Abstract
Host diet is a major determinant of the composition and function of the intestinal microbiome. Less understood is the importance of diet on ingested strains with probiotic significance. We investigated the population dynamics of exogenous Lactobacillus plantarum and its interactions with intestinal bacteria in mice undergoing switches between high-fat, high-sugar (HFHSD) and low-fat, plant-polysaccharide rich (LFPPD) diets. The survival and persistence of ingested L. plantarum WCFS1 was significantly improved during mouse consumption of HFHSD and was negatively associated with the numbers of indigenous Lactobacillus species. Diet also rapidly changed the composition of the indigenous microbiota, but with some taxa differentially affected between HFHSD periods. L. plantarum was not integrated into indigenous bacterial community networks according to co-occurrence patterns but still conferred distinct effects on bacterial species diversity and microbiota stability largely in a diet-dependent manner. Metagenome predictions supported the premise that L. plantarum dampens the effects of diet on the microbiome. This strain also consistently altered the predicted genetic content in the distal gut by enriching for genes encoding glyosyltransferases and bile salt hydrolases. Our findings demonstrate the interactions between ingested, transient probiotic bacteria and intestinal bacterial communities and how they can differ depending on host diet.
Similar content being viewed by others
Introduction
The human gastrointestinal (GI) tract is a microbial ecosystem subject to tremendous selective pressures from the host epithelium and immune and endocrine systems as well as dietary inputs comprised of a vast assortment of carbohydrates, proteins, fats, vitamins, toxins/anti-nutrients, and microorganisms. Although the indigenous bacterial colonists of the GI tract show remarkable stability, these organisms are still vulnerable to GI perturbations and “sweeps” due to external insults. Disease, antibiotics, and some medications can each result in rapid shifts to the gut microbiome as well as sustained, long-term modifications to microbial composition and function1,2,3. Similarly, alterations to the abundant macronutrients of the diet confer global changes to the intestinal community structure4, 5. Studies performed in rodent models have shown that the intestinal microbiota responds robustly to diet perturbations independent of host genotype and that the abundance of certain taxa can reflect previous diet exposures6. Maternal diets high in fat and low in microbiota-accessible carbohydrates can also result in bacterial extinctions over several generations that cannot be restored through diet alone7.
Less understood is the role of host diet on the GI fitness of transient bacteria consumed in foods and beverages. For foodborne pathogens, the intestinal microbiota can influence the capacity of those organisms to cause disease in a diet-dependent manner8,9,10. For example, colonization and virulence of enterohaemorrhagic Escherichia coli was increased in a high-fiber diet background that stimulated intestinal butyrate synthesis11. The importance of host diet on the fitness of commensal or probiotic bacteria in the GI tract was shown with the mono-association of Lactobacillus plantarum WCFS1 in germ-free mice12. L. plantarum colonized the GI tract in significantly higher quantities when the mice were fed a low-fat plant-polysaccharide rich diet (LFPPD) as opposed to a diet high in fat and sugar (HFHSD). The numbers of L. plantarum corresponded well to the transcriptome of this organism in the cecum of mice fed the HFHSD, which showed evidence of nutrient starvation (e.g. down-regulation of genes required for metabolism and DNA replication and up-regulation of genes for amino acid biosynthesis and transport)12. Remarkably, the opposite result was reached in conventionally raised mice, whereby HFHSD consumption resulted in higher numbers of L. plantarum surviving GI tract transit13. This occurred even though there was still evidence of nutrient starvation among L. plantarum in HFHSD-fed animals. The mice also responded differently in that colonic IL-10 levels were higher and trinitrobenzene sulfate (TNBS)-induced colitis was significantly attenuated when L. plantarum was administered in the background of the HFHSD compared to the LFPPD13.
A primary difference between germ-free and conventionally raised mice is the presence of an indigenous microbiome. Therefore, the survival and persistence of L. plantarum in those studies might have been due to ecological constraints resulting from a fully colonized digestive tract. In that regard, the composition of the cecal microbiota was also significantly different between mice on the LFPPD and HFHSD13. Among the diet-induced differences, Lactobacillus species were severely and significantly depleted with a HFHSD13. Reduced Lactobacillus numbers were also found for other rodent studies in which obesogenic diets were used14, 15. Reductions in certain Lactobacillus species were also similarly observed in some human dietary studies in which a high-fat, refined sugar diet was consumed16, whereas other studies obtained the opposite result17, 18. However, it remains to be determined the extent to which diet-associated, intestinal behaviors of ingested and indigenous Lactobacillus species are reversible and whether transient Lactobacillus populations are integrated within the indigenous microbiome.
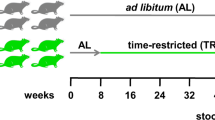
In this study, we hypothesized that the open ecological niche created for ingested L. plantarum by a HFHSD is reversible and that L. plantarum interactions with the intestinal microbiome are diet dependent. To investigate these possibilities, we exposed conventionally raised mice to dietary switches between a HFHSD and LFPPD with/without the addition of L. plantarum (Fig. 1a). The survival of L. plantarum and its associations with indigenous intestinal bacteria were investigated.
L. plantarum survived in higher numbers and persisted for longer periods in mice on HFHSD. (a) Experimental design of the mouse study. Mice starting on day 0 were switched from standard rodent chow to a HFHSD for 12 days (HFHSD1, blue), to a LFPPD for 14 days (green), and then back again to the HFHSD for a final 12 days (HFHSD2, red). L. plantarum WCFS1-R was fed orally during the last five consecutive days for each diet period (open circles). Mouse stools were collected at time points indicated in the figure for either L. plantarum enumeration (hatched circles) or 16S rRNA gene sequencing and qPCR (filled circles). (b) Culturable L. plantarum WCFS1-R in mouse stools 24 h after feeding as determined by colony enumeration. Rifampicin-resistant colonies were not detected in fecal samples collected from PBS-fed mice. The lower limit of detection was log10 3.5 CFU/g stool. Data of total 8 mice for 5 consecutive days is shown. **P < 0.01, Mann-Whitney U test. (c) L. plantarum WCFS1-R persistence in the mouse feces after the HFHSD1 period (during LFPPD consumption) and after the LFPPD period (during HFHSD2 consumption), as determined by CFU enumeration.
Results
HFHSD consistently increases the survival and persistence of L. plantarum in the GI tract
Viable L. plantarum was recovered in similar numbers from all stools 3 and 5 h after the first administration of L. plantarum WCFS1-R during each of the dietary periods (Supplementary Fig. S1a). The following day and then 24 h after each of the remaining 4 days of L. plantarum feeding showed this organism survived in significantly higher numbers in the GI tract of mice fed the HFHSD compared to LFPPD (Fig. 1b). Viable L. plantarum was recovered in 10-fold greater quantities during both HFHSD periods than during the LFPPD period (Fig. 1b). These results were supported by qPCR assessments of total L. plantarum in the stools which showed an even greater increase (up to 100-fold) when the strain was administered with the HFHSD as opposed to the LFPPD (Supplementary Fig. S1b).
Diet also affected the intestinal persistence of L. plantarum. This strain was still detected in the stools of five mice (out of eight) three days after the diet switch from LFPPD to HFHSD2 (Fig. 1c). Two mice still harbored the WCFS1-R strain six days later (Fig. 1c), and this result was not due to a “cage effect”. By comparison, L. plantarum was absent from all stools collected three days after the mice were switched from the HFHSD1 to the LFPPD (Fig. 1c).
Murine gut microbiota responds discordantly to diet switches
The diet shifts resulted in rapid and global changes to the composition of the murine GI microbiota. Principal coordinate analysis of the weighted UniFrac matrix showed a distinct separation in the fecal bacteria according to the diet being consumed (47.7% variance, HFHSD vs. LFPPD) and in a manner that was independent of L. plantarum WCFS1-R consumption (Supplementary Fig. S2). Bacteroidetes reached as low as 6.5% of the total intestinal microbiota during the HFHSD periods. These quantities were significantly lower (P < 0.0001) than at the start of the study (baseline) (38.2 ± 3.9%) and during the LFPPD (41.4% ± 2.2%) (Fig. 2a). Conversely, Firmicutes constituted 89.4% and 86.6% of the stool microbiota during HFHSD1 and HFHSD2, respectively. These percentages were significantly higher compared to levels at the start of the study and during LFPPD consumption (P < 0.0001) (Fig. 2b).
There was also evidence of discordant responses among the microbiota to the two HFHSD periods in both sham and L. plantarum – fed mice. During HFHSD1, mice harbored significantly higher proportions of the Firmicutes taxa Enterococcus and Erysipelotrichaceae relative to either LFPPD or HFHSD2 consumption (Fig. 3). During the HFHSD2 period, other Firmicutes in the Dorea, [Ruminococcus] (Lachnospiraceae family), and Anaerotruncus genera were enriched (Fig. 3) as well as Proteobacteria (Fig. 2c).
Taxonomic cladogram comparing the fecal microbiota response to different diets. Panel (a) and (b) correspond to the results from sham-treated and L. plantarum WCFS1-R fed mice respectively. Blue, green and red nodes are bacterial taxa significantly increased during the first HFHSD (HFHSD1), LFPPD and the second HFHSD (HFHSD2). Yellow nodes indicate non-significant changes in certain bacterial taxa in response to diet.
L. plantarum confers modest changes to the intestinal microbiota composition
Ingesting L. plantarum WCFS1-R resulted in alterations to the intestinal microbiota that were more limited in scope compared to the introduction of a new diet. Notable, however, were the altered alpha-diversity indices during L. plantarum and LFPPD consumption. The Shannon diversity index increased (data not shown) and significantly higher numbers of observed species were detected in the L. plantarum fed mice (Supplementary Fig. S3).
The proportions of some bacterial taxa were also modified between the L. plantarum and sham-fed controls. During HFHSD1, the relative abundance of Streptococcus was significantly higher in mice fed L. plantarum (Fig. 3). Conversely, during the LFPPD period, sham (but not L. plantarum-fed) mice were enriched with Tenericutes (Fig. 2d) and Bacteroides (Fig. 3), the former of which was enriched in sham (control)-fed animals even prior to the introduction of L. plantarum during that dietary period. Fewer changes to the gut microbiota as a consequence of L. plantarum ingestion were also found during HFHSD2. Only the sham control mice harbored elevated levels of Mucispirillum (Deferribacteres phylum), Christensenellaceae and Oscillospira (Firmicutes phylum) as well as Bilophila, a genus of sulfite-reducing bacteria in Proteobacteria phylum (Fig. 3).
Diet switches and L. plantarum alter the diversity and proportions of indigenous lactobacilli in the mouse intestine
The proportions of Lactobacillus in the mouse stools ranged from 7.3% to 37.6% of the total bacteria present in all mice. Opposite to L. plantarum WCFS1-R, the levels of indigenous Lactobacillus species (encompassing all OTUs assigned to that genus except for L. plantarum) were lowest during HFHSD consumption and increased with the LFPPD (Fig. 4). These numbers did not change with the onset of L. plantarum feeding. To this end, L. plantarum WCFS1-R did not either positively or negatively influence the levels of the indigenous Lactobacillus present. Instead, when L. plantarum was given to the animals during the two HFHSD periods, L. plantarum constituted a higher percentage of the total Lactobacillus population (41.9% and 55.9% for HFHSD1 and HFHSD2 respectively) than during the LFPPD (1.4%) (Fig. 5b).
Proportions of intestinal Lactobacillus species changed over time during diet switches. Panel (a) and (b) correspond to the results from sham-treated and L. plantarum WCFS1-R fed mice respectively. OTUs sharing 100% sequence similarity with its nearest neighbors are shown. Average values of all 8 mice in each group were plotted. Black dashes indicate the days when the mice received L. plantarum WCFS1-R.
Network analysis showed various interactions among L. plantarum WCFS1-R, indigenous Lactobacillus and other gut microbes. Intestinal microbiota during L. plantarum WCFS1-R consumption on HFHSD1 (day 10, day 12) (a), LFPPD (day 24, day 26) (b) and HFHSD2 (day 38, day 39) (c) were analyzed. Each node represents one genus with the color indicating the phylum to which the genus belongs and the size of the node proportional to the relative abundance of that organism in the bacterial community (only taxa with the average abundance over 0.001 are shown). The line between nodes indicates the Spearman correlation and the color intensity indicates the correlation coefficient (red, positively correlated and green, negatively correlated). The weight of the line corresponds to the correlation significance. Taxa in brackets are based on annotations suggested by the Greengenes database.
BLAST searches of the prominent Lactobacillus OTUs present in the mouse stools identified L. plantarum as well as other taxa highly related to L. murinus, L. reuteri, and L. gasseri and here referred to as species clusters (Supplementary Fig. S4). Lactobacilli in the L. murinus cluster were predominant (Fig. 4 and Supplementary Fig. S5a). In the sham-fed mice, the L. murinus cluster increased from 19.3% to 26.8% and then down to 8.8% of the total intestinal bacteria as the diet changed from HFHSD1 to LFPPD and then to HFHSD2 (P = 0.1 HFHSD1 vs. LFPPD; P < 0.0001 LFPPD vs. HFHSD2) (Fig. 4a). Quantification of L. murinus cell numbers by qPCR gave similar results (Supplementary Fig. S5a). These values were also similar in mice fed L. plantarum WCFS1-R (Fig. 4b and Supplementary Fig. S5a).
Bacteria highly related to L. reuteri were the second most abundant Lactobacillus species cluster in the mice (Fig. 4 and Supplementary Fig. S5b). A rapid and significant 250-fold reduction in L. reuteri occurred during the first HFHSD period compared to baseline (day 0) for all mice (Supplementary Fig. S5b). Similar to L. murinus, when the animals were fed the LFPPD, the quantities of L. reuteri increased back to levels present at the start of the study (P < 0.0001 LFPPD vs. HFHSD1). The quantities declined again when mice were given the HFHSD for the second time (P < 0.0001 HFHSD2 vs. LFPPD) (Supplementary Fig. S5b).
Lastly, proportions of the L. gasseri cluster were also diet-dependent and significantly reduced with the HFHSD in all mice compared to baseline levels (P < 0.0001 baseline vs. HFHSD1) (Fig. 4 and Supplementary Fig. S5c). However, during LFPPD consumption, the quantities of the L. gasseri cluster only increased in those mice fed L. plantarum (P < 0.0005 HFHSD1 vs LFPPD) (Supplementary Fig. S5c). Once the HFHSD2 was introduced, the numbers of L. gasseri cluster declined again (P < 0.0001 LFPPD vs HFHSD2), even with L. plantarum WCFS1-R administration (Supplementary Fig. S5c).
L. plantarum WCFS1-R has limited interactions with intestinal bacteria
We next employed network analysis to identify interactions between the mouse microbiota that changed with diet and ingestion of L. plantarum. Both diet-independent and –dependent co-occurrence patterns were found. Clostridiales (or Clostridiaceae) and Coprococcus were positively correlated in all dietary periods (r = 0.72, 0.76, 0.9 respectively), indicating the associations between these taxa are independent of the host diet (Fig. 5). Diet-dependent interactions included the positive correlations between Rikenellaceae and S24-7 (r = 0.75 and 0.7 for HFHSD1 and HFHSD2, respectively) and Bacteroidales and S24-7 (r = 0.83 and 0.9 for HFHSD1 and HFHSD2, respectively) occurring during both HFHSD periods (Fig. 5a and c). Co-occurrence patterns between native Lactobacillus species and other bacterial taxa also varied according to diet. Positive correlations were identified between Lactobacillus and Streptococcus during LFPPD and HFHSD2 (LFPPD; r = 0.78; HFHSD2, r = 0.81) and Lactobacillus and Enterococcus during HFHSD2 (r = 0.69) (Fig. 5). Conversely, negative correlations were observed between Lactobacillus and Parabacteroides during LFPPD (r = −0.83) and with Desulfovibrio during HFHSD2 (r = −0.71) (Fig. 5).
Unlike indigenous lactobacilli, L. plantarum (and presumably primarily strain WCFS1) was not well integrated into the co-occurrence networks. During both HFHSD periods, L. plantarum was only associated with those bacteria classified as unknown Lactobacillaceae (Fig. 5a and c). We defined unknown Lactobacillaceae as those OTUs that could not be assigned to the genus level. Those bacteria were present at a low abundance (ranging from 0% to 1.3% of the total bacteria present). During the LFPPD consumption, L. plantarum was additionally (negatively) associated with Parabacteroides (r = −0.71) (Fig. 5b). This result is consistent with the overall inverse correlation between L. plantarum and members of the Bacteroidetes phylum when all dietary periods were compared (Fig. 6).
Interactions between L. plantarum and the host indigenous intestinal microbes during all three dietary periods. Each node represents one genus with the color indicating the phylum to which the genus belongs and the size of the node proportional to the relative abundance of that organism in the bacterial community (only taxa with the average abundance over 0.001 are shown). The line between nodes indicates the Spearman correlation and the color intensity indicates the correlation coefficient (red, positively correlated and green, negatively correlated). The weight of the line positively corresponds to the correlation significance. Taxa in brackets are based on annotations suggested by the Greengenes database.
Predicted bacterial gene contents change upon diet switches and L. plantarum consumption
Modifications to the indigenous microbiota with the dietary switches also resulted in significant changes in the abundance of metabolic and biosynthetic pathways in the intestinal metagenomes according to PICRUSt19. A total of 9 KEGG pathways including “signal transduction”, “metabolism of cofactors and vitamins”, “membrane transport”, “cellular processes and signaling” and “cell motility” were repeatedly increased in the fecal microbiota of the sham-fed controls when the HFHSD was consumed (Supplementary Fig. S6). During LFPPD consumption, genes belonging to “glycan biosynthesis and metabolism” were enriched. Most of those diet-dependent differences in predicted metagenome pathways were absent in mice fed L. plantarum.
During each of the three dietary periods, L. plantarum ingestion instead resulted in changes to the predicted metagenomes compared to the sham-fed mice. A total of 11 KEGG pathways were enriched with L. plantarum during HFHSD1 (Supplementary Fig. S7a). Nine of those pathways were also increased during HFHSD2 and included “arachidonic acid metabolism”, “carotenoid biosynthesis” and “ion channels” (Fig. 7a and Supplementary Fig. S7). During the LFPPD period, the discriminant pathways were shifted towards genes coding for “genetic information processing” and “cellular processes” (Supplementary Fig. S7b). Lastly, L. plantarum also conferred a few diet-independent changes to the intestinal microbiome. L. plantarum fed mice were enriched for genes coding for bile acid metabolism (choloylglycine hydrolase) and glycosyltransferases across all dietary periods (Fig. 7b). Further analysis showed that most predicted reads of the choloylglycine hydrolase gene (K01442) originated from lactobacilli (Supplementary Table S3).
Pathways specifically enriched in L. plantarum WCFS1-R fed mice during all dietary periods. Metagenomics prediction was performed using PICRUSt19, a software to infer the gene content from bacterial community composition data. The whiskers of the box plot indicate the minimum and maximum value.
Discussion
Because ingested probiotic bacteria typically reside in the GI tract for only short periods of time (days to weeks), their initial interactions with the epithelium and indigenous microbiota are pivotal to determining the overall significance of these microorganisms to human health. Our results show that host diet dramatically alters the survival and persistence of ingested L. plantarum and that indigenous Lactobacillus species are oppositely and repeatedly impacted. Ingested L. plantarum WCFS1-R therefore might fill a vacant ecological niche left by the HFHSD-induced reductions to indigenous Lactobacillus species. Unlike those species, however, L. plantarum was not integrated into the microbial co-occurrence networks and therefore the indigenous microbiome. Instead, L. plantarum exerted independent, and likely more transient, effects on the intestinal community structure and function.
L. plantarum WCFS1-R survived the GI tract in 10- to 100-fold higher numbers in the current and previous studies13 when the mice were fed the HFHSD. Herein, the results were verified by three methods (colony enumeration, 16S rRNA DNA sequencing, and qPCR) and were in agreement with the higher levels of intestinal persistence when administration was stopped. There are several potential reasons why the numbers of L. plantarum were greater in the HFHS dietary background. One possibility is that nutritional preferences of L. plantarum are more suited to the HFHSD when in the presence of an indigenous microbiota. The importance of nutritional resources in the intestine was previously shown for E. coli, whereby the capacity to consume distinct sugars prevented nutritional-niche overlap and enabled co-colonization of different commensal E. coli strains20. The primary carbohydrate in the HFHSD used in this study is sucrose. Although L. plantarum WCFS1-R is able to consume sucrose, it is unlikely that sucrose utilization conferred a nutritional advantage to L. plantarum because sucrose is rapidly absorbed in the small intestine21. To this regard, we were unable to detect sucrose in the stools of mice fed the same HFHSD in a different study (data not shown).
Global restructuring of the indigenous microbiota in response to dietary shifts is another factor that could explain L. plantarum GI fitness. Consistent with numerous other studies in humans and mice, we found that the host diet is a significant determinant of the gut microbiome. Diet-induced changes can even overrule host genotype and inter-personal variation6, 22. Within three days after the switch to the HFHSD, the intestinal microbiota composition was altered to the extent that the ratio of the dominant phyla Firmicutes and Bacteroidetes was significantly increased as shown in some human studies23,24,25. Several of the HFHSD-enriched taxa (e.g. Erysipelotrichaceae, Enterococcus, and Bilophila) were also present in greater proportions in obese humans4, 26, 27. The predicted metagenomes of the sham-fed mice indicate that bacteria enriched with HFHSD possess higher numbers of genes for “xenobiotics degradation”, “lipid metabolism”, and “cell motility”. By comparison, LFPPD enriched for bacterial taxa specialized for “glycan biosynthesis and metabolism”. This finding is consistent with prior metagenome analyses of diet-induced changes to the murine gut microbiota28. However, it is notable that there were some differences between the two HFHSD periods. Changes in certain taxa and predicted metagenome pathways between HFHSD1 and HFHSD2 were indicative of recently reported hysteresis effects in the microbiota observed following serial dietary switches6.
Predominant and repeatable changes with HFHSD were reductions to the quantities of indigenous Lactobacillus species. The absolute (by qPCR) and relative (by DNA sequencing) numbers of each Lactobacillus species cluster were lower during both HFHSD periods. The abundant species were highly related to L. murinus, L. gasseri, and L. reuteri, lactobacilli recognized to be indigenous inhabitants of rodent intestines29, 30 and the human GI tract31. Reduced quantities of these bacteria during HFHSD2 confirmed that losses to indigenous Lactobacillus cell numbers were not due to an initial response of the mouse to the altered diet, but rather caused by robust, ecosystem-level changes in the intestine.
Therefore, increases in L. plantarum survival and persistence during HFHSD consumption could be largely the consequence of an open ecological niche in the intestine left by reductions to the indigenous Lactobacillus species. In other words, survival of L. plantarum might have been supported within the HFHSD but not LFPPD backgrounds due to reduced competition with indigenous Lactobacillus species. This possibility is in agreement with prior findings showing that dietary Lactobacillus survives longer in ampicillin-treated mice containing no detectable indigenous lactobacilli32. The intestinal persistence of Lactococcus lactis CNCM I-1631 in both animal models and human subjects was also positively associated with a resident gut microbiota containing a low prevalence of closely related species33. Similarly, prolonged persistence of Bifidobacterium longum AH1206 in human subjects was correlated to initially low levels of resident B. longum and an under-representation of carbohydrate utilization genes in the resident GI tract microbiota34.
However, this competitive exclusion hypothesis apparently does not apply to all ingested bacteria. Instead, there appears to be circumstances or certain functional relationships that fit the “like will to like” rule observed for some enteric pathogens and L. reuteri 35. Although we did not find such a relationship between L. plantarum and the indigenous lactobacilli, the quantities of L. plantarum were strongly and positively correlated with other members of the Firmicutes phylum, including other lactic acid bacteria such as Streptococcus (Fig. 6). Moreover, it is notable that during the LFPPD period the L. gasseri cluster was only enriched on the days in which WCFS1-R was consumed. This finding is consistent with a prior study in which Lactobacillus species diversity increased in mice fed L. plantarum and L. casei in combination with a regular rodent chow diet36. Such associations indicate that certain Lactobacillus species/strains might be compatible in the intestine and could be confirmed in studies in which different Lactobacillus strains are deliberately paired to evaluate co-colonization potential based on predicted cooperative or antagonistic cell traits.
Pivotal to predicting Lactobacillus survival and persistence is understanding the diet-induced, selective pressures in the intestine. Lactobacillus can be grouped as either host- or environment- associated37, 38. Host-associated species include L. murinus, L. gasseri, L. johnsonii, and L. reuteri; whereas L. plantarum, an organism that is essential to numerous food fermentations, is regarded to have a broader distribution39. A difference between these species is that unlike host-associated lactobacilli, L. plantarum and other broad-host range Lactobacillus species tend to harbor more genes dedicated to oxidative stress tolerance38, 40, 41. This distinction was supported by the larger numbers of genes required for tolerance to reactive oxygen species (ROS) in the genome of L. plantarum WCFS1 compared to the genomes of representative strains of L. murinus, L. reuteri and L. gasseri (Supplementary Table S4). Resistance to ROS might be an important ecological fitness determinant in the intestine during exposure to obesogenic diets. It is understood that HFHSD consumption elevates ROS concentrations at localized sites in the GI tract15, 42. Such altered intestinal redox conditions can result in the enrichment of aero-tolerant bacteria at the expense of oxygen-sensitive populations43, including some indigenous Lactobacillus species15, 42. Therefore, ROS-tolerant L. plantarum WCFS1-R might have benefited in the HFHSD background by gaining access to otherwise limited resources previously utilized by indigenous lactobacilli.
Beyond the compensatory changes to Lactobacillus numbers, ingestion of L. plantarum WCFS1-R appears to have also exerted independent effects on the composition of the intestinal microbiota. Overall, L. plantarum conferred a stabilizing influence and there were fewer significantly-changing, diet-responsive taxa in mice fed L. plantarum WCFS1-R. This result is in agreement with other studies indicating that ingested Lactobacillus could serve to maintain gut microbiota stability in individuals with intestinal diseases, allergies, or on antibiotics44,45,46. For example, mice consuming L. plantarum WCFS1-R did not harbor significantly increased proportions of Bilophila, a pro-inflammatory pathobiont enriched in diets high in saturated fat4, 47. L. plantarum WCFS1-R was also negatively correlated with Parabacteroides during the LFPPD period. Parabacteroides levels were elevated in patients with colorectal cancer48 and in elderly people in long-term residential care compared to the community-dwelling elders and young healthy subjects49. Lastly, it is notable that the proportions of the low-abundant phylum Tenericutes increased in sham-fed animals with LFPPD in a manner that was independent of L. plantarum exposure within that dietary period. It is possible that this difference between the sham- and L. plantarum- fed groups was due to the previous contact of those animals with L. plantarum WCFS1 during HFHSD1.
Mice fed L. plantarum also contained altered metagenomes according to predictions with PICRUSt. L. plantarum WCFS1-R resulted in significant changes to multiple KEGG pathways, some in a diet-dependent manner. Moreover, there were multiple, diet-associated KEGG pathways that were no longer changed within the indigenous microbiota upon L. plantarum WCFS1-R consumption. Independent of host diet, L. plantarum consistently enriched bile acid metabolism genes and glycosyltransferase genes. This could be caused by the introduction of the WCFS1-R strain, the genome of which contains multiple copies of bile salt hydrolase and glycosyltransferase genes50, 51. Lastly, during the LFPPD period, even though L. plantarum constituted a small percentage of the bacteria present in the distal gut, mice fed L. plantarum WCFS1-R were predicted to contain increases in numbers of genes coding for “cell motility” and “carbohydrate metabolism”. These genes were also enriched in human studies upon probiotic consumption, suggesting that there are certain core responses to exogenous Lactobacillus by the host microbiome52, 53. Because the accuracy of PICRUSt is based on the availability and annotation of known bacterial genomes, other methods such as metagenomics and metabolomics are needed to confirm and further understand the functional significance of L. plantarum in vivo.
The intestinal ecosystem is remarkably resistant to exogenous microorganisms54, 55. Commensal or probiotic bacteria usually result in minor disturbances in the distal gut in human adult populations56. Such changes might not be trivial, however, because we showed that even though L. plantarum WCFS1-R was not well integrated within the indigenous microbiota, ingestion of this strain did have consequences on the gut microbiota structure in a diet-dependent manner. The high-fat and high- sugar diet diminished the indigenous Lactobacillus population while supported L. plantarum survival in the mouse model tested here, suggesting the consumption of exogenous Lactobacillus species might provide compensatory effects on the microbiota which are augmented in obesogenic Western diets. This is of special interest because Lactobacillus is increasingly recognized to have pivotal roles in host development, immune tolerance, and drug sensitivity57,58,59,60. Provision of exogenous (probiotic) Lactobacillus strains, with appropriate stress-tolerance, nutritional, and effector capacities, could be usefully applied to replenish lactobacilli lost due to the consumption of obesogenic diets. Alternatively, symbiotic approaches might be needed for probiotic Lactobacillus when administered under unfavorable dietary (or other) conditions.
Methods
L. plantarum WCFS1-R preparation and enumeration
L. plantarum WCFS1 is a single colony isolate of L. plantarum NCIMB 8826, a strain able to survive in the human GI tract61. A spontaneous rifampicin-resistant mutant of L. plantarum WCFS1 (WCFS1-R) was isolated and used throughout the study as previously described13. For each day of oral administration, L. plantarum was grown in de Mann Rogosa and Sharpe (MRS) medium (BD, Franklin Lakes, NJ, USA) until exponential phase was reached (an optical density (OD) at 600 nm of approximately 0.5). The cells were collected by centrifugation at 4,000 rpm for 10 min, washed twice with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), and then suspended in a volume of 20 μl PBS (approximately 109 cells) for mice to drink off of the tip of a gavage bulb following procedures described in Lee et al.62. This method provides a quantitative and reproducible compromise between controlled invasive orogastric gavage and provision of the strain in drinking water. Enumeration of rifampicin-resistant L. plantarum from mouse feces was performed as previously described62. Briefly, feces were diluted with PBS, homogenized by mixing with 2 mm-glass beads for 10 sec at 4 m/sec twice in 1 min intervals in a Fast Prep 24 instrument (MP Biomedicals, Solon, OH, USA). Serial dilutions of the fecal suspensions were then plated onto MRS agars containing 50 μg ml−1 Rifampicin (Thermo Fisher Scientific, Waltham, MA, USA) and incubated at 37 °C for two days prior to enumeration.
Mouse study design
All procedures were performed under the protocol approved by the UC Davis Animal Care and Use Committee (protocol # 17899). A total of 16 female BALB/c mice (Harlan, Livermore, CA, USA) were housed four per cage and given free access to food and water on a 12 h light/dark cycle. During the first 5 days, mice were fed the 2018 Teklad Global 18% Protein Rodent Diet (Harlan, Livermore, CA, USA), a standard rodent chow that is enriched in plant-polysaccharides. Animals then received a high fat and high refined sugar diet (HFHSD) (Research Diet D12079B, Research Diet, New Brunswick, NJ, USA) for 12 days (HFHSD1, Fig. 1a). Next, the mice were switched onto a low- fat and plant-polysaccharide rich diet (LFPPD) (Teklad 2014 Global 14% Protein Rodent Maintenance Diet, Harlan, Livermore, CA, USA) for the subsequent 14 days. Lastly, mice were switched back to the HFHSD for 12 days prior to sacrifice (HFHSD2). Fat and sucrose contributed 43% and 41% of the energy content of the HFHSD respectively (Supplementary Table S1), and for the LFPPD, 13% energy was from fat and 67% energy from carbohydrates, mainly consisting of plant-derived polysaccharides (Supplementary Table S2).
For comparisons to L. plantarum feeding, eight (out of 16) mice consumed 109 L. plantarum WCFS1-R per day for the last 5 days during each diet period and the remaining animals (n = 8) were sham fed with PBS in the same volume as used to deliver the bacterial culture (Fig. 1a). Freshly expelled stools were examined for culturable L. plantarum prior to L. plantarum administration, at 0 h, 3 h and 5 h after the first feeding, and then every 24 h for the remainder of the dietary period (Fig. 1a). Fresh stools were also collected at five different time points during each diet, flash frozen in liquid nitrogen, and stored at −80 °C for gut microbiota analysis (Fig. 1a). The body weights of all animals were measured every two to three days for the duration of the study; however, no differences in body weights were found between the sham and L. plantarum fed mice (data not shown).
16S rRNA gene sequencing and data analysis
Total bacterial DNA was extracted from 240 frozen mouse stool samples and the V4 region of community 16S rRNA genes were amplified by PCR as previously described63. Pooled PCR amplicons were sequenced according to the paired-end protocol on the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) at the UC Davis Genome Center (http://dnatech.genomecenter.ucdavis.edu/).
Raw fastq files from both ends were assembled, demultiplexed and analyzed in the QIIME 1.8.0 (Quantitative Insights Into Microbial Ecology) software package64. An average of 25,876 high quality reads were obtained for each sample, and 544 Operational Taxonomic Units (OTUs) with an abundance over 0.005% of the total reads were identified with 97% similarity using a closed reference OTU picking method. Representative sequences from each OTU were assigned to their corresponding taxonomy based on the GreenGenes database (version 13_8)65. Beta diversity was calculated using the UniFrac distance between samples66 and visualized based on principle coordinate analysis (PCoA). Linear discriminant analysis (LDA) effect size analysis (LEfSe)67 was used to identify discriminant bacterial species for each diet with/without L. plantarum feeding. Pair-wise Spearman correlation with FDR (Benjamini & Hochberg method) adjusted P value was performed for bacterial network analysis. Correlation networks were visualized in an open source software platform Cytoscape (version 3.2.1)68. Bacterial gene contents were predicted through Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt, version 1.0.0)19. Discriminant gene categories were identified using LEfSe67. When indicated, discriminant genes were further filtered through the fold-change analysis with a cut-off of over a 2-fold change to be considered significant. The DNA sequences were deposited in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) with the accession number SRP057487.
Lactobacillus species were identified by blasting OTU sequences to NCBI 16S rRNA gene database (http://blast.ncbi.nlm.nih.gov). A neighbor-joining phylogenetic tree was constructed in MEGA669 with Lactobacillus and Lactobacillaceae OTU sequences and 16S rRNA gene sequences of their representative closest neighbor downloaded from NCBI (Supplementary Fig. S4). Due to the high sequence similarity of 16S rRNA gene V4 region among Lactobacillus species, OTUs were classified to species clusters, composed of Lactobacillus with identical V4 regions. The clusters were designated as follows: Lactobacillus murinus cluster includes L. murinus, Lactobacillus faecis, Lactobacillus apodemi and Lactobacillus animalis; Lactobacillus reuteri cluster includes L. reuteri, Lactobacillus antri, Lactobacillus frumenti, Lactobacillus panis and Lactobacillus oris; Lactobacillus gasseri cluster includes L. gasseri, Lactobacillus hominis, Lactobacillus johnsonii and Lactobacillus taiwanensis; Lactobacillus plantarum cluster includes L. plantarum, Lactobacillus fabifermentans, Lactobacillus paraplantarum and Lactobacillus pentosus.
Lactobacillus species-specific, quantitative PCR
Genomic DNA of L. plantarum WCFS1-R, Lactobacillus murinus ASF 361, Lactobacillus reuteri ATCC 23272 and Lactobacillus gasseri ATCC 19992 were extracted using the DNeasy Blood & Tissue Kit (Qiagen Inc., Valencia, CA, USA), quantified with the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies, Carlsbad, CA, USA). The DNA was then serially diluted and used to construct standard curves ranging from 10 to 107 of 16S rRNA gene copies per reaction for absolute quantification. A total of five 16S rRNA genes for each L. plantarum genome and six 16S rRNA genes for each L. murinus, L. reuteri and L. gasseri genome70, 71 was used to estimate corresponding Lactobacillus cell numbers and 16S rRNA gene copy numbers in fecal samples.
L. plantarum, L. murinus, L. reuteri and L. gasseri - specific PCR primer sequences were either selected from the literature72,73,74,75 or designed using Primer 376 (Table 1). These primers were also selective for closely related Lactobacillus species according to Primer-BLAST77. Primers for L. reuteri were found to be distinct for members of the L. reuteri species. Primers for L. plantarum were predicted to also target L. pentosus. Those for L. murinus were specific for L. murinus, L. animalis and L. crispatus, and for L. gasseri, the primers were limited to L. gasseri, L. johnsonii, L. taiwanensis and L. hominis. Real-time, quantitative PCR (qPCR) was performed in an ABI 7500 Fast Real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). Each reaction contained SsoFast EvaGreen supermix with low ROX (2X) (Bio-Rad, Hercules, CA, USA), 250 nM of each primer (Life Technologies, Carlsbad, CA, USA) and approximately 30 ng of fecal bacterial genomic DNA. PCR amplification was initiated at 98 °C for 3 min, followed by 40 cycles of 98 °C for 5 sec and either 60 °C for 30 sec for total bacteria, L. plantarum and L. murinus, 57 °C for 30 sec for L. reuteri, and 60 °C for 45 sec for L. gasseri. Each reaction was performed in duplicate. A melting curve was added at the final stage to confirm the amplification specificity. PCR amplification efficiency was calculated each time based on the standard curve and data were only considered to be valid when the amplification efficiency was within 90–100%.
Statistical analysis
Statistical analyses and plots were generated in Graphpad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). For the heat map image of the metagenomics prediction, R studio (Version 0.98.1091, RStudio, Inc., Boston, MA, USA) was used. Unless specified, non-parametric statistical comparisons were performed, including Mann-Whitney U test and Kruskal-Wallis one-way analysis of variance with Dunn’s post hoc tests.
References
Clemente, J., Ursell, L., Parfrey, L. & Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 148, 1258–1270, doi:10.1016/j.cell.2012.01.035 (2012).
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 108, 4554–4561, doi:10.1073/pnas.1000087107 (2011).
David, L. A. et al. Gut microbial succession follows acute secretory diarrhea in humans. mBio 6, e00381–00315, doi:10.1128/mBio.00381-15 (2015).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563, doi:10.1038/nature12820 (2014).
Xu, Z. & Knight, R. Dietary effects on human gut microbiome diversity. Br J Nutr 113, S1–S5, doi:10.1017/S0007114514004127 (2015).
Carmody, R. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84, doi:10.1016/j.chom.2014.11.010 (2015).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215, doi:10.1038/nature16504 (2016).
Martinez-Medina, M. et al. Western diet induces dysbiosis with increased E. coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 63, 116–124, doi:10.1136/gutjnl-2012-304119 (2014).
Stecher, B. The roles of inflammation, nutrient availability and the commensal microbiota in enteric pathogen infection. Microbiol Spectr 3, doi:10.1128/microbiolspec.MBP-0008-2014 (2015).
Villarino, N. F. et al. Composition of the gut microbiota modulates the severity of malaria. Proc Natl Acad Sci USA 113, 2235–2240, doi:10.1073/pnas.1504887113 (2016).
Zumbrun, S. D. et al. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc Natl Acad Sci USA 110, E2126–E2133, doi:10.1073/pnas.1222014110 (2013).
Marco, M. L. et al. Lifestyle of Lactobacillus plantarum in the mouse caecum. Environ Microbiol 11, 2747–2757, doi:10.1111/j.1462-2920.2009.02001.x (2009).
Tachon, S., Lee, B. & Marco, M. L. Diet alters probiotic Lactobacillus persistence and function in the intestine. Environ Microbiol 16, 2915–2926, doi:10.1111/1462-2920.12297 (2014).
Lam, Y. Y. et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One 7, e34233, doi:10.1371/journal.pone.0034233 (2012).
Qiao, Y., Sun, J., Ding, Y., Le, G. & Shi, Y. Alterations of the gut microbiota in high-fat diet mice is strongly linked to oxidative stress. Appl Microbiol Biotechnol 97, 1689–1697, doi:10.1007/s00253-012-4323-6 (2013).
Santacruz, A. et al. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity 17, 1906–1915, doi:10.1038/oby.2009.112 (2009).
Armougom, F., Henry, M., Vialettes, B., Raccah, D. & Raoult, D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS One 4, e7125, doi:10.1371/journal.pone.0007125 (2009).
Million, M. et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes 36, 817–825http://www.nature.com/ijo/journal/v36/n6/suppinfo/ijo2011153s1.html (2012).
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotech 31, 814–821, doi:10.1038/nbt.2676 (2013).
Maltby, R., Leatham-Jensen, M. P., Gibson, T., Cohen, P. S. & Conway, T. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One 8, e53957, doi:10.1371/journal.pone.0053957 (2013).
Dahlqvist, A. & Borgström, B. Digestion and absorption of disaccharides in man. Biochem J 81, 411–418 (1961).
Scott, K. P., Gratz, S. W., Sheridan, P. O., Flint, H. J. & Duncan, S. H. The influence of diet on the gut microbiota. Pharmacol Res 69, 52–60, doi:10.1016/j.phrs.2012.10.020 (2013).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: Human gut microbes associated with obesity. Nature 444, 1022–1023, doi:10.1038/4441022a (2006).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 107, 14691–14696, doi:10.1073/pnas.1005963107 (2010).
Jumpertz, R. et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr 94, 58–65, doi:10.3945/ajcn.110.010132 (2011).
Zhang, H. et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA 106, 2365–2370, doi:10.1073/pnas.0812600106 (2009).
Zuo, H. J. et al. Gut bacteria alteration in obese people and its relationship with gene polymorphism. World J Gastroenterol 17, 1076–1081, doi:10.3748/wjg.v17.i8.1076 (2011).
Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1, 6ra14–16ra14, doi:10.1126/scitranslmed.3000322 (2009).
Dewhirst, F. E. et al. Phylogeny of the defined murine microbiota: Altered Schaedler flora. Appl Environ Microbiol 65, 3287–3292 (1999).
De Waard, R. et al. Comparison of faecal Lactobacillus populations in experimental animals from different breeding facilities and possible consequences for probiotic studies. Lett Appl Microbiol 34, 105–109, doi:10.1046/j.1472-765x.2002.01051.x (2002).
Reuter, G. The Lactobacillus and Bifidobacterium microflora of the human intestine: Composition and succession. Curr Issues Intest Microbiol 2, 43–53 (2001).
Laura, P., Lam, C., Conway, P. L. & O’Riordan, K. Gastrointestinal microbial community shifts observed following oral administration of a Lactobacillus fermentum strain to mice. FEMS Microbiol Ecol 43, 133–140, doi:10.1111/j.1574-6941.2003.tb01052.x (2003).
Zhang, C. et al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J 10, 2235–2245, doi:10.1038/ismej.2016.13 (2016).
Maldonado-Gómez, M. et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe 20, 515–526, doi:10.1016/j.chom.2016.09.001 (2016).
Stecher, B. et al. Like will to like: Abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 6, e1000711, doi:10.1371/journal.ppat.1000711 (2010).
Fuentes, S. et al. Administration of Lactobacillus casei and Lactobacillus plantarum affects the diversity of murine intestinal lactobacilli, but not the overall bacterial community structure. Res Microbiol 159, 237–243 (2008).
O’Sullivan, O. et al. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol 9, 50–50, doi:10.1186/1471-2180-9-50 (2009).
Sun, Z. et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6, doi:10.1038/ncomms9322 (2015).
Walter, J. Ecological role of lactobacilli in the gastrointestinal tract: Implications for fundamental and biomedical research. Appl Environ Microbiol 74, 4985–4996, doi:10.1128/aem.00753-08 (2008).
Siezen, R. J. & van Hylckama Vlieg, J. E. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb Cell Fact 10, 1–13, doi:10.1186/1475-2859-10-s1-s3 (2011).
Martino, M. E. et al. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ Microbiol 18, 4974–4989 (2016).
Sun, J. et al. High-fat-diet–induced obesity is associated with decreased antiinflammatory Lactobacillus reuteri sensitive to oxidative stress in mouse Peyer’s patches. Nutrition 32, 265–272, doi:10.1016/j.nut.2015.08.020 (2016).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129, doi:10.1016/j.chom.2007.08.002 (2007).
Kajander, K. et al. Clinical trial: Multispecies probiotic supplementation alleviates the symptoms of irritable bowel syndrome and stabilizes intestinal microbiota. Aliment Pharmacol Ther 27, 48–57, doi:10.1111/j.1365-2036.2007.03542.x (2008).
Kubota, A. et al. Lactobacillus strains stabilize intestinal microbiota in Japanese cedar pollinosis patients. Microbiol Immunol 53, 198–205, doi:10.1111/j.1348-0421.2009.00113.x (2009).
Engelbrektson, A. et al. Probiotics to minimize the disruption of faecal microbiota in healthy subjects undergoing antibiotic therapy. J Med Microbiol 58, 663–670, doi:10.1099/jmm.0.47615-0 (2009).
Devkota, S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 487, 104–108, doi:10.1038/nature11225 (2012).
Wang, T. et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 6, 320–329, doi:10.1038/ismej.2011.109 (2012).
Claesson, M. J. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184, doi:10.1038/nature11319 (2012).
Kleerebezem, M. et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA 100, doi:10.1073/pnas.0337704100 (2003).
Lambert, J. M., Bongers, R. S., de Vos, W. M. & Kleerebezem, M. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl Environ Microbiol 74, 4719–4726, doi:10.1128/aem.00137-08 (2008).
McNulty, N. P. et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3, 106ra106–106ra106, doi:10.1126/scitranslmed.3002701 (2011).
Eloe-Fadrosh, E. A. et al. Functional dynamics of the gut microbiome in elderly people during probiotic consumption. mBio 6, doi:10.1128/mBio.00231-15 (2015).
Lawley, T. D. & Walker, A. W. Intestinal colonization resistance. Immunology 138, 1–11, doi:10.1111/j.1365-2567.2012.03616.x (2013).
Gibson, M. K., Pesesky, M. W. & Dantas, G. The yin and yang of bacterial resilience in the human gut microbiota. J Mol Biol 426, 3866–3876, doi:10.1016/j.jmb.2014.05.029 (2014).
Derrien, M. & van Hylckama Vlieg, J. E. T. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 23, 354–366, doi:10.1016/j.tim.2015.03.002 (2015).
Kamdar, K. et al. Genetic and metabolic signals during acute enteric bacterial infection alter the microbiota and drive progression to chronic inflammatory disease. Cell Host Microbe 19, 21–31, doi:10.1016/j.chom.2015.12.006 (2016).
Schwarzer, M. et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351, 854–857, doi:10.1126/science.aad8588 (2016).
Li, J. et al. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sci USA 113, E1306–E1315, doi:10.1073/pnas.1518189113 (2016).
An, J. & Ha, E. M. Combination therapy of Lactobacillus plantarum supernatant and 5-fluouracil increases chemosensitivity in colorectal cancer cells. J Microbiol Biotechnol 26, 1490–1503, doi:10.4014/jmb.1605.05024 (2016).
Vesa, P. & Marteau Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment Pharmacol Ther 14, 823–828, doi:10.1046/j.1365-2036.2000.00763.x (2000).
Lee, B., Yin, X., Griffey, S. M. & Marco, M. L. Attenuation of colitis by Lactobacillus casei BL23 is dependent on the dairy delivery matrix. Appl Environ Microbiol 81, 6425–6435, doi:10.1128/aem.01360-15 (2015).
Yin, X., Yan, Y., Kim, E. B., Lee, B. & Marco, M. L. Short communication: Effect of milk and milk containing Lactobacillus casei on the intestinal microbiota of mice. J Dairy Sci 97, 2049–2055, doi:10.3168/jds.2013-7477 (2014).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth 7, 335–336, doi:10.1038/nmeth.f.303 (2010).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72, 5069–5072, doi:10.1128/aem.03006-05 (2006).
Lozupone, C. & Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71, 8228–8235, doi:10.1128/aem.71.12.8228-8235.2005 (2005).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60, doi:10.1186/gb-2011-12-6-r60 (2011).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, doi:10.1101/gr.1239303 (2003).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30, 2725–2729, doi:10.1093/molbev/mst197 (2013).
Lee, C. M., Sieo, C. C., Abdullah, N. & Ho, Y. W. Estimation of 16S rRNA gene copy number in several probiotic Lactobacillus strains isolated from the gastrointestinal tract of chicken. FEMS Microbiol Lett 287, 136–141, doi:10.1111/j.1574-6968.2008.01305.x (2008).
Sarma-Rupavtarm, R. B., Ge, Z., Schauer, D. B., Fox, J. G. & Polz, M. F. Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl Environ Microbiol 70, 2791–2800, doi:10.1128/aem.70.5.2791-2800.2004 (2004).
Belenguer, A. et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72, 3593–3599, doi:10.1128/aem.72.5.3593-3599.2006 (2006).
Klocke, M. et al. Monitoring Lactobacillus plantarum in grass silages with the aid of 16S rDNA-based quantitative real-time PCR assays. Syst Appl Microbiol 29, 49–58, doi:10.1016/j.syapm.2005.06.001 (2006).
Byun, R. et al. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J Clin Microbiol 42, 3128–3136, doi:10.1128/jcm.42.7.3128-3136.2004 (2004).
Bindels, L. B. et al. Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. PLoS One 7, e37971, doi:10.1371/journal.pone.0037971 (2012).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol, 365–386, doi:10.1385/1-59259-192-2:365 (1999).
Ye, J. et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134, doi:10.1186/1471-2105-13-134 (2012).
Acknowledgements
This project was funded by the Agriculture and Food Research Initiative Grant 2012-67017-30219 from the USDA National Institute of Food and Agriculture.
Author information
Authors and Affiliations
Contributions
M.M. and X.Y. initiated and designed the study. X.Y., B.L. and J.Z. performed the experiment. X.Y. analyzed the data. X.Y. and M.M. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yin, X., Lee, B., Zaragoza, J. et al. Dietary perturbations alter the ecological significance of ingested Lactobacillus plantarum in the digestive tract. Sci Rep 7, 7267 (2017). https://doi.org/10.1038/s41598-017-07428-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07428-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.