLT11 (pCZ1) and SLT12 (pCZ1) efficiently expressed the heterologous O-antigen. In the presence of arabinose, SLT16 (pCZ1) expressed both the homologous and heterologous O-antigens, whereas in the absence of arabinose, SLT16 (pCZ1) mainly expressed the heterologous O-antigen. We deleted the crp/cya genes in SLT12 (pCZ1) and SLT16 (pCZ1) for attenuation purposes, generating the recombinant vaccine strains SLT17 (pCZ1) and SLT18 (pCZ1). Immunization with either SLT17 (pCZ1) or SLT18 (pCZ1) induced specific IgG against the heterologous O-antigen, which mediated significant killing of Salmonella Choleraesuis and provided full protection against a lethal homologous challenge in mice. Furthermore, SLT17 (pCZ1) or SLT18 (pCZ1) immunization resulted in 83% or 50% heterologous protection against Salmonella Choleraesuis challenge, respectively. Our study demonstrates that heterologous O-antigen expression is a promising strategy for the development of multivalent Salmonella vaccines.
Similar content being viewed by others
Introduction
Non-typhoidal Salmonella (NTS), a group of Gram-negative and facultative intracellular bacteria, are important zoonotic pathogens that cause foodborne gastrointestinal disease in humans and animals and pose a public health burden worldwide1, 2. It is estimated that 93.8 million cases of gastroenteritis occur due to NTS worldwide, leading to 155,000 diarrheal deaths each year3. NTS can also cause invasive diseases, such as bacteremia, septicemia and meningitis, with high morbidity and mortality in high risk individuals in industrialized and developing countries and in young children in sub-Saharan Africa4, 5. However, there is no vaccine currently available against NTS serovars in humans, although some O-antigen-based conjugate vaccines, live attenuated vaccines and outer membrane protein (OMP)-based subunit vaccines are in development.
Exposure to animal and animal food products is one of the major risk factors resulting in human salmonellosis6,7,8. NTS comprise more than 2500 different serovars; however, relatively few restricted serovars are associated with bacterial transmission between animals and humans. These serovars include Salmonella enterica serovar Typhimurium (Salmonella Typhimurium), Salmonella Enteritidis and Group C Salmonella, such as Salmonella Choleraesuis and Salmonella Newport2, 9, 10. Hence, a significant burden is placed on both the animal industry and the healthcare system due to these bacteria. Furthermore, with the increasing frequencies of multi-drug resistant Salmonella strains, antibiotic treatment in human patients is becoming increasingly difficult11, 12. Thus, there is growing recognition that an ideal multivalent vaccine with broad coverage of epidemic serovars is needed to control NTS infections in both animals and humans.
The lipopolysaccharide (LPS) O-antigen, which is expressed on the outer surface of the bacterium and consists of oligosaccharide repeats, is the basis for the typing system of Salmonella serovars together with the flagellar antigen and determines the serogroup13, 14. The O-antigen is highly immunogenic and has become an important target of protective immunity15. Passive transfer of monoclonal IgG specific to the O-antigen of Salmonella Typhimurium conferred protection against virulent Salmonella Typhimurium challenge16, 17. A protective monoclonal IgA (named Sal4) against the O-antigen of Salmonella Typhimurium could impair type 3 secretion and outer membrane integrity, rendering the bacteria avirulent18. The key role of the O-antigen in protective immunity is to promote the development of O-antigen-based conjugate vaccines, such as O:9-flagellin19, 20, O:4,12-TT21, O:4,5/O:9-CRM19722 and Os-po23. All these conjugate vaccines have provided protection against virulent challenge in preclinical studies; nevertheless, they protect against only serovars with the same O-antigen specificity. Although cross-protection may be achieved by mixing these conjugate vaccines, the increasing cost of this approach is a disadvantage for application of the glycoconjugate technology24. Additionally, some attenuated Salmonella vaccines have exhibited cross-protective efficacy against different serovars25, 26; however, the cross-efficacy is limited, and only partial protection is achieved after challenge with virulent strains of heterologous serovars. Thus, there is an urgent need for novel approaches for developing a multivalent vaccine.
Live attenuated Salmonella Typhimurium has been extensively used as a vaccine vehicle to deliver heterologous antigens to the immune system and stimulate a protective immune response against a variety of targeted pathogens at a low cost27. A series of technologies, including the balanced-lethal vector-host systems and the regulated delayed in vivo attenuation, have been developed in Salmonella to improve the potency of recombinant vaccines28. We hypothesized that immunization with a live attenuated Salmonella Typhimurium expressing heterologous O-antigens of other serovars in the outer membrane would provide both homologous and heterologous protection. The O-antigen gene cluster of Salmonella serovars responsible for O-antigen biosynthesis has been identified13. Here, we introduced the O-antigen gene cluster of Salmonella Choleraesuis, which resides on the Asd+ plasmid pCZ1, into three Salmonella Typhimurium Δasd strains: SLT11 (Δasd ΔrfbP), SLT12 (Δasd ΔrmlB-rfbP) and the arabinose-regulated strain SLT16 (Δasd ΔrfbP ∆pagL::TT araCPBAD rfbP), and we then verified the expression of the homologous and heterologous O-antigens in the three recombinant strains. Because Salmonella O-antigen also plays a vital role in bacterial survival and virulence29,30,31, we first determined the effects of heterologous O-antigen expression on several phenotypes, including swimming, sensitivities to polymyxin B and sodium deoxycholate (DOC) and colonization. We then introduced crp and cya gene mutations into the SLT12 (pCZ1) and SLT16 (pCZ1) strains for attenuation, and finally, we measured the immunogenicity and protective efficacy of the recombinant attenuated vaccine strains of Salmonella Typhimurium in BALB/c mice.
Results
Construction and characterization of recombinant Salmonella Typhimurium strains
The asd-based balanced-lethal vector-host system32 was used to stably express heterologous O-antigen in Salmonella Typhimurium. The Asd+ plasmid pCZ1 was constructed by overlapping DNA elements, including the pSC101 origin, asd gene cassette, TIT2 terminator, Ptrc promoter, kanamycin-resistance cassette from the plasmid pQK66433 and the whole O-antigen gene cluster of Salmonella Choleraesuis. Moreover, to block the synthesis of the Salmonella Typhimurium O-antigen, the rfbP gene (responsible for the addition of the Gal-1-phosphate residue to UndP to initiate O-unit synthesis) or the whole O-antigen gene cluster (rmlB-rfbP) was deleted from the Salmonella Typhimurium strain SLT10 (S100 Δasd), generating SLT11 (SLT10 ΔrfbP) or SLT12 (SLT10 ΔrmlB-rfbP), respectively. In addition, we used arabinose-dependent regulated delayed attenuation systems34, 35 to construct strain SLT16 (SLT10 ΔrfbP ∆pagL::TT araCPBAD rfbP) by replacing the rfbP promoter with the araCPBAD promoter to ensure that the expression of homologous Salmonella Typhimurium O-antigen was activated by exogenous arabinose provided during in vitro growth34. Then, pCZ1 was introduced into SLT11, SLT12 and SLT16 to complete the construction of the three recombinant Salmonella strains SLT11 (pCZ1), SLT12 (pCZ1) and SLT16 (pCZ1). The control strains SLT11 (pQK664), SLT12 (pQK664) and SLT16 (pQK664) were also constructed.
LPS analysis by silver staining and Western immunoblotting was employed for the parent and recombinant strains, which were cultured and adjusted to the same optical density before analysis. Silver staining showed that the Salmonella Typhimurium parent strain SLT10 and the Salmonella Choleraesuis wild-type strain S340 produced complete LPS ladders (Fig. 1A, lanes 1 and 6). As expected, SLT11 (pCZ1) and SLT12 (pCZ1) displayed smooth LPS phenotypes (Fig. 1A, lanes 3 and 5), whereas SLT11 (pQK664) and SLT12 (pQK664) displayed rough LPS phenotypes (Fig. 1A, lanes 2 and 4). Western immunoblotting with O:4- and O:7-specific antisera demonstrated that SLT12 (pCZ1) expressed the heterologous O-antigen but not the homologous O-antigen (Fig. 1B and C, lane 5); however, unexpectedly, SLT11 (pCZ1) expressed not only the heterologous O-antigen but also short-length homologous O-antigen (Fig. 1B and C, lane 3). SLT16 (pCZ1) expressed O-antigens in an arabinose-dependent manner (Fig. 1D,E and F). In the presence of arabinose, SLT16 (pCZ1) expressed both the complete homologous and heterologous O-antigens (Fig. 1E and F, lane 4), whereas in the absence of arabinose, SLT16 (pCZ1) expressed the heterologous O-antigen and short-length homologous O-antigen (Fig. 1E and F, lane 5). Thus, SLT16 (pCZ1) displayed a chimeric O-antigen profile. In contrast, the control strain SLT16 (pQK664) expressed homologous O-antigen but not heterologous O-antigen when grown with arabinose (Fig. 1E and F, lane 2) and did not express either the homologous or heterologous O-antigen when grown without arabinose (Fig. 1E and F, lane 3). Because SLT12 (pCZ1) expressed more heterologous antigen than SLT11 (pCZ1) (Fig. 1A, lane 3 vs lane 5), SLT12 (pCZ1) and SLT16 (pCZ1) were used for further biological analyses.
LPS analysis by silver-stain and Western immunoblotting. (A,B and C). LPS extracted from Salmonella Typhimurium SLT10 (pQK664) (lane 1), SLT11 (pQK664) (lane 2), SLT11 (pCZ1) (lane 3), SLT12 (pQK664) (lane 4), SLT12 (pCZ1) (lane 5) and Salmonella Choleraesuis S340 (lane 6) was subjected to SDS-PAGE followed by silver staining (A) and immunoblotting analysis using O:4-specific antisera (B) and O:7-specific antisera (C). (D,E and F) LPS extracted from SLT10 (pQK664) (lane 1), SLT16 (pQK664) grown with arabinose (lane 2) or without arabinose (lane 3), SLT16 (pCZ1) grown with arabinose (lane 4) or without arabinose (lane 5) and S340 (lane 6) was subjected to SDS-PAGE followed by silver staining (D) and immunoblotting using O:4-specific antisera (E) and O:7-specific antisera (F). Each lane corresponds to LPS from 108 bacteria.
Effects of heterologous O-antigen expression on bacterial phenotypes
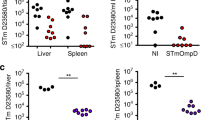
Because the O-antigen plays an important role in Salmonella Typhimurium colonization and survival processes31, we evaluated whether the altered O-antigen profiles could affect bacterial phenotypes, including swimming, resistance to polymyxin B and DOC and colonization in vivo. As shown in Fig. 2A, the rough strains SLT12 (pQK664) and SLT16 (pQK664) (grown without arabinose) showed a defective swimming phenotype, while SLT12 (pCZ1), SLT16 (pQK664) (grown with arabinose) and SLT16 (pCZ1) (grown with or without arabinose) with the smooth LPS phenotype showed a similar motility capacity as the parent strain SLT10 (pQK664). A similar result was observed in experiments examining resistance to polymyxin B and DOC. The strains SLT12 (pCZ1), SLT16 (pCZ1) and SLT16 (pQK664) (grown with arabinose) exhibited comparable resistance to polymyxin B and DOC as the parent strain (Fig. 2B and C), while SLT12 (pQK664) and SLT16 (pQK664) (grown without arabinose) exhibited decreased resistance to polymyxin B and DOC (Fig. 2B and C). In addition, SLT12 (pCZ1), SLT16 (pCZ1) and SLT16 (pQK664) displayed colonization levels in Peyer’s Patches (PP), liver and spleen of BALB/c mice that were comparable to the parent strain (Fig. 2D,E and F). Unexpectedly, no bacteria were detected in the three tissues after infection with the rough strain SLT12 (pQK664) (Fig. 2D,E and F); thus, SLT12 (pQK664) colonization was significantly decreased compared to that of the parent strain. Therefore, our results demonstrated that heterologous O-antigen expression does not affect bacterial swimming, resistance to polymyxin B and DOC or colonization.
Effects of heterologous O-antigen expression on Salmonella Typhimurium biological activities. (A) Swimming assay. SLT10 (pQK664), SLT12 (pQK664), SLT12 (pCZ1) and the two arabinose-regulated strains SLT16 (pQK664) and SLT16 (pCZ1) were grown in LB broth with or without 0.1% arabinose. Then, 6 μl of the bacterial suspension (approximately 1 × 106 CFU) was spotted onto the middle of LB plates containing 0.3% agar with or without arabinose, and the diameter of the colonies was measured 6 h after incubation. (B and C) Resistance to polymyxin and DOC. The same Salmonella Typhimurium strains described in A were cultured in LB broth with or without 0.1% arabinose. One-hundred microliters of the cell suspension (approximately 1 × 106–5 × 106 CFU) was inoculated with or without 0.12 μg/ml polymyxin B (B) or 4 mg/ml DOC (C) for 1 h at 37 °C. The bacteria were diluted and plated onto LB plates, and the survival rate was calculated the following day. (D,E and F) Colonization. Groups of BALB/c mice (n = 4/group) were orally inoculated with 1 × 109 CFU of each indicated strain. Viable bacteria were recovered from PP (D), liver (E) and spleen (F) 6 days after infection. The bacterial number in each tissue was calculated as log10 CFU/g. The asterisk above the error bar indicates significance compared to the SLT10 (pQK664) group. ***p < 0.001.
Construction of the recombinant vaccine strains and virulence evaluation
The crp and cya genes were deleted from SLT12 (pQK664), SLT12 (pCZ1), SLT16 (pQK664) and SLT16 (pCZ1) to attenuate virulence, generating the two recombinant vaccine strains SLT17 (pCZ1) and SLT18 (pCZ1) and the two control vaccine strains SLT17 (pQK664) and SLT18 (pQK664). Silver staining and Western immunoblotting showed that SLT17 (pCZ1) or SLT18 (pCZ1) displayed a similar LPS phenotype as SLT12 (pCZ1) or SLT16 (pCZ1) (Figs 1 and 3 and Supplementary Fig. S1); the control vaccine strains SLT17 (pQK664) and SLT18 (pQK664) (grown without arabinose) showed rough LPS phenotypes (Fig. 3 and Supplementary Fig. S1, lanes 2 and 5). The silver staining also showed that SLT17 (pCZ1) expressed more heterologous O-antigens than SLT18 (pCZ1) (grown without arabinose) (Fig. 3, lane 3 vs lane 7). Furthermore, the 50% lethal dose (LD50) and colonization for each of the four vaccine strains were determined in BALB/c mice. All vaccine strains were avirulent, and their LD50 values were more than 2 × 109 CFU, which was > 104-fold higher than that of the wild-type strain S100 (LD50 of 5.4 × 105 CFU) (Supplementary Table S1). The colonization levels of all of the vaccine strains in PP, liver and spleen were significantly decreased compared with those of the wild-type S100 strain (Supplementary Fig. S2). The vaccine strains SLT17 (pCZ1), SLT18 (pQK664) and SLT18 (pCZ1) retained similar capacities to efficiently colonize mouse tissues. In contrast, no bacteria were detected in the three tissues after infection with the control strain SLT17 (pQK664) (Supplementary Fig. S2).
LPS phenotypes of the vaccine strains. LPS samples extracted from Salmonella Typhimurium wild-type S100 (lane 1), SLT17 (pQK664) (lane 2), SLT17 (pCZ1) (lane 3) and SLT18 (pQK664) grown with arabinose (lane 4) or without arabinose (lane 5), SLT18 (pCZ1) grown with arabinose (lane 6) or without arabinose (lane 7) and Salmonella Choleraesuis S340 (lane 8) were visualized by silver staining following PAGE. Each lane corresponds to LPS from 108 bacteria.
Immunogenicity and protective efficacy of the recombinant attenuated vaccine strains
Seven-week-old BALB/c mice were orally immunized with PBS or approximately 109 CFU of each vaccine strain twice at an interval of four weeks. The mice were then challenged orally with the virulent strains Salmonella Typhimurium S10036 and Salmonella Choleraesuis S34036 at a dose of at least 100 × LD50 one month after the second immunization. Serum IgG and fecal IgA antibodies against either the homologous Salmonella Typhimurium or heterologous Salmonella Choleraesuis LPS were detected by enzyme-linked immunosorbent assay (ELISA) after immunization. No specific serum IgG antibody against either the homologous or heterologous LPS was induced after the first immunization in any of the immunized groups (Fig. 4A and B). All vaccine strains except SLT17 (pQK664) stimulated significantly higher levels of serum IgG and fecal IgA against Salmonella Typhimurium LPS than the PBS control after the second immunization (Fig. 4A and C), and the IgG levels induced by the vaccine strains SLT18 (pQK664) and SLT18 (pCZ1) were significantly higher than that of the vaccine strain SLT17 (pCZ1) (Fig. 4A). Furthermore, only the recombinant vaccine strains SLT17 (pCZ1) and SLT18 (pCZ1) induced a specific serum IgG or fecal IgA response against Salmonella Choleraesuis LPS after the second immunization, and the IgG or IgA level induced by SLT17 (pCZ1) was significantly higher than that of SLT18 (pCZ1) (Fig. 4B and C).
Antibody responses induced by the recombinant attenuated vaccines. (A and B) Serum IgG responses. Groups of BALB/c mice were immunized with 1 × 109 CFU of SLT17 (pQK664), SLT17 (pCZ1), SLT18 (pQK664), SLT18 (pCZ1) or PBS twice at an interval of 4 weeks. Serum samples were collected from 6 mice from each group 3 weeks and 7 weeks after the first immunization. The serum IgG specific to Salmonella Typhimurium LPS (A) and Salmonella Choleraesuis LPS (B) was measured by quantitative ELISA in each group (C). Fecal IgA responses. The fecal samples were collected from 6 mice of each group 3 weeks after the second immunization. Fecal IgAs against Salmonella Typhimurium LPS and Salmonella Choleraesuis LPS were detected by indirect ELISA. The asterisk above the error bar indicates significance compared with the PBS control group. The asterisk above the line indicates significance between the two indicated groups. *p < 0.05. **p < 0.01. ***p < 0.001.
To determine whether each vaccine strain elicited bactericidal antibodies, we used the serum from each group at week 3 post-second immunization and measured the relative bactericidal activities against the homologous Salmonella Typhimurium strain S100 and the heterologous Salmonella Choleraesuis strain S340. The bacteria were incubated with 25% heat-inactivated serum plus active guinea pig complement or no complement for 1.5 h, and the relative survival was then calculated as the percent CFU in each serum with active complement compared to the CFU of the same serum with no complement. Salmonella O:4-specific and O:7-specific antisera were included as positive controls. As shown in Fig. 5A, the antibodies elicited by either the recombinant strains SLT17 (pCZ1) and SLT18 (pCZ1) or the control strains SLT17 (pQK664) and SLT18 (pQK664) were able to kill the homologous S100 strain at a similar level, which was significantly better than the antibodies from the PBS group. The ability of positive O:4-specific antisera to mediate killing was comparable to that of the antisera induced by the SLT17 (pCZ1), SLT18 (pQK664) and SLT18 (pCZ1) groups but better than that of the serum induced by SLT17 (pQK664) (Fig. 5A). Furthermore, the antibody from the SLT17 (pCZ1) and SLT18 (pCZ1) groups resulted in significant killing against the heterologous S340 strain, which was much better than that of the PBS group and comparable to that of the positive O:7-specific antisera (Fig. 5B). In contrast, no significant killing against the heterologous S340 strain was observed in the control SLT17 (pQK664) or SLT18 (pQK664) groups. Notably, the relative survival of S340 in the SLT17 (pCZ1) group was significantly lower than that of the SLT18 (pCZ1) group (Fig. 5B), indicating that SLT17 (pCZ1) immunization induces enhanced serum bactericidal effects.
Serum bactericidal assay. Salmonella Typhimurium S100 (A) or Salmonella Choleraesuis S340 (B) was incubated with the serum collected from each immunized group at 3 weeks post-second immunization. Active guinea pig complement was added to or omitted from the mixture. The relative survival was calculated at 1.5 h post-incubation. The asterisk above the error bar indicates significance compared to the PBS control group. The asterisk above the line indicates significance between the two indicated groups. *p < 0.05. ***p < 0.001.
All 12 mice in the SLT18 (pQK664), SLT17 (pCZ1) and SLT18 (pCZ1) immunized groups survived the Salmonella Typhimurium S100 challenge, suggesting 100% protection. In contrast, only 2 of 12 mice in the SLT17 (pQK664) group survived the challenge, and all 12 mice of the PBS group succumbed to the challenge (Fig. 6A). In the presence of the Salmonella Choleraesuis S340 challenge, the SLT17 (pCZ1) immunization resulted in 83% survival, which was better than the 50% survival observed for the SLT18 (pCZ1) group (Fig. 6B, log-rank test, p < 0.01). All of the mice in the control SLT17 (pQK664), SLT18 (pQK664) and PBS groups succumbed to the challenge (Fig. 6B). Thus, the recombinant vaccine strains SLT17 (pCZ1) and SLT18 (pCZ1) provided full protection against lethal homologous challenge and 83% and 50% heterologous protection against Salmonella Choleraesuis lethal challenge, respectively.
Discussion
NTS serovars Typhimurium, Enteritidis and Group C Salmonella are important causes of foodborne salmonellosis worldwide3. There is urgent need for an effective vaccine with broad serovar coverage for the prevention of salmonellosis in humans and animals. Although some types of vaccines, such as live attenuated vaccines and O-antigen-based conjugate vaccines, are in development37, there is no or only partial cross-protection achieved by these vaccines. Here, we developed two bivalent vaccines, SLT17 (pCZ1) and SLT18 (pCZ1), by recombinant expression of Salmonella Choleraesuis O-antigen (Group C1) in attenuated Salmonella Typhimurium strains. The reason for the selection of Salmonella Typhimurium as the vaccine vector rather than other serovars is that Salmonella Typhimurium is the most well-studied NTS serovar, and a set of recombinant vaccine technologies has been developed for this serovar28. Here, we used the Asd-based balanced-lethal vector-host system32 to maintain the expression of the heterologous O-antigen and an arabinose-dependent regulated attenuation system34 to achieve expression of the homologous O-antigen dependent on exogenously supplied arabinose during in vitro growth.
O-antigens are characterized as immunodominant and protective antigens in Salmonella as well as in several other infectious bacteria. Recombinant attenuated Salmonella vaccines expressing Shigella sonnei O-antigen or Burkholderia mallei O-antigen have been constructed and exhibit protection against lethal challenge38, 39. Surprisingly, a similar strategy has not been used for the development of bivalent or multivalent Salmonella vaccines. A potential advantage of O-antigen-based Salmonella vaccines is that they should offer cross-protection against other serovars within the same serogroup because all serovars in one serogroup express the same dominant O-antigen epitopes24 (group C1 serovars all express O:6,7 antigen13). Antigen level is an important factor influencing the magnitude of the immune response. To maximize the expression of the heterologous Salmonella Choleraesuis O-antigen, synthesis of the Salmonella Typhimurium O-antigen (Group B1) was blocked by deletion of either the rfbP gene or the whole O-antigen gene cluster. The pCZ1 plasmid containing the whole O-antigen gene cluster of Salmonella Choleraesuis was then introduced into the rough mutant strains. This strategy for gene deletion and introduction was based on the finding that the Group C1 O-antigen and its gene cluster are quite distinct from the Group B1 O-antigen13. As expected, the two recombinant strains, SLT11 (pCZ1), with deletion of the rfbP gene, and SLT12 (pCZ1), with deletion of the whole Salmonella Typhimurium O-antigen gene cluster, efficiently expressed the heterologous O-antigen, whereas unexpectedly, SLT11 (pCZ1) also expressed the homologous O-antigen, as indicated by the immunoblotting results (Fig. 1B and C, lanes 3 and 5). Because the genes for initiating O-unit synthesis in Groups C1 and B1 were different (wecA for C1 O-antigen and rfbP for B1 O-antigen)13 and the homologous O-antigen expression was not observed in SLT11 (pQK664) (Fig. 1B and C, lane 2), it was impossible to express the complete Salmonella Typhimurium O-antigen structure in SLT11 (pCZ1). According to the O-antigen structures and the required glycosyltransferase genes between Groups B1 and C113, we speculate that a branching sugar (abequose) was synthesized and attached to D-mannose in the backbone of heterologous O-antigen in SLT11 (pCZ1) by the abe and wbaV genes, which reside in the Salmonella Typhimurium O-antigen gene cluster, conferring immunodominant epitope 4 specificity15, which can be recognized by the Salmonella O:4 specific antisera that was employed. In contrast, the band specifying the Salmonella Typhimurium O-antigen was not observed in SLT12 (pCZ1) (Fig. 1B, lane 5) because the entire Salmonella Typhimurium O-antigen gene cluster was deleted. Therefore, it was possible that the SLT11 (pCZ1) expressed the O:4 epitope rather than the complete homologous O-antigen. Given that the antibodies specific to the homologous O-antigen play important roles in immune protection against Salmonella Typhimurium31, we constructed another recombinant strain, SLT16 (pCZ1), in which the heterologous O-antigen and complete homologous O-antigen were expressed when arabinose was available; the heterologous O-antigen and short-length homologous (probably the O:4 epitope) were expressed when grown without arabinose, similar to that displayed by the SLT11 (pCZ1) (Fig. 1D,E and F). Because arabinose is not present in tissues upon the invasion of Salmonella into the gut-associated lymphoid tissue40, the expression of complete homologous O-antigen was activated before the bacteria invaded these cells and ceased after invasion due to cell division.
The O-antigen is an important virulence factor, which is involved in bacterial motility, colonization and survival in the host29, 31. Loss of the entire O-antigen attenuates swimming motility and intestinal colonization and renders bacteria highly susceptible to antimicrobial agents. In our constructed recombinant strains SLT12 (pCZ1) and SLT16 (pCZ1), the wild-type Salmonella Typhimurium O-antigen was replaced by the heterologous O-antigen or the chimeric O-antigen. We determined whether the altered O-antigen structures influenced swimming motility, resistance to polymyxin B and DOC and colonization of Salmonella Typhimurium. The results showed that none of the examined phenotypes were affected by heterologous O-antigen expression (Fig. 2), indicating that O-antigen expression itself, rather than O-antigen variety, was vital for these Salmonella phenotypes. Additionally, the rough strains SLT12 (pQK664) and SLT16 (pQK664) (grown without arabinose) showed decreased swimming and increased sensitivity to polymyxin B and DOC, which was consistent with previous studies29. Surprisingly, no detectable bacteria were recovered from the PP, liver or spleen tissues 6 days after oral inoculation with STL12 (pQK664) (Fig. 2D,E and F). The attenuated colonization is consistent with previous results showing that deletion of genes responsible for O-antigen synthesis, such as rfaH and rfbP, results in rough LPS and decreased but not undetectable colonization31. This result suggested that deletion of the whole O-antigen cluster further attenuates colonization.
Next, the crp and cya genes, which have been identified as the mutation targets of live vaccines41, 42, were deleted from the recombinant and control strains for attenuation and to evaluate the immunogenicity and protective efficacy. The derived vaccine strains showed similar LPS phenotypes as their parent strains (Figs 3 and S1), suggesting that deletions of crp and cya do not affect LPS structure, in agreement with previous reports43. Both the recombinant vaccine strains SLT17 (pCZ1) and SLT18 (pCZ1) induced significantly higher serum IgG and fecal IgA responses against the homologous and heterologous LPS than the PBS control at 3 weeks post-second immunization (Fig. 4). The bactericidal effects exerted by sera induced by the two recombinant vaccine strains were comparable to those of the homologous S100 strain with the O:4-specific antisera (Fig. 5A) and the heterologous S340 strain with the O:7-specific antisera (Fig. 5B). The control strain SLT17 (pQK664) did not produce antibodies immunoreactive to either the homologous or heterologous LPS (Fig. 4), possibly due to a combination of the rough LPS phenotype (Supplementary Fig. S1) and undetectable colonization in mouse tissues (Supplementary Fig. S2); however, the serum from the SLT17 (pQK664)-immunized group mediated significantly increased killing of the homologous S100 strain compared with the PBS group. Combined with the finding that immunization with SLT17 (pQK664) induced low-level homologous protection (Fig. 6A), this result suggested that SLT17 (pQK664) was still immunogenic and induced a specific antibody response against other Salmonella Typhimurium antigens with high immunogenicity rather than the rough LPS. Furthermore, the other control strain, SLT18 (pQK664), which expressed the homologous O-antigen only when arabinose was present during in vitro growth, produced significant serum IgG and fecal IgA specific to the homologous LPS but not to the heterologous LPS (Fig. 4). In addition, serum produced from SLT18 (pQK664) exhibited killing against the homologous S100 strain but not the heterologous S340 strain (Fig. 5A and B), suggesting that there was less cross-immunoreactivity between serum antibodies specific to Salmonella Typhimurium and Salmonella Choleraesuis. However, SLT17 (pCZ1), which expressed only the heterologous O-antigen, elicited detectable anti-homologous LPS IgG and IgA (Fig. 4A and C), similar to the phenomenon that was observed in immunization experiments based on the Salmonella Typhimurium ΔrfbP mutant with a rough LPS phenotype31. We speculate that the antibodies detected were in response to the core region or lipid A of Salmonella Typhimurium LPS, which were contained in the ELISA coating antigens.
The potent antibody responses specific to the heterologous O-antigen and its functional bactericidal effects against Salmonella Choleraesuis induced by the two recombinant vaccines SLT17 (pCZ1) and SLT18 (pCZ1) demonstrated the high immunogenicity of the delivered heterologous O-antigen. To determine whether the induced antibody responses conferred heterologous protection, the protective efficacy was further investigated. The vaccine strains SLT17 (pCZ1), SLT18 (pQK664) and SLT18 (pCZ1) provided full homologous protection against lethal challenge with Salmonella Typhimurium (Fig. 6A), which was in line with the previous vaccine studies based on the crp/cya mutant and validated the immunization and challenge conditions of our study. The full homologous protection conferred by the SLT17 (pCZ1), which did not express the homologous O-antigen, was in line with the finding observed in previous studies, which showed that attenuated Salmonella Typhimurium ΔrfaH or ΔrmlB mutant with rough LPS phenotypes also exhibited protection to homologous lethal challenge30, 36. Combined with the finding that the serum induced by SLT17 (pCZ1) exhibited significant bactericidal effects to the homologous S100 strain, the result suggested that the O-antigen-mediated antibody response was not indispensable for preventing Salmonella infections, and immune responses specific for other Salmonella antigens, such as OMPs, support the efficacy of these live Salmonella vaccines with rough LPS phenotypes36. In contrast, the control SLT17 (pQK664) strain mediated inefficient homologous protection (Fig. 6A), which is likely due to the undetectable colonization (Supplementary Fig. S2) and the resulting poor immunogenicity. In comparison with the full homologous protection provided by SLT18 (pQK664), no heterologous protection was observed in the SLT18 (pQK664) control groups (Fig. 6B), indicating little cross-protection between Salmonella Typhimurium and Salmonella Choleraesuis. This finding is in agreement with the corresponding poor cross-immunoreactivity of serum antibodies between Salmonella Typhimurium and Salmonella Choleraesuis. Furthermore, the recombinant vaccines SLT17 (pCZ1) and SLT18 (pCZ1) resulted in 83% and 50% heterologous protection against lethal challenge with Salmonella Choleraesuis, respectively. The differences in the protection efficacy was correlated with differences in immunogenicity: SLT17 (pCZ1) immunization stimulated higher serum IgG and fecal IgA responses against the heterologous O-antigen and resulted in better killing of Salmonella Choleraesuis than SLT18 (pCZ1). We speculate that the higher amount of heterologous O-antigen expressed by SLT17 (pCZ1) than SLT18 (pCZ1) (Fig. 3) resulted in the differences in immunogenicity and protective efficacy.
Because only partial heterologous protection was achieved by the developed recombinant vaccines, considerable efforts should be made to improve the protective efficacy in future studies. One strategy is to increase the expression level of heterologous O-antigen, which might be achieved by constructing a new recombinant plasmid with a higher copy number, such as a pBR322-derived plasmid, or inserting the Salmonella Choleraesuis O-antigen gene cluster into a bacterial chromosome to replace the Salmonella Typhimurium O-antigen gene cluster, resulting in stable expression of heterologous O-antigen initiated by the native promoter39, 44. The other strategy is the construction of more effective recombinant Salmonella vectors via well-developed regulated delayed attenuation45. Importantly, our study provides a new approach: heterologous expression of O-antigen to develop multivalent Salmonella vaccines. Some issues remain to be addressed in subsequent studies, and these include whether Salmonella can simultaneously express two or more types of heterologous O-antigens, and ensuring the expression of sufficient amounts of heterologous O-antigens to stimulate a protective immune response when multiple O-antigens are expressed at the same time. Nevertheless, a mixture of several recombinant Salmonella strains, each of which express only one type of heterologous O-antigen, can be used as multivalent vaccine candidates and subjected to evaluations of immunogenicity and heterologous protection efficacy in future studies.
In summary, we developed two bivalent vaccines, SLT17 (pCZ1) and SLT18 (pCZ1), by stable recombinant expression of the heterologous Salmonella Choleraesuis O-antigen in attenuated Salmonella Typhimurium. Expression of the heterologous O-antigen had no adverse effects on bacterial swimming, resistance to polymyxin B and DOC or colonization. Immunization with SLT17 (pCZ1) and SLT18 (pCZ1) provided full protection against Salmonella Typhimurium infection and exhibited improved cross-protection against Salmonella Choleraesuis infection. The ability of the two vaccines to induce strong antibody-mediated immunity represents a promising step towards the development of live multivalent vaccines with broad serovar coverage.
Methods
Ethics statement
All animal experiments in this study were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China. All animal procedures were approved by the Animal Ethics Committee of the Sichuan Agricultural University.
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids utilized in this study are described in Table 1. Salmonella enterica and E. coli were grown in Luria-Bertani (LB) broth or on LB agar with or without 0.1% arabinose. When required, antibiotics and diaminopimelic acid (DAP) were added to the medium at the following concentrations: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; DAP, 50 μg/ml. LB agar containing 10% sucrose was used for sacB gene-based counter selection in the allelic exchange experiments. The transformation of Salmonella Typhimurium was performed via electroporation. Transformants were selected on LB agar plates containing the appropriate antibiotics.
Plasmids and mutant strain construction
The primers used to delete the rfbP gene have been described previously31. Other primers used in this study are listed in Supplementary Table S2. The introduction of gene mutations in Salmonella Typhimurium was carried out by allelic exchange using the suicide T-vector pYA4278 as previously described31. For deletion of the asd gene, the primers Dasd-1F/Dasd-1R and Dasd-2F/Dasd-2R were used to amplify approximately 450 bp upstream and downstream segments from the S100 genome, respectively. The two fragments were then joined by PCR using primers Dasd-1F and Dasd-2R. The PCR product was then ligated to AhdI-digested pYA4278 to generate plasmid pCZ2, which was introduced into Salmonella Typhimurium S100 for the asd deletion mutation. The selection and characterization of the asd mutants were carried out by PCR using the primers Dasd-1F and Dasd-2R. The same method was applied to delete the rfbP, pagL, crp and cya genes and the whole O-antigen gene cluster (rmlB-rfbP). To construct the arabinose-regulated mutants, the primers rfbP-F and rfbP-R were used to amplify the rfbP gene from the S100 genome, and the PCR product was then inserted into plasmid pSS24536 between the NheI and KpnI sites, generating the pCZ10 plasmid. The primers TT-F and rfbP-R were then used to amplify the TTaraCPBAD rfbP fragment from pCZ10. The product was inserted into NotI and SbfI double-digested pCZ5 (pYA4278-ΔpagL), generating the plasmid pCZ8, which was introduced into the strain SLT13 (Δasd ΔrfbP ∆pagL) for TTaraCPBAD rfbP insertion in the pagL gene site, generating the strain SLT16 (Δasd ΔrfbP ∆pagL::TT araC PBAD rfbP).
To express the heterologous O-antigen in Salmonella Typhimurium mutant strains, the recombinant plasmid pCZ1 was constructed. The primer pair C1-O-antigen-F and C1-O-antigen-R was used to amplify the whole O-antigen gene cluster (8870 bp) of Salmonella Choleraesuis from the S340 genome, and the primers pQK664-C1F and pQK664-C1R were used to amplify a DNA fragment containing the pSC101 origin, asd gene cassette, TIT2 terminator, Ptrc promoter and kanamycin-resistance cassette from the plasmid pQK66433. Next, the two PCR products were joined by the Gibson Assembly Kit (NEB, Beverley, MA, USA), generating the plasmid pCZ1. Then, the pCZ1 or the control plasmid pQK664 was transformed into Salmonella Typhimurium SLT11 (Δasd ΔrfbP), SLT12 (Δasd ΔrmlB-rfbP) and SLT16 (Δasd ΔrfbP ∆pagL::TT araCPBAD rfbP), generating the three recombinant strains SLT11 (pCZ1), SLT12 (pCZ1) and SLT16 (pCZ1) and the three control strains SLT11 (pQK664), SLT12 (pQK664) and SLT16 (pQK664).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), silver staining and Western immunoblotting
To confirm the expression of Salmonella Typhimurium and Salmonella Choleraesuis O-antigens, SDS-PAGE, silver staining and Western blot analyses were performed as previously described46, 47. In brief, each bacterial strain was cultured and adjusted to an OD600 = 1.0. Two microliters of each suspension were then centrifuged and resuspended in 100 ml of lysis buffer containing proteinase K. Ten microliters of the solution were then separated by SDS-PAGE in a 12% (w/v) acrylamide gel and then silver-stained or subjected to Western immunoblotting with Salmonella O:4- or O:7-specific antisera diluted to 1:200 (Tianjin Biochip Corporation, Tianjin, China).
Detection of bacterial biological activities
The bacteria were grown to an OD600 = 0.8–0.9 in LB broth with or without 0.1% arabinose, harvested and resuspended in LB. For the swimming assay, 6 μl of a bacterial suspension (approximately 1 × 106 CFU) was spotted onto the middle of LB plates containing 0.3% agar with or without arabinose. The diameter of the colonies was measured 6 h after incubation at 37 °C. To measure the resistance to polymyxin B and DOC, 100 μl of the cell suspension (approximately 1 × 106–5 × 106 CFU) was inoculated with or without polymyxin B at a final concentration of 0.12 μg/ml or DOC at a final concentration of 4 mg/ml for 1 h at 37 °C. The bacteria were diluted to the appropriate concentration and plated onto LB plates. The survival rate was calculated as the CFU per ml of the polymyxin B- or DOC-treated group divided by the CFU of the non-treated group.
Determination of bacterial colonization and virulence (LD50) in mice
Six-week-old female BALB/c mice were obtained from Dashuo Experimental Animal Ltd. (Chengdu, China). The mice were acclimated for 7 days after arrival before the experiments were initiated. The colonization and the LD50 of Salmonella Typhimurium strains were measured as previously described31. In brief, for the colonization experiments, BALB/c mice were inoculated orally with 1 × 109 CFU of each strain. Six days after oral inoculation, tissues, including PP, spleen and liver, were collected from three animals, weighed and homogenized in 1 ml of PBS. Then, appropriate dilutions were plated onto MacConkey agar or LB agar to determine the number of viable bacteria. The colonization value was calculated as CFU per gram of tissue (CFU/g). For determination of the LD50, stepwise increasing doses of Salmonella Typhimurium strains were orally inoculated into groups of BALB/c mice. The mortality of the animals was monitored over a period of 1 month after infection. The LD50 was calculated using the method of Reed and Muench.
Immunization and challenge
The Salmonella vaccine strains SLT17 (pQK664), SLT17 (pCZ1), SLT18 (pQK664) and SLT18 (pCZ1) were grown statically overnight in LB broth (0.1% arabinose was added for culture of the arabinose-regulated vaccine strains) at 37 °C. The next day, the overnight culture was diluted 1:100 into 100 ml of LB broth (0.1% arabinose was added for culture of the arabinose-regulated vaccine strains) and grown with shaking at 37 °C to an OD600 value of 0.8 to 0.9. The cells were harvested by centrifugation and resuspended in 1 ml of phosphate buffer saline (PBS). Groups of mice (12 mice per group) were orally inoculated with 20 µl of PBS containing 1 × 109 CFU of each strain or with PBS without bacteria twice at an interval of 4 weeks. The serum was collected three weeks after each immunization. The feces used for the analysis of fecal IgA were collected three weeks after the second immunization and suspended in 100 µl of PBS, and the supernatant was collected after centrifugation at 12,000 × g and 4 °C for 5 min. One month post-second immunization, mice in each immunization group were orally challenged with at least 100 × LD50 of the Salmonella Typhimurium virulent strain S100 and the Salmonella Choleraesuis virulent strain S340, respectively. The challenged mice were monitored, and mortality was recorded daily for 15 days.
ELISA
Salmonella Typhimurium LPS was purchased from Sigma (St. Louis, MO, USA), and Salmonella Choleraesuis LPS was extracted and purified as described48. The serum IgG concentration was determined by quantitative ELISA, and the fecal IgA was measured by indirect ELISA, as previously described36, 49. In brief, a 96-well ELISA microplate was coated with 100 ng/well LPS or 100 ng/well goat anti-mouse Ig(H + L) (BD, San Diego, CA, USA) and then blocked with 2% BSA (BD) after overnight incubation at 4 °C. For the detection of serum IgG, the serum samples were diluted to 1:100 in PBS containing 2% BSA, and 100 μl of this solution was added to the LPS-coated wells in duplicate. Meanwhile, 100 μl of mouse IgG (BD) with serial two-fold dilutions from 0.5 mg/ml were added to the goat anti-mouse Ig(H + L)-coated wells for the standard curve. For the detection of fecal IgA, the fecal supernatant was diluted 1:4 and added to the LPS-coated wells in duplicate. After 1 h of incubation at 37 °C, 100 μl of biotinylated goat anti-mouse IgG (Southern Biotech, Birmingham, AL, USA) or biotinylated goat anti-mouse IgA (Southern Biotech) and 100 μl of streptavidin-alkaline phosphatase conjugate (Southern Biotech) were added to each well sequentially, followed by p-nitrophenylphosphate substrate in diethanolamine buffer. Finally, the plate was read at 405 nm using a microplate reader (Bio-Rad Laboratories, California, USA). The standard curve was drawn using Curve Expert (Hyams DG, Starkville, MS, USA), and the concentration of serum IgG antibodies was calculated according to the standard curve.
Serum bactericidal assay
The serumcollected from mice of each immunized group at week 3 post-second immunization was pooled for the serum bactericidal assay as described previously50. The commercial Salmonella O:4-specific and O:7-specific antisera were used as positive controls. All serum samples were heated at 56 °C for 30 min to inactivate endogenous complement. Salmonella Typhimurium S100 or Salmonella Choleraesuis S340 were grown in LB medium to log phase and were diluted in SBA buffer (50 mM phosphate, 0.041% MgCl2·6H2O, 33 mg/ml CaCl2, 0.5% BSA) to approximately 3 × 103 CFU/ml. The bacteria were incubated with 25% heat-inactivated serum supplemented with or without active guinea pig complement (Sigma) for 1.5 h. The relative survival was calculated as the percent CFU counted in each pooled serum with active complement compared to the CFU of the same serum with no complement. Each sample and control was tested in triplicate.
Statistical analysis
The data are shown as the means ± SD. One-way ANOVA followed by Tukey’s multiple-comparison test were used to evaluate statistical significance. A probability value of P < 0.05 was considered statistically significant. The survival curve post-challenge was analyzed using the log-rank test. All in vitro experiments were repeated at least three times, and the in vivo experiments were repeated at least twice.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Feasey, N. A., Dougan, G., Kingsley, R. A., Heyderman, R. S. & Gordon, M. A. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379, 2489–2499, doi:10.1016/S0140-6736(11)61752-2 (2012).
Zhang, J. et al. Serovars and antimicrobial resistance of non-typhoidal Salmonella from human patients in Shanghai, China, 2006–2010. Epidemiology and infection 142, 826–832, doi:10.1017/S0950268813001659 (2014).
Majowicz, S. E. et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 50, 882–889, doi:10.1086/650733 (2010).
MacLennan, C. A. & Levine, M. M. Invasive nontyphoidal Salmonella disease in Africa: current status. Expert review of anti-infective therapy 11, 443–446, doi:10.1586/eri.13.27 (2013).
Maclennan, C. A. Out of Africa: links between invasive nontyphoidal Salmonella disease, typhoid fever, and malaria. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 58, 648–650, doi:10.1093/cid/cit803 (2014).
Sandt, C. H. et al. A comparison of non-typhoidal Salmonella from humans and food animals using pulsed-field gel electrophoresis and antimicrobial susceptibility patterns. PloS one 8, e77836, doi:10.1371/journal.pone.0077836 (2013).
Sanchez-Vargas, F. M., Abu-El-Haija, M. A. & Gomez-Duarte, O. G. Salmonella infections: an update on epidemiology, management, and prevention. Travel medicine and infectious disease 9, 263–277, doi:10.1016/j.tmaid.2011.11.001 (2011).
Centers for Disease, C. & Prevention. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food - 10 states, 2009. MMWR. Morbidity and mortality weekly report 59, 418–422 (2010).
Galanis, E. et al. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerging infectious diseases 12, 381–388, doi:10.3201/eid1205.050854 (2006).
Chiu, C. H., Su, L. H. & Chu, C. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clinical microbiology reviews 17, 311–322 (2004).
Hall, R. M. Salmonella genomic islands and antibiotic resistance in Salmonella enterica. Future microbiology 5, 1525–1538, doi:10.2217/fmb.10.122 (2010).
Liang, Z. et al. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009–2012. BMC infectious diseases 15, 53, doi:10.1186/s12879-015-0784-4 (2015).
Liu, B. et al. Structural diversity in Salmonella O antigens and its genetic basis. FEMS microbiology reviews 38, 56–89, doi:10.1111/1574-6976.12034 (2014).
Brenner, F. W., Villar, R. G., Angulo, F. J., Tauxe, R. & Swaminathan, B. Salmonella nomenclature. Journal of clinical microbiology 38, 2465–2467 (2000).
Simon, R. & Levine, M. M. Glycoconjugate vaccine strategies for protection against invasive Salmonella infections. Human vaccines & immunotherapeutics 8, 494–498, doi:10.4161/hv.19158 (2012).
Colwell, D. E., Michalek, S. M., Briles, D. E., Jirillo, E. & McGhee, J. R. Monoclonal antibodies to Salmonella lipopolysaccharide: anti-O-polysaccharide antibodies protect C3H mice against challenge with virulent Salmonella typhimurium. J Immunol 133, 950–957 (1984).
Michetti, P., Mahan, M. J., Slauch, J. M., Mekalanos, J. J. & Neutra, M. R. Monoclonal secretory immunoglobulin A protects mice against oral challenge with the invasive pathogen Salmonella typhimurium. Infect Immun 60, 1786–1792 (1992).
Forbes, S. J. et al. Association of a protective monoclonal IgA with the O antigen of Salmonella enterica serovar Typhimurium impacts type 3 secretion and outer membrane integrity. Infect Immun 80, 2454–2463, doi:10.1128/IAI.00018-12 (2012).
Simon, R. et al. Sustained protection in mice immunized with fractional doses of Salmonella Enteritidis core and O polysaccharide-flagellin glycoconjugates. PloS one 8, e64680, doi:10.1371/journal.pone.0064680 (2013).
Simon, R. et al. Salmonella enterica serovar enteritidis core O polysaccharide conjugated to H:g,m flagellin as a candidate vaccine for protection against invasive infection with S. enteritidis. Infect Immun 79, 4240–4249, doi:10.1128/IAI.05484-11 (2011).
Watson, D. C., Robbins, J. B. & Szu, S. C. Protection of mice against Salmonella typhimurium with an O-specific polysaccharide-protein conjugate vaccine. Infect Immun 60, 4679–4686 (1992).
Micoli, F. et al. A scalable method for O-antigen purification applied to various Salmonella serovars. Analytical biochemistry 434, 136–145, doi:10.1016/j.ab.2012.10.038 (2013).
Svenson, S. B., Nurminen, M. & Lindberg, A. A. Artificial Salmonella vaccines: O-antigenic oligosaccharide-protein conjugates induce protection against infection with Salmonella typhimurium. Infect Immun 25, 863–872 (1979).
MacLennan, C. A., Martin, L. B. & Micoli, F. Vaccines against invasive Salmonella disease: current status and future directions. Human vaccines & immunotherapeutics 10, 1478–1493, doi:10.4161/hv.29054 (2014).
Hassan, J. O. & Curtiss, R. 3rd Development and evaluation of an experimental vaccination program using a live avirulent Salmonella typhimurium strain to protect immunized chickens against challenge with homologous and heterologous Salmonella serotypes. Infect Immun 62, 5519–5527 (1994).
Dueger, E. L., House, J. K., Heithoff, D. M. & Mahan, M. J. Salmonella DNA adenine methylase mutants elicit protective immune responses to homologous and heterologous serovars in chickens. Infect Immun 69, 7950–7954, doi:10.1128/IAI.69.12.7950-7954.2001 (2001).
Roland, K. L. & Brenneman, K. E. Salmonella as a vaccine delivery vehicle. Expert review of vaccines 12, 1033–1045, doi:10.1586/14760584.2013.825454 (2013).
Wang, S., Kong, Q. & Curtiss, R. 3rd New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microbial pathogenesis 58, 17–28, doi:10.1016/j.micpath.2012.10.006 (2013).
Toguchi, A., Siano, M., Burkart, M. & Harshey, R. M. Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. Journal of bacteriology 182, 6308–6321 (2000).
Nagy, G. et al. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect Immun 74, 5914–5925, doi:10.1128/IAI.00619-06 (2006).
Kong, Q. et al. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect Immun 79, 4227–4239, doi:10.1128/IAI.05398-11 (2011).
Galan, J. E., Nakayama, K. & Curtiss, R. 3rd Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 94, 29–35 (1990).
Xiao, K. et al. Identification of the Avian Pasteurella multocida phoP Gene and Evaluation of the Effects of phoP Deletion on Virulence and Immunogenicity. Int J Mol Sci 17, doi:10.3390/ijms17010012 (2016).
Kong, Q., Liu, Q., Jansen, A. M. & Curtiss, R. 3rd Regulated delayed expression of rfc enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Vaccine 28, 6094–6103, doi:10.1016/j.vaccine.2010.06.074 (2010).
Wang, S. et al. Salmonella vaccine vectors displaying delayed antigen synthesis in vivo to enhance immunogenicity. Infect Immun 78, 3969–3980, doi:10.1128/IAI.00444-10 (2010).
Huang, C. et al. Regulated delayed synthesis of lipopolysaccharide and enterobacterial common antigen of Salmonella Typhimurium enhances immunogenicity and cross-protective efficacy against heterologous Salmonella challenge. Vaccine 34, 4285–4292, doi:10.1016/j.vaccine.2016.07.010 (2016).
Simon, R., Tennant, S. M., Galen, J. E. & Levine, M. M. Mouse models to assess the efficacy of non-typhoidal Salmonella vaccines: revisiting the role of host innate susceptibility and routes of challenge. Vaccine 29, 5094–5106, doi:10.1016/j.vaccine.2011.05.022 (2011).
Moustafa, D. A. et al. Recombinant Salmonella Expressing Burkholderia mallei LPS O Antigen Provides Protection in a Murine Model of Melioidosis and Glanders. PloS one 10, e0132032, doi:10.1371/journal.pone.0132032 (2015).
Dharmasena, M. N., Hanisch, B. W., Wai, T. T. & Kopecko, D. J. Stable expression of Shigella sonnei form I O-polysaccharide genes recombineered into the chromosome of live Salmonella oral vaccine vector Ty21a. International journal of medical microbiology: IJMM 303, 105–113, doi:10.1016/j.ijmm.2013.01.001 (2013).
Kong, W. et al. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proceedings of the National Academy of Sciences of the United States of America 105, 9361–9366, doi:10.1073/pnas.0803801105 (2008).
Tacket, C. O. et al. Comparison of the safety and immunogenicity of delta aroC delta aroD and delta cya delta crp Salmonella typhi strains in adult volunteers. Infect Immun 60, 536–541 (1992).
Chu, C. Y. et al. Heterologous protection in pigs induced by a plasmid-cured and crp gene-deleted Salmonella choleraesuis live vaccine. Vaccine 25, 7031–7040, doi:10.1016/j.vaccine.2007.07.063 (2007).
Roland, K., Curtiss, R. 3rd & Sizemore, D. Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis 43, 429–441 (1999).
Lin, I. Y., Van, T. T. & Smooker, P. M. Live-Attenuated Bacterial Vectors: Tools for Vaccine and Therapeutic Agent Delivery. Vaccines 3, 940–972, doi:10.3390/vaccines3040940 (2015).
Curtiss, R. 3rd et al. Salmonella enterica serovar typhimurium strains with regulated delayed attenuation in vivo. Infect Immun 77, 1071–1082, doi:10.1128/IAI.00693-08 (2009).
Hitchcock, P. J. & Brown, T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. Journal of bacteriology 154, 269–277 (1983).
Xu, D. Q., Cisar, J. O., Ambulos, N. Jr., Burr, D. H. & Kopecko, D. J. Molecular cloning and characterization of genes for Shigella sonnei form I O polysaccharide: proposed biosynthetic pathway and stable expression in a live salmonella vaccine vector. Infect Immun 70, 4414–4423 (2002).
Rezania, S. et al. Extraction, Purification and Characterization of Lipopolysaccharide from Escherichia coli and Salmonella typhi. Avicenna journal of medical biotechnology 3, 3–9 (2011).
Zhu, L. et al. Mucosal IgA and IFN-gamma+ CD8 T cell immunity are important in the efficacy of live Salmonella enteria serovar Choleraesuis vaccines. Scientific reports 7, 46408, doi:10.1038/srep46408 (2017).
Rondini, S. et al. Design of glycoconjugate vaccines against invasive African Salmonella enterica serovar Typhimurium. Infect Immun 83, 996–1007, doi:10.1128/IAI.03079-14 (2015).
Liu, Q. et al. Immunogenicity and Cross-Protective Efficacy Induced by Outer Membrane Proteins from Salmonella Typhimurium Mutants with Truncated LPS in Mice. Int J Mol Sci 17, 416, doi:10.3390/ijms17030416 (2016).
Acknowledgements
We thank Sheng Liang, Pei Li, Hai-Yan Du, Xin-Yu Lei and Yu-Xin Liu in our laboratory for generous assistance during the study. This research was supported by the National Natural Science Foundation of China (31502106), the Applied Basic Research Programs of Science and Technology Department of Sichuan Province (2015JY0244), the National Natural Science Foundation of China (31570928) and the National Science and Technology Support Program (2015BAD12B05).
Author information
Authors and Affiliations
Contributions
R.-Y.J., Q.-K.K. and X.-X.Z. conceived and designed the experiments in detail. X.-X.Z. and Q.-L.D. performed the research. D.-K.Z., M.-F.L., and S.C. participated in the animal experiments. K.-F.S., Y.Q. and Y.W. contributed reagents and analysis tools. X.-X.Z. analyzed the data and wrote the paper, and R.-Y.J. revised the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, X., Dai, Q., Zhu, D. et al. Recombinant attenuated Salmonella Typhimurium with heterologous expression of the Salmonella Choleraesuis O-polysaccharide: high immunogenicity and protection. Sci Rep 7, 7127 (2017). https://doi.org/10.1038/s41598-017-07689-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07689-5
This article is cited by
-
Immunogenicity and protection of a Pasteurella multocida strain with a truncated lipopolysaccharide outer core in ducks
Veterinary Research (2022)
-
Protective effects of a food-grade recombinant Lactobacillus plantarum with surface displayed AMA1 and EtMIC2 proteins of Eimeria tenella in broiler chickens
Microbial Cell Factories (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.