Abstract
Dental caries is closely associated with the microbial dybiosis between acidogenic/aciduric pathogens and alkali-generating commensal bacteria colonized in the oral cavity. Our recent studies have shown that arginine may represent a promising anti-caries agent by modulating microbial composition in an in vitro consortium. However, the effect of arginine on the oral microbiota has yet to be comprehensively delineated in either clinical cohort or in vitro biofilm models that better represent the microbial diversity of oral cavity. Here, by employing a clinical cohort and a saliva-derived biofilm model, we demonstrated that arginine treatment could favorably modulate the oral microbiota of caries-active individuals. Specifically, treatment with arginine-containing dentifrice normalized the oral microbiota of caries-active individuals similar to that of caries-free controls in terms of microbial structure, abundance of typical species, enzymatic activities of glycolysis and alkali-generation related enzymes and their corresponding transcripts. Moreover, we found that combinatory use of arginine with fluoride could better enrich alkali-generating Streptococcus sanguinis and suppress acidogenic/aciduric Streptococcus mutans, and thus significantly retard the demineralizing capability of saliva-derived oral biofilm. Hence, we propose that fluoride and arginine have a potential synergistic effect in maintaining an eco-friendly oral microbial equilibrium in favor of better caries management.
Similar content being viewed by others
Introduction
Dental caries is one of the most prevalent infectious diseases worldwide1, 2, and is closely associated with the microbial disequilibrium between acidogenic/aciduric pathogens and alkali-generating commensal bacteria colonized in the oral cavity3,4,5,6. Novel strategies that suppress virulent species within a pathogenic biofilm could be effective alternatives to conventional antimicrobials that indiscriminately kill commensal bacteria7,8,9,10. Nourishing the dental plaque biofilm with substrates that encourage alkali production may inhibit tooth demineralization and favorably modulate the microbial metabolism and composition within dental plaque, thus being linked to the ecological management of dental caries7, 11,12,13. Metabolism of urea and arginine provide two major sources of alkali in dental biofilm7, 13. Arginine in the mouth is catabolized primarily by the microbial arginine deiminase system (ADS), which is presented in several streptococcal species, particularly Streptococcus sanguinis and Streptococcus gordonii 7. Previous work revealed a correlation between high salivary arginine concentration and caries resistance14. In addition, numerous cross-sectional studies found that caries-active or -experienced individuals exhibited significantly lower ADS activity compared with caries-free individuals15,16,17,18,19, further supporting the hypothesis that alkali-generation by oral bacteria may halt the development of dental caries.
Arginine has been incorporated into oral hygiene products for years. The Colgate-Palmolive Company has developed a Pro-Argin technology that contains 8% arginine in its toothpaste against tooth hypersensitivity20. In addition, the arginine-containing dentifrice has shown its potential in caries prevention. Oral hygiene products containing arginine bicarbonate (CaviStat®)21, 22 and toothpaste containing 1.5% arginine23,24,25,26,27, were proven to be highly effective against the initiation and progression of dental caries. The anti-caries effect of the arginine-containing dentifrice is attributed to its ecological effect on oral microbiota10, 18. In addition, a potential synergism between arginine and fluoride has also been indicated in our previous studies10, 28. The aim of the current study is to validate the microbiota-modulating effect of arginine with both clinical cohort and ex vivo oral biofilm model. We found that arginine treatment could modulate the oral microbiota of caries-active (CA) individuals to a consortium similar to that of caries-free (CF) individuals, characterized by enriched alkali-generating species and suppressed acidogenic species. In addition, the combinatory use of arginine with fluoride could synergistically suppress the demineralizing capability of saliva-derived oral biofilm.
Results
Treatment with arginine-containing dentifrice alters oral microbial composition
We recruited in total a cohort of 21 CF individuals with no clinical evidence of caries experience [decayed, missing and filled teeth (DMFT) = 0] and 21 CA individuals (DMFT ≥ 6) to investigate the effect of arginine-containing dentifrice on oral microbiota (Fig. 1 and Supplementary Table S1). Each individual was instructed to use 8% arginine toothpaste for two weeks. Saliva samples were collected before and after treatment, and microbial profiles were analyzed by bacterial 16S rDNA sequencing (n = 21 in each group). CA and CF groups exhibited distinct microbial community structure as demonstrated by Principal Component Analysis (PCA; Fig. 2a, Supplementary Table S2). After treatment, the salivary microbiome in both groups was more coherent (Fig. 2b, Supplementary Table S2).
Treatment with arginine-containing dentifrice alters oral microbial composition. Principal Component Analysis (PCA) of saliva microbiome before (a) and after (b) 2-week arginine-containing toothpaste treatment (n = 21 in each group). Abundance of S. mutans (S. m) (c) and S. sanguinis (S. s) (d) in all types of samples before and after the treatment. Bacterial counts were determined by species-specific qPCR, and normalized with the total bacterial load. Data are presented as standard box plot, with the boxes presenting the first and third quartiles and the whiskers representing the 5th and 95th percentiles. (n = 15; Kruskal-Wallis test followed by Dunn’s multiple comparison test; *p < 0.05, **p < 0.01, ***p < 0.001). (e) Representative images of in situ plaques labeled by S. m- and S. s-specific fluorescent in situ hybridization (FISH) probe. (f) Quantitative analysis of S. m/S. s ratio in the in situ plaques, bacterial loads were measured based on integral optical density (IOD). Data are presented as standard box plot. (n = 3; Kruskal-Wallis test followed by Dunn’s multiple comparison test; **p < 0.05, **p < 0.01). Pre-CF and Post-CF = caries-free group before and after arginine-containing toothpaste treatment respectively; Pre-CA and Post-CA = caries-active group before and after arginine-containing toothpaste treatment respectively.
We further examined the modification of microbial composition after the treatment. Operational Taxonomic Units (OTUs) classified at 3% dissimilarity were blasted with the oral “CORE” database, and relative abundance of each OTU was compared. We noticed that 4 OTUs (i.e., OTU_238, OTU_631, OTU_647 and OTU_398) belonging to the genus Streptococcus were repressed after arginine treatment (Supplementary Table S3). Species-specific qPCR was further used to investigate the effects of arginine on taxa belonging to the genus Streptococcus. We found that 2-week application of 8% arginine toothpaste reduced the abundance of Streptococcus mutans in supra-/sub-gingival plaque and saliva collected from the CA group (Fig. 2c). Meanwhile, the arginolytic species, S. sanguinis was enriched in supragingival plaque and saliva samples from CA group after treatment (Fig. 2d). The proportions of other arginolytic species (i.e. S. gordonii), a typical urealytic species (Actinomyces naeslundii) and a periodontal anaerobe (Porphyromonas gingivalis) were not altered (Supplementary Fig. S1).
Since the S. mutans/S. sanguinis ratio is believed to be correlated with dental caries29, 30, we further employed species-specific fluorescent in situ hybridization (FISH) to visualize the compositional alteration of these two bacteria in samples collected by a custom-designed in situ plaque collector (Fig. 2e, Supplementary Fig. S2). In parallel to the qPCR data, we observed that the S. mutans/S. sanguinis ratio was higher in the CA group (median = 5.10) relative to the CF group (median = 0.52; Fig. 2e and f) before the treatment, while the treatment with arginine-containing toothpaste substantially reversed this S. mutans/S. sanguinis disequilibrium in CA group (Fig. 2e and f).
Treatment with arginine-containing dentifrice alters the enzymatic activity and expression of genes involved in the acid-base metabolism of oral microbiome
Since the metabolic activities of oral bacteria are correlated with their cariogenicity15,16,17,18,19, 31, we holistically examined the acidogenicity, and arginolytic/urealytic activities of the oral microbiota. Microbiota obtained from CF individuals exhibited higher ADS and urease activities but a lower lactate dehydrogenase (LDH) activity relative to the CA individuals before the treatment (Fig. 3a–f). Treatment with arginine-containing dentifrice up-regulated the ADS/urease activities (Fig. 3a–d) and suppressed the LDH activities (Fig. 3e and f) in both supra- and sub-gingival plaques obtained from CA group, to an equivalent level of CF group after treatment. A similar trend was also observed in the saliva ADS activity of CA group, while the urease and LDH activities in saliva didn’t alter after treatment (Supplementary Fig. S3). Meanwhile, treatment with arginine-containing dentifrice did not alter the ADS/urease and LDH activities of plaque and saliva samples collected from CF group (Fig. 3a–f, Supplementary Fig. S3).
Treatment with arginine-containing dentifrice alters the enzymatic activity and expression of genes involved in microbial acid-base metabolism. Arginine deiminase system (ADS) (a,b), urease (c,d) and lactate dehydrogenase (LDH) (e,f) activities in supra- (a,c,e) and subgingival plaque (b,d,f). Data are presented as mean ± standard deviation (s.d.). (n = 15; one-way ANOVA test followed by Tukey’s test; *p < 0.05, **p < 0.01). The relative fold changes of ldh, arcA and ureC expression levels in supragingival plaques after treatment of every subject in caries free (CF) group (g) and caries-active (CA) group (h) are plotted. Data are presented as the results after log2 transformation, with the black horizontal lines representing the mean values (n = 15; paired sample t-test; **p < 0.01, ***p < 0.001). Pre-CF and Post-CF = caries-free group before and after arginine-containing toothpaste treatment respectively; Pre-CA and Post-CA = caries-active group before and after arginine-containing toothpaste treatment respectively.
We further investigated the gene expression levels of ldh (LDH), arcA (arginine deiminase in ADS) and ureC (α-subunit of urease) in supragingival plaque samples (see primers in Supplementary Table S4). Consistent with observed changes in the enzymatic activities of supragingival plaque samples, arcA and ureC transcripts within the supragingival microbiome were up-regulated, and ldh was suppressed in CA individuals after arginine treatment (Fig. 3h). A similar trend in gene expression was also observed in the CF group (Fig. 3g), whereas no significant changes in corresponding enzyme activities were found (Fig. 3a,c and e). There were no significant differences between CF and CA group regarding the changes of ldh, arcA and ureC expression levels after the arginine toothpaste treatment.
Arginine enriches S. sanguinis within dual-species biofilms
Treatment with 2.5% arginine could inhibit the formation of S. mutans biofilms without suppressing bacterial growth, while 5% and 10% exerted more significant inhibition against both planktonic growth and biofilm formation of this acidogenic species (Fig. 4a and b; Supplementary Fig. S4). Nevertheless, the biofilm formation of S. sanguinis was almost unaffected by 0.625–5% arginine (Fig. 4a and c), indicating that the S. mutans biofilm was more vulnerable to high concentrations of arginine compared to S. sanguinis biofilm.
The destabilizing effect of arginine on S. mutans (S. m) and S. sanguinis (S. s) biofilms. (a) Representative stereomicroscope images of crystal violet stained 24-hour S. m/S. s biofilms treated with different concentrations of arginine (Arg). Quantitative analysis of S. m (b) and S. s (c) biofilms exposed to arginine. OD595nm = optical density at 595 nm. Data are presented as mean ± s.d. (n = 3; one-way ANOVA test followed by Dunnett’s test to compare experimental groups with PBS-treated control group; **p < 0.01, ***p < 0.001). (d) Representative images of 24-hour dual-species biofilms labeled by S. m- and S. s-specific fluorescent in situ hybridization (FISH) probe. Quantitative analysis of S. m/S. s percentage (e) and number (f) based on integral optical density (IOD). Data are presented as mean ± s.d. (n = 3; one-way ANOVA test followed by Dunnett’s test to compare experimental groups with PBS-treated control group; significant difference in S. m number are indicated with *p < 0.05, ***p < 0.001, significant differences in S. s number are indicated with #p < 0.05, ###p < 0.001).
The inhibitory effect of arginine on biofilm formation was further investigated in the context of dual-species biofilms with species-specific FISH. Treatment with 0.625% and 1.25% arginine had no obvious effects on microbial composition within the biofilm (Fig. 4d–f). Interestingly, 2.5% and 5% arginine could reverse the S. mutans/S. sanguinis ratio in distinct manners (Fig. 4d–f). 2.5% arginine could suppress S. mutans, and increased the number of S. sanguinis in the dual-species biofilm compared to the PBS-treated control (Fig. 4d and f). On the other hand, 5% arginine could suppress S. mutans, but had no impact on the number of S. sanguinis compared to the PBS-treated control (Fig. 4d and f).
Combinatory use of arginine augments the anti-demineralization effect of fluoride against saliva-derived biofilm
Since most arginine-containing oral hygiene products also contain fluoride, we further test whether arginine could augment the inhibitory effect of fluoride against microbial demineralizing activity. We established a saliva-derived biofilm model, which better represent the diversity and overall metabolic functionality of human oral microbiome8, 32, 33, to evaluate the anti-demineralization effect of arginine alone or together with NaF. Biofilms grown on human enamel discs for 10 days, and were treated 3 times per day, with PBS, 2.5% arginine alone, 500 ppm NaF alone or 2.5% arginine and 500 ppm NaF in combination. Resulted lesions of enamel discs were examined with transverse microradiography (TMR). The severities of saliva-derived biofilm-induced lesions differed among different groups, with the arginine and NaF combinatory treatment exhibiting the least demineralization on the enamel surface (Fig. 5a–c).
Combinatory use of fluoride augments the anti-demineralization effect of arginine against saliva-derived biofilm. (a) Representative Transverse Microradiography (TMR) images of human enamel discs exposed to 10-day biofilm-induced experimental demineralization. The high-density regions represent the sound enamel tissues, while the low-density shadows indicate the caries-like lesions. Lesion depth (b) and integrated mineral loss (c) were calculated. Data are presented as mean ± s.d. (n = 3; one-way ANOVA test followed by Student-Newman-Keuls test; different letters indicate significant inter-group differences, p < 0.05). Quantitative data of S. mutans (S. m)/S. sanguinis (S. s) ratio (d) and number (e) in the 24-hour human saliva-derived multispecies biofilm. Bacterial numbers were determined by qPCR. Data are presented as mean ± s.d. [n = 6; one-way ANOVA test followed by Student-Newman-Keuls test; different red letters (S. m number) and black letters (S. m/S. s ratio and S. s number) indicate significant inter-group differences, p < 0.05]. Arg = arginine; CFU = colony-forming unit.
Bacterial quantification by qPCR revealed that application of 2.5% arginine alone could reduce the S. mutans/S. sanguinis ratio in the saliva-derived biofilms (Fig. 5d), consistent with the data in dual-species biofilm model (Fig. 4d–f). Moreover, addition of NaF to the arginine treatment further decreased the S. mutans/S. sanguinis ratio by suppressing S. mutans and enriching S. sanguinis at the same time (Fig. 5d and e). Treatment of 500 ppm NaF alone repressed both bacteria in the biofilms (Fig. 5e), and have no impact on the S. mutans/S. sanguinis ratio (Fig. 5d).
Discussion
Human tooth surfaces are colonized by hundreds of bacterial species. Bacteria with cariogenic potential are also frequently detected in healthy individuals34,35,36,37,38, suggesting an ecological etiology of dental caries. Frequent exposure of the microbial biofilm to carbohydrates leads to acid accumulation and subsequent pH declination, which selectively enrich acidogenic/aciduric species (e.g. mutans streptococci and lactobacilli) within biofilms and suppress those less aciduric commensal residents (e.g. S. sanguinis), and consequently drive the shift of microbial community to a more acidogenic/cariogenic consortium. This positive feedback mechanism leads to a continuous pH decline to the critical pH, below which tooth hard tissue demineralization begins and dental caries gradually occurs4,5,6. In the current study, we found that CF and CA subjects present distinct saliva microbiome, and CA individuals exhibited depressed alkali-generation capability, suggesting a microbial dysbiosis within cariogenic biofilms. Moreover, several metagenomic39, metatranscriptomic40, 41 and metaproteomic42 studies also showed that the oral microbiome of CA individuals exhibited distinct compositions and functions.
Due to the poly-microbial etiology of caries, an ecological management aiming to restore the microbial dysbiosis could be more promising. Arginine-containing oral hygiene products, which were previously used in the treatment of tooth hypersensitivity20, have been indicated to be effective in caries control21,22,23, 43. Its anti-caries effects have been attributed to its major impacts on the ecology of oral microbial communities by moderating the pH through ammonia production7, 11, 13. A similar study by Nascimento et al. found that, after 4-week treatment of 1.5% arginine dentifrice, the plaque microbial community of CA individuals and CF individuals became more coherent18. More recently, a study by Koopman et al. analyzed both salivary and plaque microbiomes of nine healthy individuals following 8-week application of 8% arginine toothpaste, and found a shift in the salivary microbial composition and function, but not in the plaque microbiome44. Here, we found that 2-week application of 8% arginine dentifrice favorably shifted the salivary microbiota of CA individuals to a community similar to that of CF individuals, characterized by increased alkali-generation and decreased acid production. These findings are consistent with the data reported by Nascimento et al.18. Interestingly, the S. mutans/S. sanguinis composition, as well as ADS, urease and LDH activities of dental plaque and saliva samples collected from CF group were unchanged after arginine treatment, indicating that arginine is likely to rescue a metabolic disequilibrium rather than to build a novel equilibrium of oral microbiome.
Although arginine is not commonly recognized as an antimicrobial agent, 8% arginine toothpaste could significantly suppress S. mutans biofilm formed on dentin discs45. Sustained millimolar concentrations of L-Arginine (>100 mM, about 1.7%) also reduced biofilm mass under flowing condition, and 500 mM of arginine even altered the microbial composition46. Though the mechanism of arginine being able to destabilize biofilm is not clear, previous data suggest that arginine may negatively influence the production of biofilm extracellular polysaccharides matrix9, 47, which plays a major role in the organization and cohesion of bacterial biofilms, especially that of S. mutans 48. In our in vitro dual-species biofilm assays, 2.5% of arginine remarkably down-regulated the S. mutans/S. sanguinis ratio, possibly due to the varied vulnerability of these two species to the biofilm-inhibition capability of arginine observed in the current study. Consistently, we also found 8% arginine treatment (approximately corresponding to a concentration of 2.67% arginine in vitro) significantly reduced the abundance of S. mutans (in supra-, subgingival plaque, saliva and in situ plaque samples) and enriched S. sanguinis (in supragingival plaque, saliva and in situ plaque samples) in CA individuals.
Most arginine-containing oral hygiene products also contain fluoride, a widely recognized anti-caries agent. Fluoride exerts its anti-caries effects mainly by improving the acid resistance of tooth hard tissue49. Recently, several clinical trials have shown that a dentifrice with arginine and fluoride could better prevent the initiation and progression of caries than fluoride alone24,25,26,27, 50. However, the mechanism of the synergism is yet unknown. In the current study, by utilizing a saliva-derived biofilm-induced caries model, we demonstrated that the superior anti-demineralization effect of the arginine/fluoride combination was possibly attributed to the synergism between these two agents in modulating the acidogenic/arginolytic species composition of biofilm. The observation that arginine and fluoride could synergistically modulate the microbial ecology of oral biofilm is consistent with our previous in vitro work10. In addition, our previous study also showed that arginine was able to promote fluoride uptake into artificial carious lesions, and subsequently enhanced the remineralization effect of fluoride on established lesions28. This may also partially explain the enhanced anti-caries effect of arginine and fluoride combination observed in the current study and other clinical studies.
Some cautions should be taken when interpreting data from this study. The lack of paralleled 16S rRNA sequencing data of plaque samples may hamper a comprehensive understanding of the ecological effect of arginine on the oral microbiome. Whether the benefits of arginine treatment were derived from specific reduction of S. mutans/S. sanguinis ratio or non-specific fix of a global dysbiosis still needs further study. Another limitation of the current study is that the enzymatic assays were carried out on frozen/thawed samples, which may not accurately reflect the instant enzymatic activity of the oral bacteria after arginine treatment.
Based on our current findings and previous data in vitro 9, 10, 28, we propose that arginine has a favorable ecological effect on oral microbiome, and thus represents a promising approach to caries control. Moreover, combinatory application of arginine and fluoride could synergistically attenuate enamel demineralization induced by saliva-derived multispecies biofilm, thus favoring better caries management.
Methods
Ethical Aspects
The study protocol was reviewed and approved by the Institutional Review Board of West China Hospital of Stomatology, Sichuan University (statement no. WCHSIRB-D-2013-067-R2), and was carried out in accordance with the guidelines. Written informed consent was obtained from all participants in the study. The trial was registered at the US National Institutes of Health (ClinicalTrials.gov) # NCT02988349, on November 24, 2016.
Experimental Design and Sample Collection
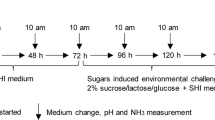
21 CF individuals with no clinical evidence of caries experience (DMFT = 0) and 21 CA individuals (DMFT ≥ 6) were recruited (by 2 times) in the clinical trial (Fig. 1). The demographic data of the volunteers are listed in Supplementary Table S1. The exclusion criteria were applied: smoker or former smoker, presence of any systemic disease that could alter the production or composition of saliva, treatment with antibiotics, steroids or any medication known to cause dry mouth in the last 3 months, presence of dental prostheses or orthodontic devices, and presence of gingivitis or periodontitis. The clinical trial had a total duration of 4 weeks, comprised of washout phase of 2 weeks and a treatment phase of 2 weeks (Fig. 1). During the washout phase, subjects were instructed to brush their teeth twice daily for 3 min using Colgate® Total® toothpaste (containing 1450 ppm NaF), whereas during the treatment phase, subjects used Colgate® Sensitive Pro-Relief® toothpaste (containing 8% arginine and 1450 ppm NaF). Additionally, 3 randomly chosen subjects in each group in the second recruitment (see information in Supplementary Table S1) were asked to wear novel in situ plaque acquisition devices during both phases (patent no. 2014100507567; Supplementary Fig. S2; see details about the preparation of this device in Supplementary Information). The in situ plaque formed on hydroxyapatite discs (4 mm × 4 mm × 2 mm). Volunteers were asked to refrain from brushing and flossing their teeth, eating, and drinking anything other than water for 12 h prior to sample collection visits at the end of both phases. In the first recruitment, supra- and subgingival plaque (n = 15 in each group) and stimulated saliva (n = 15 in each group) samples were collected. In the second recruitment, stimulated saliva (n = 6 in each group) and in situ plaque samples (n = 3 in each group) were collected (Fig. 1). Scraped plaque was immediately transferred to and dispersed in sterile micro-centrifuge tubes containing 1 × PBS, and stored at −80 °C till further analysis. Saliva sample from each subject was evenly divided into 3 portions and stored at −80 °C. The in situ plaque samples were transferred into 4% paraformaldehyde, kept under 4 °C for 16 h, and then stored in 50% (v/v) ethanol at −20 °C.
Salivary Microbiome Analysis
Genomic DNA of saliva samples (collected in the first and second recruitment, n = 21 from each group) were isolated using TIANamp bacteria DNA kit (Tiangen biotech, Beijing, China) according to the manufacturer’s instructions. The DNA quality was evaluated with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and final concentration was quantified via the Pico-Green kit (Invitrogen, Carlsbad, CA, USA). The barcoded 16S rRNA amplicon sequencing was performed through Illumina MiSeq technology where on average 330-bp-long reads were produced. Details on microbial 16S rRNA gene amplification, highly paralleled DNA sequencing, preprocessing of reads are included in the Supporting Information. The pre-processed sequencing data was further analyzed with statistical methods as following: (1) PCA was used to compare the phylogenetic structures between groups. Moreover, two non-parametric analyses for multivariate data, including analysis of similarities (ANOSIM)51 and non-parametric multivariate analysis of variance (Adonis) using distance matrices52 were used to examine the community difference between the two groups. (2) Taxonomic annotations were assigned to each OUT’s representative sequence by blasting with the oral “CORE” reference database53. The relative abundances of bacterial taxa at genus or species levels were analyzed using Student’s t-test.
qPCR Analysis of Oral Samples
Genomic DNA of supragingival, subgingival and saliva samples (collected in the first recruitment, n = 15 from each group) were isolated using TIANamp bacteria DNA kit (Tiangen biotech) according to the manufacturer’s instructions. S. mutans, S. sanguinis, S. gordonii, Actinomyces naeslundii, Porphyromonas gingivalis and all bacterial counts were quantified using qPCR as previously described54 (see details in Supplementary Information; primers and probes were listed in Supplementary Table S4). Each sample was examined in triplicate.
FISH
FISH was performed on the fixed in situ plaque samples (collected in the second recruitment, n = 3 from each group) as described previously30. Species-specific probes (Supplementary Table S4) were utilized to label S. mutans and S. sanguinis in the samples (see details in Supplementary Information). Pictures from at least 3 randomly selected positions of each labeled sample were captured using an Olympus BX3-CBH fluorescence microscope (Olympus Corp., Japan) equipped with an Andor iXon3 camera (Andor, Concord, MA, USA), the ISO (=400) and exposure time (=0.1S) were kept constant. Images were processed using Cell Sens Dimension (Olympus Corp.) without any qualitative changes to the raw images. The amount of bacteria was analyzed based on integral optical density (IOD) with Image pro plus 6.0 (Media Cybernetics, Silver Spring, MD, USA).
Determination of Enzyme Activity of Microbial Samples
Enzyme activities were measured in supragingival, subgingival and saliva samples (collected in the first recruitment, n = 15 from each group). Urease and ADS activities were determined as described by Nascimento et al.16. In brief, ammonia generated from incubation (37 °C, 120 min) of 125 μl oral samples (suspended plaque and saliva samples) and a 500 μl mixture [50 mM urea (Sigma-Aldrich, St. Louis, MO, USA) or L-arginine-HCL (Sigma-Aldrich), 0.5 mM Tris-maleate buffer (pH = 6.0)] were measured by Nessler’s reagent (Sigma-Aldrich) with ammonium sulfate as a standard. Meanwhile, protein content in each sample was determined by Bradford’s Assay with bovine serum albumin as a standard. Urease and ADS activities were expressed as μmol ammonia produced per min and were normalized to mg of protein (μmol/min/mg). LDH activities in plaque and saliva samples were measured by the LDH Activity Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. One unit of LDH was defined as the amount of the enzyme that catalyzed the conversion of lactate into pyruvate to generate 1.0 μmol of NADH per min. LDH activity was expressed as the amount of enzyme (U) per g of protein (U/g).
Gene Expression Levels in Supragingival Plaque Samples
Total bacterial RNA from supragingival samples (collected in the first recruitment, n = 15 from each group) were isolated and purified according to the manufacturer’s instructions of RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and reverse transcribed (500 ng RNA) using Prime Script RT Reagent Kit (TaKaRa, Japan) with random hexamer primers. qPCR amplification was performed on the CFX96 system (Bio-Rad, Hercules, CA, USA). The reaction mixture (25 μl) contained 1 × SYBR Premix Ex Taq (TaKaRa), 2 μl template cDNA, forward and reverse primers (500 nM each; sequences were listed in Supplementary Table S4). Expression level alterations of arcA, ureC and ldh after arginine-dentifrice treatment were calculated by Bio-Rad CFX Manager software (version 2.0) according to the 2−∆∆ CT method.
Bacterial Strains and Growth Media
S. mutans UA159 and S. sanguinis ATCC 10556 were commercially obtained from the American Type Culture Collection (ATCC). S. mutans and S. sanguinis were routinely grown at 37 °C under aerobic condition (5% CO2) in brain heart infusion broth (BHI; Difco, Sparks, MD). The potential inhibitory effects of L-arginine-HCL on the planktonic growth of S. mutans or S. sanguinis were monitored by measurement of the optical density of the cell culture at 600 nm (OD600nm). The effects of L-arginine-HCL on the biofilm formation of S. mutans or S. sanguinis were determined by crystal violet (CV) staining as described previously55. Each well of the 96-well microtiter plate contained overnight cultures of S. mutans or S. sanguinis (~1 × 106 CFU/ml), 200 μl BHI containing 1% (m/v) sucrose, and L-arginine-HCL (0–10%). The images of CV stained 24 h biofilms were captured by EZ4-HD stereomicroscopy (Leica, Germany), and representative pictures were shown. The CV stain on the abiotic surface was de-stained with 95% ethanol and the biofilm biomass was determined by measuring the OD595nm value of the collected corresponding de-stained solution. The data (normalized by the mean of non-treated control) are reported as the mean of 3 separate tests.
In vitro Dual-Species Biofilm Model
Dual-species biofilm was established as described previously10. Briefly, overnight culture of S. mutans and S. sanguinis (~1 × 106 CFU/mL each) were inoculated into 2 ml BHI containing 1% (w/v) sucrose and 0.625–10% (w/v) L-arginine-HCL (medium pH was adjusted to 7.0 prior to inoculation). Each well of the 24-well plate contained a piece of saliva-coated hydroxyapatite disc. The plate was incubated aerobically for 24 h. Then the biofilms on discs were labeled by species-specific FISH probes (listed in Supplementary Table S4). Biofilm images capturing, processing and bacteria quantification were performed as mentioned above.
Human Saliva-Derived Multispecies Biofilm-Induced Caries Model
Multispecies biofilm was established by seeding pooled saliva (collected from 5 CA subjects in the current study, see information in Supplementary Table S1) into SHI medium, which is able to sustain the growth of an in vitro microbial community with a high diversity and similar microbial profiles to original salivary microflora8, 32, 33. Saliva-coated human enamel discs were prepared as described previously28 (see details in Supplementary Information), and were transferred aseptically into 24-well plate containing 30 μl pooled saliva and 1.5 ml SHI medium. Biofilms were let grow onto the discs under anaerobic conditions (90% N2, 5% CO2, 5% H2) at 37 °C for 24 h. Then discs with biofilm were exposed to PBS, 2.5% L-arginine-HCL alone, 500 ppm NaF alone or 2.5% L-arginine-HCL + 500 ppm NaF for 5 min three times per day (at 8:30, 12:30 and 16:30). To minimize the variation in baseline mineral level, enamel discs obtained from the same tooth were evenly distributed to each test group. The pH of all experimental solutions was adjusted to 7.0 prior to treatment. After exposure, specimens were washed with PBS and repositioned in the plate. SHI medium was refreshed after the third exposure in every day. After 10-day incubation, biofilms were detached and discs were washed. The result enamel discs were prepared as described by Eversole et al.56. X-ray films of experimental lesions were acquired by an X-ray generator (Softex, Japan) equipped with a microradiography camera, and then were further examined using Zeiss AXIO Imager A2 microscope (Carl Zeiss, Germany). Quantitative data was acquired by a calibrated analysis system TMR2006 (Inspektor Research Systems BV, Netherlands) (see details in Supplementary Information). Data are presented as the mean of 3 separate tests.
Bacterial Quantification in Human Saliva-Derived Multispecies Biofilms
Aforementioned 24 h multispecies biofilms were scrapped using a sterile knife from saliva-coated human enamel discs into 1 ml PBS, sonicated to prepare the cell suspensions. Genomic DNA was isolated using a TIANamp bacteria DNA kit (Tiangen biotech) according to the manufacturer’s instruction. S. mutans and S. sanguinis numbers were determined by qPCR as mentioned above (see details in Supplementary Information; primers and probes used were listed in Supplementary Table S4). Data are presented as the mean of 6 separate tests.
Statistical Analysis
Statistical analysis of data other than 16S rRNA sequencing was performed with SPSS (version 16.0 for Windows). qPCR and FISH quantitative data in clinical trial were analyzed by Kruskal-Wallis test to compare the means of all groups, and followed by Dunn’s multiple comparison test to test all pairs of groups. Gene expression data were analyzed by paired sample t-test. Other non-sequencing data were done with one-way ANOVA test, followed by Tukey’s test or Student-Newman-Keuls test to compare all pairs of groups, or Dunnett’s test to compare experimental groups with control group. Data were considered significantly different if the two-tailed p value was <0.05.
Data Availability
The 16S rRNA sequencing raw data have been deposited in public database Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) under accession number SRP082293.
Clinical Trial Registration
The trial was registered at the US National Institutes of Health (ClinicalTrials.gov) # NCT02988349, on November 24, 2016.
References
Kassebaum, N. J. et al. Global Burden of Untreated Caries A Systematic Review and Metaregression. J Dent Res 94, 650–658 (2015).
Selwitz, R. H., Ismail, A. I. & Pitts, N. B. Dental caries. The Lancet 369, 51–59 (2007).
Kleinberg, I. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit Rev Oral Biol Med 13, 108–125 (2002).
Marsh, P. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent Res 8, 263–271 (1994).
Marsh, P. Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294 (2003).
Takahashi, N. & Nyvad, B. The role of bacteria in the caries process ecological perspectives. J Dent Res 90, 294–303 (2011).
Burne, R. A. & Marquis, R. E. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett 193, 1–6 (2000).
Guo, L. et al. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proc Natl Acad Sci USA 112, 7569–7574 (2015).
He, J. et al. L-arginine modifies the exopolysaccharides matrix and thwarts Streptococcus mutans outgrowth within mixed-species oral biofilms. J Bacteriol, JB. 00021–00016 (2016).
Zheng, X. et al. Combinatorial effects of arginine and fluoride on oral bacteria. J Dent Res 94, 344–353 (2015).
Burne, R. et al. Progress dissecting the oral microbiome in caries and health. Adv Dent Res 24, 77–80 (2012).
Huang, X., Schulte, R. M., Burne, R. A. & Nascimento, M. M. Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res 49, 165–176 (2015).
Liu, Y.-L., Nascimento, M. & Burne, R. A. Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int J Oral Sci 4, 135–140 (2012).
van Wuyckhuyse, B. et al. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J Dent Res 74, 686–690 (1995).
Gordan, V. et al. Could alkali production be considered an approach for caries control? Caries Res 44, 547–554 (2010).
Nascimento, M., Gordan, V., Garvan, C., Browngardt, C. & Burne, R. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol Immunol 24, 89–95 (2009).
Nascimento, M. et al. Oral arginine metabolism may decrease the risk for dental caries in children. J Dent Res 92, 604–608 (2013).
Nascimento, M. M. et al. The effect of arginine on oral biofilm communities. Mol Oral Microbiol 29, 45–54 (2014).
Reyes, E. et al. Caries-free subjects have high levels of urease and arginine deiminase activity. J Appl Oral Sci 22, 235–240 (2014).
Cummins, D. Clinical evidence for the superior efficacy of a dentifrice containing 8.0% arginine and calcium carbonate in providing instant and lasting relief of dentin hypersensitivity. J Clin Dent 22, 97 (2011).
Acevedo, A. M. et al. Clinical evaluation of the ability of Cavistat® in a mint confection to inhibit the development of dental caries in children. J Clin Dent 19, 1–8 (2008).
Acevedo, A. M., Machado, C., Rivera, L. E., Wolff, M. & Kleinberg, I. The inhibitory effect of an arginine bicarbonate/calcium carbonate CaviStat-containing dentifrice on the development of dental caries in Venezuelan school children. J Clin Dent 16, 63–70 (2005).
Cummins, D. The development and validation of a new technology, based upon 1.5% arginine, an insoluble calcium compound and fluoride, for everyday use in the prevention and treatment of dental caries. J Dent 41, S1–S11 (2013).
Li, X. et al. Randomized clinical trial of the efficacy of dentifrices containing 1.5% arginine, an insoluble calcium compound and 1,450 ppm fluoride over two years. J Clin Dent 26, 7–12 (2015).
Srisilapanan, P. et al. Comparison of the efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride to a dentifrice containing 1450 ppm fluoride alone in the management of early coronal caries as assessed using quantitative light-induced fluorescence. J Dent 41, S29–S34 (2013).
Souza, M. et al. Comparing the efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride to a dentifrice containing 1450 ppm fluoride alone in the management of primary root caries. J Dent 41, S35–S41 (2013).
Yin, W. et al. The anti-caries efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride as sodium monofluorophosphate assessed using quantitative light-induced fluorescence (QLF). J Dent 41, S22–S28 (2013).
Cheng, X. et al. Arginine promotes fluoride uptake into artificial carious lesions in vitro. Aust Dent J 60, 104–111 (2015).
Becker, M. R. et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 40, 1001–1009 (2002).
Zheng, X. et al. Involvement of gshAB in the interspecies competition within oral biofilm. J Dent Res 92, 819–824 (2013).
Shu, M. et al. The relationship between dental caries status and dental plaque urease activity. Oral Microbiol Immunol 22, 61–66 (2007).
Edlund, A. et al. An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome 1, 1 (2013).
Tian, Y. et al. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol 25, 357–367 (2010).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Consortium, T. H. M. P. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Teng, F. et al. Prediction of Early Childhood Caries via Spatial-Temporal Variations of Oral Microbiota. Cell Host & Microbe 18, 296–306 (2015).
Xu, X. et al. Oral cavity contains distinct niches with dynamic microbial communities. Environmental Microbiology 17, 699–710 (2015).
Simón-Soro, A. & Mira, A. Solving the etiology of dental caries. Trends in Microbiology 23, 76–82 (2015).
Belda-Ferre, P. et al. The oral metagenome in health and disease. ISME J 6, 46–56 (2012).
Benitez-Paez, A., Belda-Ferre, P., Simon-Soro, A. & Mira, A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics 15, 311 (2014).
Simon-Soro, A., Guillen-Navarro, M. & Mira, A. Metatranscriptomics reveals overall active bacterial composition in caries lesions. J Oral Microbiol 6, 25443 (2014).
Belda-Ferre, P. et al. The human oral metaproteome reveals potential biomarkers for caries disease. Proteomics 15, 3497–3507 (2015).
Sullivan, R. et al. Evaluation of a dentifrice containing 8% arginine, calcium carbonate, and sodium monofluorophosphate to repair acid-softened enamel using an intra-oral remineralization model. J Clin Dent 25, A14–19 (2013).
Koopman, J. E. et al. Changes in the oral ecosystem induced by the use of 8% arginine toothpaste. Arch Oral Biol 73, 79–87 (2017).
Fu, D. et al. Effect of desensitising paste containing 8% arginine and calcium carbonate on biofilm formation of Streptococcus mutans in vitro. J Dent 41, 619–627 (2013).
Kolderman, E. et al. L-arginine destabilizes oral multi-species biofilm communities developed in human saliva. PloS one 10, e0121835 (2015).
Sharma, S. et al. Nanoscale characterization of effect of L-arginine on Streptococcus mutans biofilm adhesion by atomic force microscopy. Microbiology 160, 1466–1473 (2014).
Koo, H., Falsetta, M. & Klein, M. The exopolysaccharide matrix a virulence determinant of cariogenic biofilm. J Dent Res 92, 1065–1073 (2013).
Koo, H. Strategies to enhance the biological effects of fluoride on dental biofilms. Adv Dent Res 20, 17–21 (2008).
Kraivaphan, P. et al. Two-year caries clinical study of the efficacy of novel dentifrices containing 1.5% arginine, an insoluble calcium compound and 1,450 ppm fluoride. Caries Res 47, 582–590 (2013).
Clarke, K. R. Non-parametric multivariate analyses of changes in community structure. Austral ecology 18, 117–143 (1993).
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral ecology 26, 32–46 (2001).
Griffen, A. L. et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PloS one 6, e19051 (2011).
Yoshida, A. et al. Development of a 5′ nuclease-based real-time PCR assay for quantitative detection of cariogenic dental pathogens Streptococcus mutans and Streptococcus sobrinus. J Clin Microbiol 41, 4438–4441 (2003).
Xu, X., Zhou, X. D. & Wu, C. D. The tea catechin epigallocatechin gallate suppresses cariogenic virulence factors of Streptococcus mutans. Antimicrob Agents Chemother 55, 1229–1236 (2011).
Eversole, S., Saunders-Burkhardt, A. & Faller, R. Erosion Prevention Potential of an Over-the-Counter Stabilized SnF2 Dentifrice Compared to 5000 ppm F Prescription-Strength Products. J Clin Dent 26, 44–49 (2015).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81371135 to JL, 81430011 to XZ), Brilliant Young Investigator Award of Sichuan University (2015SCU04A16 to XX), the State Key Laboratory of Oral Diseases (Sichuan University) Open Fund (SKLOD2016OF03), and Sichuan Province Science and Technology Innovation Team Program (2017TD0016).
Author information
Authors and Affiliations
Contributions
X.Z., J.H., L.W. and S.Z. performed the majority of the experiments, analyzed the data and drafted the manuscript. S.H. and J.X. helped the bioinformatics on 16S rRNA sequencing data. L.Z., X.P. and L.C. helped to design the study and draft the manuscript. Y.H., J.L., J.X. and X.Z. provided suggestions for the project and critically reviewed the manuscript. X.X. supervised the project and wrote most of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zheng, X., He, J., Wang, L. et al. Ecological Effect of Arginine on Oral Microbiota. Sci Rep 7, 7206 (2017). https://doi.org/10.1038/s41598-017-07042-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07042-w
This article is cited by
-
How probiotics, prebiotics, synbiotics, and postbiotics prevent dental caries: an oral microbiota perspective
npj Biofilms and Microbiomes (2024)
-
Effectiveness of fluoride-containing toothpastes associated with different technologies to remineralize enamel after pH cycling: an in vitro study
BMC Oral Health (2022)
-
The effect of different dietary ratios of lysine and arginine in diets with high or low methionine levels on oxidative and epigenetic DNA damage, the gene expression of tight junction proteins and selected metabolic parameters in Clostridium perfringens-challenged turkeys
Veterinary Research (2020)
-
Gingival solitary chemosensory cells are immune sentinels for periodontitis
Nature Communications (2019)
-
Insights into the human oral microbiome
Archives of Microbiology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.