Abstract
Trihelix gene family is an important transcription factor (TF) family involved in plants’ growth and development. This extensive study of trihelix genes from Arabidopsis thaliana to Brassica rapa could shed light on the evolution in plants and support crop breeding. In this study, a total of 52 trihelix genes were identified in B.rapa. Whole-genome annotation, molecular-evolution and gene-expression analyses of all known trihelix genes were conducted. By statistics of the number of trihelix genes in each species, we found the expansion of trihelix gene family started with angiosperm evolution. And SIP1 was more preferentially retained than other subgroups (GT-1, GT-2, GTγ, SH4), consistent with the gene dosage hypothesis. Then we investigated the evolutionary patterns, footprints and conservation of trihelix genes in selected plants. The putative trihelix proteins were highly conserved, but their expression patterns varied. Half of these genes were highly expressed in all the selected organs but some showed tissue-specific expression patterns. Furthermore, among six abiotic stresses (Cold, Heat, PEG, NaCl, ABA and GA), most trihelix genes were activated by salt and ABA treatment. In summary, the phylogenetic, evolution and expression analyses of trihelix gene family in B.rapa establish a solid foundation for future comprehensive functional analysis of BraTHs.
Similar content being viewed by others
Introduction
The transcriptional regulation of genes plays important roles in both plant growth and in response to environmental stresses. Various classes of transcriptional factors (TFs) control the processes by interacting with cis-acting elements, or with other TFs involved in gene expression1, 2. Trihelix DNA-binding factors are a family of plant-specific transcription factor, which are classified as GT factors because they were discovered as proteins that bind specifically to GT elements3,4,5,6. The DNA-binding domain of GT factors features a typical trihelix (helix-loop-helix-loop-helix) structure. This is not a completely new domain as it has similarities to the individual repeats of the MYB family from which the trihelix may have been derived5. Taken together, with a degenerate core sequence of 5′-G-Pu-(T/A)-A-A-(T/A)-3′, the domain forms a specific binding site of GT elements4, 6,7,8.
Early studies suggested that trihelix factors are involved in regulating plant responses to light4. However, with more trihelix factors cloned and characterized in the past decade, this plant-specific transcription factor family has also been found to play important roles in a variety of developmental processes and stress responses, such as: morphogenesis control of manifold flower organs, seed scattering during crop domestication, responses to salt and drought stresses and the regulation of late embryogenesis9,10,11,12,13,14,15,16,17. A. thaliana was used to study most of the functions involved in plant development. The first discovered was the GT-1 factor of pea (Pisum sativum), which specifically binds to the light-induced gene rbcS-3A 4. Some other members of the GT-1 subfamily were later identified in rice, Arabidopsis, and tobacco9, 18,19,20. Recently, an important trihelix gene, SHA1, identified in rice, was found to be involved in regulating the seed scattering process21. Another two trihelix genes, ASIL1 and ASIL2, have been reported to involve in chlorophyll accumulation in A. thaliana 22 (Table 1). Although most trihelix genes participate in plant developmental programs, two recent studies indicate that some are involved in plants stress-tolerance, especially salt tolerance16, 17 (Table 1). Loss-of-function mutations in A. thaliana GT-2 Like 1 (AtGTL1) gene, which negatively regulates water use efficiency by modulating stomatal density, led to increased plant tolerance to water deficit23. In addition, the involvement of two soybean trihelix factors [GmGT-2A (Glyma04g39400) and GmGT-2B (Glyma10g30300)] in abiotic stress tolerance has recently been proposed, following heterologous expression in Arabidopsis 17. Overexpression of these two genes could increase the tolerance to salt, drought, and cold. OsGT γ -1, another gene found in γ clade, could regulate salt resistance with different expression level16, 17.
The trihelix family had previously been classified into three distinctive subfamilies (GTα, GTβ, and GTγ), using Arabidopsis and rice genes24. Then, Kaplan-levy et al. classified trihelix genes from rice and Arabidopsis into five clades, named GT-2, GT-1, SH4, SIP1, and GTγ, with the name of each clade based on the first member identified9.
The trihelix DNA-binding proteins are unique to plants, suggesting that they would be implicated in plant-specific gene regulation, as suggested for other plant lineage-specific factors8. There are 30 trihelix genes in Arabidopsis and 31 in rice. Compared with some of the big plant transcription factor families, such as the MYB, AP2/EREBP, NAC, and bHLH, all with more than 100 members in Arabidopsis, the number of trihelix genes is relatively modest2. Although trihelix genes have been identified in Arabidopsis and rice, the evolutionary and functional information of this family in Chinese cabbage are still unknown. Thus a more thorough systematic analysis is needed to uncover these mysteries.
The Chinese cabbage genome (Chiifu-401–42) has recently been sequenced and assembled25. Data suggested B. rapa was closely related to A. thaliana, and has experienced a whole genome triplication since its divergence from A. thaliana 26, 27. In this work, for distinguishing trihelix genes from different clades, they were abbreviated as TH. We systematically and comprehensively describe the TH transcription factors in B. rapa through a comparative genome analysis. The main objectives of our study were as follows: (i) identify and characterize the TH transcription factors in the B. rapa genome; (ii) analyze the copy number variation of trihelix genes and expansion following WGT in B. rapa; (iii) investigate the evolution of the trihelix gene family in the plant kingdom and construct its evolution model; (iv) construct TH transcription factor interaction networks, and analyze TH transcription factor expression patterns through comparative genomics.
Results
Identification of Trihelix proteins in plants and comparative analyses
We identified all the putative trihelix genes in B.rapa through HMM search. This search resulted in the identification of 52 trihelix proteins. Subsequently, all these protein sequences were subjected to Pfam and SMART analyses, and named BraTH01 to BraTH52 (Supplementary Table 2). For comparative genomic analyses, we searched for trihelix protein coding sequences in the representative genomes of 25 plants (Fig. 1) and identified a total of 1106 trihelix proteins (Supplementary Table 5). These proteins represent the major evolutionary lineages of the species for the analysis of the trihelix transcription factors. Interestingly, all of those transcription factors were only found in higher plants, none of them were found in lower plants. This phenomenon shows that the trihelix proteins may have expanded after the divergence of the higher plant from the lower plant species, and strongly suggests that this family is land plant-specific, consistent with previous studies.
Cumulatively, the number of trihelix genes in B.rapa (52) exceeded that in most other plants in our analyses. In terms of the density of trihelix proteins in the whole B.rapa genome (0.183), we found that it was more than that in most species used in our analyses. Although Glycine max contained 104 trihelix proteins, its trihelix protein density (G. max, 0.055) was lower than that in B.rapa due to its large genome size. This suggested that the trihelix proteins might play a very important role in plant evolution. Since several whole-genome duplication (WGD) events happened during angiosperm evolution, it is likely that this higher number is caused by an elevated duplication frequency, in combination with an increased retention of trihelix genes. Thus, the number and density of trihelix proteins increased as plants evolved, possibly because of genome duplication.
Copy number variation and collinearity analysis of Trihelix genes
We then investigated the copy number variation of trihelix genes in A. thaliana and B. rapa during the Brassica-specific WGT event. There are 30 trihelix genes identified in A. thaliana and 52 in B. rapa (BraTH01 to BraTH52) (Table 1 and Supplementary Table 2). The collinear relationships of the gene pairs in the Trihelix family in B.rapa are shown in Fig. 2. We totally identified 15 pairs (pairs and groups of three) of highly similar orthologous that shared a high degree of identity through the BRAD database. The B.rapa genome was divided into three sub-genomes according to their fractionation degree, namely the least fractionated (LF), medium fractionated (MF1), and most fractionated (MF2). In this study, 45 (87%) trihelix genes were identified in the three B. rapa sub-genomes and located in the syntenic regions (Fig. 2, Supplementary Fig. 1a and Supplementary Table 3). Then, we specifically compared the retention of trihelix genes by counting the number of gene copies and the different distributions of the three sub-genomes. It was found that the majority of SIP1 (22%) genes were retained in two or three copies, which is higher than the retention of other subfamily trihelix genes (Supplementary Fig. 1b).
The syntenic trihelix genes between Brassica rapa and Arabidopsis thaliana. The ten Chinese cabbage chromosomes (Br01-Br10) and the five A. Thaliana chromosomes (At1-At5) are shown in different random colors. The green lines represent the syntenic genes pairs between Chinese cabbage and Arabidopsis, the yellow lines represent the syntenic genes in Chinese cabbage.
Expansion and Structural Characteristics of Trihelix Genes in Brassica rapa
To verify the extent of the lineage-specific expansion of the trihelix genes in B. rapa and A. thaliana, we performed a joint phylogenetic analysis of all the trihelix genes, and the homologous genes were marked on the tree (Fig. 3). All the trihelix genes were divided into five groups (SIP1, SH4, GTγ, GT-1 and GT-2), consistent with the previous reports in A. thaliana 9. Overall, almost no GT-2s was lost. After the split, B. rapa gained 9 and 1 genes and lost 8 and 4 genes in classes SIP1 and GTγ, respectively, resulting in the different expansion of these trihelix genes. Because of the Brassica-specific WGT event, the gene number of these two classes in B. rapa was greater than that in A. thaliana. (Fig. 3 and Supplementary Table 3).
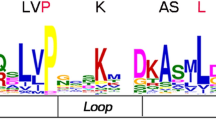
Furthermore, the sequence features of B. rapa trihelix proteins were also analyzed through MEME program, which can predict the conserved motifs among the B.rapa and Arabidopsis trihelix proteins. We identified 10 motifs in each comparison and named motif 1 to motif 10 (Fig. 3). Besides, the LOGO of these protein motifs was also obtained by MEME (Supplementary Fig. 2). Trihelix proteins often have similar motifs and intron/exon structure if they belong to the same group. All of the BraTHs contain motif 1, 2, 7, indicating that they all have a highly conserved domain. Additionally, besides the common motifs, 22 GT-2 clade trihelix members contain several specific motifs, such as motif 6, 10 that were shared in this subgroup. Interestingly, by comparing the genomic and cDNA sequences, we found that all the GTγ genes just have one exon and do not have intron, which is different from other clades. The average exon length of GTγ is greater than that of the SIP1, SH4, GT-1 and GT-2 (Fig. 4c). Furthermore, the number of exons in GTγ is the fewest, and in GT-1 is the most (Fig. 4b). Notably, the gene length of SIP1 was shorter but the exon length was longer than other subgroups (Fig. 4b,c).
Chromosome distribution, Ks and duplication Analysis of the Trihelix Genes in B.rapa
All BraTH genes were positioned on the ten B.rapa chromosomes with a non-random distribution (Supplementary Fig. 3). Chromosome 07 and Chromosome 02 each contains the most BraTH genes (22%), whereas chromosomes 01, 04 and 10 each contains the fewest (4%) (Supplementary Fig. 3b). Additionally, according to the previous reports, we reconstructed the 24 conserved chromosomal blocks (labeled A–X) in B. rapa genome and the color coding of these blocks depended on their positions in a proposed ancestral karyotype (AK1–8)25, 28. AK1 and AK3 each contains most of the BraTH genes (21%), followed by AK6 (13%), while AK7 contains only 4% of BraTH genes (Supplementary Fig. 3c). Specifically, we also observed that some BraTH genes clustered together in a region of the chromosome. For example, 4 genes clustered in the end of chromosome 9, and two of them belonged to SIP1clade (BraTH14 and BraTH15).
Furthermore, the duplication types were identified by the MCScanX program and the divergence time of the duplicated genes were estimated by calculating the number of synonymous substitutions (Ks) and Ka (nonsynonymous substitution rates). A total of 22 trihelix duplicated gene pairs were analyzed (Supplementary Table 4). SIP1, SH4, GTγ, GT-1 and GT-2 duplicated gene pairs belonged to segmental duplication, and all the duplicated BraTH gene pairs had a Ka/Ks ratio less than 1, indicating the purifying selection of these genes. Ranging from 0.3 to 0.5 and focusing on approximately 0.34 (~11 Myr), the Ks values of the BraTH genes were used to estimate the divergence time (Supplementary Fig. 4).The divergence time of BraTH duplicated gene pairs was 8 MYA, which indicates that their divergence occurred during the Brassica triplication events (5~9 MYA).
Evolution footprint of Trihelix genes in plants
To investigate the evolution of the trihelix gene family in the plant kingdom, we selected 8 representative plant species (Brassica rapa, Arabidopsis thaliana, Carica papaya, Populus trichocarpa, Vitis vinifera, Amborella trichopoda, Phscomitrella. patens and Selaginella. moellendorffii) for comparative analysis (Fig. 5). The reason is that V. vinifera, P. trichocarpa, and C. papaya did not undergo α and β duplications and A. trichopoda, a basal angiosperm, did not undergo the γ duplication event29,30,31,32,33. We constructed 8 phylogenetic trees of the trihelix genes to analyze the evolutionary relationships of these species (Supplementary Fig. 5). The phylogenetic trees showed that the trihelix gene family formed five distinct clades (SIP1, SH4, GTγ, GT-1 and GT-2), which is consistent with the result for B. rapa and A. thaliana. Trihelix genes were found exist in A. trichopoda, which indicates that these five groups originated from duplication events prior to the γ event. Meanwhile, no GTγ were detected in S. moellendorffii. Furthermore, we found that there were more trihelix genes existing in P. trichocarpa and B. rapa than in other species.
The analysis of trihelix genes evolution (a)Phylogenetic relationships among trihelix genes; (b) genetic distance among the different groups of trihelix genes; (c) comparison of the percentage of trihelix genes and copy numbers of trihelix genes, trihelix genes in representative species. (d) The rounded red box represents the trihelix domain, the rounded bule box represents the Central a-helica domain, the rounded green box represents the SH4 a-helica domain, the rounded yellow box represents the GTγ a-helica domain, the rounded purple box represents the SIP1 a-helica domain and the noncolored rounded box represents the lost domain.
To further determine the relationship among the five subgroups, the analysis of genetic distance was performed with the box plot (Fig. 5b). It was shown that the genetic distance between GT-1 and GTγ was shorter than GT-1 with other groups (Fig. 5b). Notably, the genetic distance between the SIP1 and SH4 was shorter than that between the SH4 and GT-2, SH4 and GTγ, SH4 and GT-1. These results indicated that SH4 has a closer relationship with SIP1, which means SIP1 and SH4 may share a common evolutionary origin. Subsequently, the family size and the percentage of trihelix genes in eight plant species suggested that trihelix genes expanded rapidly during evolution and further expanded in the Brassicaceae (Fig. 5c). WGD is known to have significant impact on the expansion and evolution of gene families in plant genomes. However, along with the gradual increase in the trihelix percentage, the genes of GTγ were completely lost in S. moellendorffii (Supplementary Fig. 5). During the course of evolution, the expansion of SIP1 was relatively more stable when compared with other subgroups, and it appeared most recently and expanded most rapidly. Here, we proposed a possible evolutionary footprint or model of the trihelix gene family in plants (Fig. 5d). GT-2 contains two trihelix domains and one central a-helical domain. GT-1 is related to GT-2 but possess only one trihelix domain and one entral a-helical domain, possibly originated from GT-2 by losting one trihelix domain during the evolution; alternatively, it might be that GT-2 originated from GT-1 by gaining one trihelix domain.
Tissue-specific expression Trihelix genes in Brassica rapa and Arabidopsis Thaliana
Since no trihelix factors in B.rapa has been previously documented, and to investigate the divergence of homologs and putative functions of trihelix genes in B.rapa growth and development, we analyzed the expression patterns of trihelix genes in five tissues (roots, stems, leaves, flowers, and siliques) of A. thaliana and B. rapa (Fig. 6 Supplementary Tables 6, 7). The results showed high alterations in expression levels among different TH group genes in B.rapa. Among 75 trihelix genes (including 23 AtTHs and 52 BraTHs), 1 (BraTH37) has no expression and 2 (BraTH35 and BraTH40) have slight expression in any tissues. The rest of AtTHs and BraTHs were expressed in at least one tissue. Many proteins did not show striking differences in their expression levels among different organs or tissues. Half of (26) BraTH genes were highly expressed in all the five tissues and most of them belong to SIP1 subfamily. However, a small number of genes were detected selectively expressed highly in a specific tissue. Among them, 4 genes (BraTH03, 15, 18, 36) showed preferential expression patterns in the stem. Similarly, BraTH17 predominantly expressed in the flower, whereas BraTH42 has a relatively high expression level in the siliques (Fig. 6). Therefore, these genes may mainly function in organ- or tissue-specific development in B.rapa. Interestingly, several homologs showed highly similar expression patterns in five tissues. Meanwhile, most BraTH genes presented quite different expression profiles to their homologs in Arabidopsis. For instance, At1G13450 and At3G25990 had higher expression in stem than that of other organs (Fig. 6), whereas BraTH28, 39, 47 were constitutively expressed in nearly all the organs with high abundance. The divergences in expression profiles between homologs revealed that some of them may acquire new functions after duplication in the evolutionary process.
We next investigated the expression trends among 13 duplicated gene groups (Supplementary Fig. 6). These duplicated genes showed different expression pattern types in five tissues. Seven pairs of duplicated BraTH genes were expressed in the same trend, suggesting that duplicated genes might have similar functions. Among them, BraTH45/46, BraTH23/36, BraTH35/40 and BraTH05/15 had an expression peak in steam, whereas BraTH11/33 and BraTH13/19/41 had the highest expression in root, besides, BraTH28/39 was high expressed in flower (Supplementary Fig. 6c,g,h,m,d,j,f). Although they had similar expression trend, the expression abundance was differentiated. The duplicated gene groups are highly similar in their amino acid and nucleotide sequences, but that does not mean they all have the same expression trend and they may not be involved in the same pathway or do not have similar functions. For example, BraTH12/42 showed a totally contrary expression trend and BraTH01/10 exhibited relatively high transcript abundance in the stem and flower, respectively. Notably, BraTH17/26/07/16/06/25 exhibited different transcript abundance in all the five tissues (Supplementary Fig. 6a). These two types of expression patterns suggested that the functions of these duplicated genes might have diverged in the course of evolution.
Expression profiling and Coregulatory Networks of Trihelix genes in response to abiotic stresses and hormone
The examination of trihelix genes in function is now at an accelerating pace but the full functions of this family may not yet have been uncovered. Table 1 showed scattered examples of trihelix genes in responding to environmental stimuli. In recent two studies, some trihelix factors are reported to involve in the basic resistance to abiotic stresses, especially salt-resistence16, 17. To understand the expression profiles of trihelix genes under different environmental conditions, the expression patterns of 31 selected trihelix genes were studied in response to various abiotic stresses and hormone treatments using qRT-PCR experiment (Supplementary Table 8). Heat map representation for transcript expression fold change in response to abiotic stresses and hormone treatments was shown in (Fig. 7, Supplementary Fig. 8).
Under four abiotic stresses (cold, heat, PEG, NaCl), most of BraTH genes were upregulated by NaCl and downregulated by PEG treatment, just as previous reports in Arabidopsis (Table 1). Under the two hormone treatments (ABA and GA), more genes were induced by ABA treatment compared to the GA treatment. Meanwhile, the expression profiling of the five clades was also different from each other. It seems that the GT-1 and GT-2 clade genes are more sensitive to stresses. All the SH4 genes were downregulated after heat and PEG treatments and the majorities of GT-1(except BraTH-10 and BraTH12) were downregulated after cold treatment. By contrast, the SIP1 clade was significantly induced in response to NaCl and ABA treatments and slightly induced by Cold treatment but showed repression after heat, PEG and GA treatments. In GTγ clade, all the five B.rapa genes (except BraTH03 and BraTH27 at 12 h) (Fig. 7d), were significantly induced under NaCl treatment, as well as heat and ABA treatment. It is worth mentioning that some homologous genes among B.rapa and Arabidopsis showed quite different expression patterns under the same stress conditions. For example, it was reported that the At5g28300 was induced by salinity, drought, cold and ABA in two-week-old seedlings (Table 1)9. However, we did not find its homologs (BraTH20 and BraTH29) in B.rapa had the same expression pattern. There are three GTγ group genes in Arabidopsis (Table 1)24, but their expression seems not show the similar trends34, and it will be of interest to further study the extent of stress-related functions. Regulatory subfunctionalization may have contributed to the transcriptional divergence among the genes in B.rapa and Arabidopsis, as it is not very likely these genes evolved all of the functions independently in chilling, hormone, and salt tolerance.
To further understand the connection between these trihelix genes, we established the correlation and co-regulatory networks based on the PCCs of the relative expression of genes (Fig. 8, Supplementary Table 9). Some genes showed close correlations, such as BraTH28 and BraTH31 and BraTH38 and BraTH39. Additionally, a number of genes exhibited inverse correlations, such as BraTH29 and almost all of other BraTHs, except BraTH10, 17, 21 (Fig. 8a). BraTH gene pairs with PCC values that were significant at the 0.05 significance level and were greater than 0.5 were collected and visualized to construct hormones and abiotic stresses coregulatory networks (Fig. 8b). All the gene pairs with positive significant correlations were shown in the co-regulatory network, a total of 29 nodes. A close relationship was observed between GT-1 genes and other subfamilies. Meanwhile, most of the duplicated genes seem to have no correlation, except BraTH11/33 and BraTH13/19/41. The divergence trend of the duplicated genes was reflected by the networks. The networks depicted the expansion of the gene family, which could help plants adapt to the diversified living environment by increasing cooperation or obtaining new functions.
Correlations and co-regulatory networks of 31 trihelix genes under stress treatments. (a) Correlation analysis by using the R package program. Each correlation is shown by the shades of blue and orange. Blue and orange indicate a positive correlation and negative correlation, respectively. (b) Co-regulatory networks. The co-regulatory networks of 31 trihelix genes under stress treatments were established based on the Pearson correlation coefficients (PCC) of these gene pairs using transformed qPCR data The PCC of co-regulatory gene pairs was considered significant at the 0.05 significance level (p-value), and different colour line styles indicate the different significance levels of the co-regulated gene pairs.
Moreover, to research how BraTH genes interact with other genes, an interaction network associated with BraTH genes was built according to Arabidopsis orthologs (Supplementary Fig. 8). The green and yellow lines stand for positive correlation (Pearson correlation coefficient >0) with 98 pairs of interacting genes, negative correlation (Pearson correlation coefficient <0) with nine pairs of interacting genes. The interaction network of BraTH genes showed a very complicated correlation with other genes in Chinese cabbage, which may indicate BraTH genes involve in many fundamental mechanisms by regulating many downstream factors or being regulated by many upstream genes.
BraTH28 targeted to the nucleus
Sequence analysis showed the existence of a putative nuclear localization signal in BraTH28. To test whether BraTH28 is targeted to the nucleus, a BraTH28-GFP fusion construct under the control of the CaMV 35S promoter was introduced into onion epidermal cells. The GFP alone is located throughout the cell, while BraTH28 specifically targets the nucleus and possesses both transcriptional activation and DNA-binding abilities, implicating its function as a nuclear transcription factor (Fig. 9).
Nuclear localization of BraTH28 in onion epidermal cells. Onion epidermal cells were transfected by 35S::GFP (a–c) or 35S::BraTH28-GFP (d–f) and photographed under a confocal microscope at 488 nm after 48 h (a,d). (b and e) are respectively the transmission image of (a and d). (c and f) are merged images of (a and b or d and e) respectively.
Discussion
During the course of evolution, abundant genetic materials and bulk genetic variations have been provided by genome duplication, which supports plants to adapt better to diversified environments, such as drought, high salinity, and extreme temperatures. Transcriptional regulation of gene expression plays a major role in both plant development and in response to environmental stimuli. Trihelix transcriptional factors are involved, directly or indirectly, in diverse physiological processes associated with stresses, the development of perianth organs, trichomes, stomata and the seed abscission layer, and the regulation of late embryogenesis11, 17,18,19. In this study, 52 trihelix genes were identified in the B. rapa genome, and they contained a high number of gene copies. This finding suggests that these genes had a high degree of retention following WGD. Thus, the central issue in the evolution of duplicated genes is why BraSIP1 were retained more than other subgroups. One possible explanation is that the functional requirement plays important roles in both plants’ developmental processes and defenses35, 36. This finding is consistent with the gene dosage hypothesis that genes encoding members of macromolecular complexes were preferentially retained following polyploidy and underrepresented in copy number variants, thus keeping the network stability37, 38. In addition, we found that BraTHs diverged 8 MYA during the Brassica-specific WGT event. We inferred that there may have been a stronger selective pressure on BraTHs that made them duplicate early to meet their survival needs, reflecting that the functions of BraTHs were more varied and complex.
In previous reports, the trihelix family is apparently limited to land plants9, although a report of their presence in humans and Drosophila39 needs to be studied further. They do not exist in the green algae (Chlorophyta)40,41,42, and have undergone large scale expansion in the lineage of the last common ancestor of land plants42. The presumed origin of the trihelix domain from a MYB-like gene carrying only one repeat5, and their relationship to other divergent MYB-like genes, needs to be examined in further detail. In this work, a phylogenetic tree of trihelix transcription factors from B.rapa and the dicotyledonous model plant Arabidopsis was constructed. The result was consistent with domain and Trihelix type classifications of B.rapa trihelix transcription factors. Basing on the current genomic data, we built a model diagram for the origin and evolution of trihelix family transcription factors. Among all motifs, motif 1, 2, and 4 contained a (F/Y)- (F/Y)-X-X-(L/I/M)-X-X-(L/I/M) sequence. Motif 9 and 10 were present in GT-2 members, while motif 1 and motif 4 was found in other subgroups. In addition, comparative structural analysis of BraTHs revealed that BraTHs in the same group shared similar exon–intron structures. The analysis on structures of BraTH genes may provide a way to find out which group of trihelix genes might be of a more ancient origin. The Ks values supported that trihelix genes did not have significant difference among the three subgenomes (LF, MF1, MF2). Actually, they may have similar gene structures, the similar intron and exon numbers of each subfamily also supported that. Taken all the results together, our study offers significant insights into the unique features and roles of this family in eukaryotic organisms. The fairly high conservation in gene structure observed here between genes identified by phylogenetic reconstruction was to be expected in genes of an ancient family which play a key metabolic role in virtually all living organisms.
Most land plants have undergone polyploidization during their long evolutionary histories43, 44. Polyploidy not only led to WGD but also offered chances for duplicated genes to diverge subsequently in three broad evolutionary ways: subfunctionalization, neofunctionalization, and nonfunctionalization (pseudogenization or deletion)44. Some duplicated genes could also have completely redundant functions45. Through the analyses of (i) phylogenetic relationships, (ii) gene structures, (iii) synteny analysis and (iv) nucleotide distance, we found that SIP1 had a close relationship with SH4, and we constructed the evolutionary model of trihelix family (Fig. 5d). We estimated the cleavage trihelix domain mechanisms, of which the domain may be modified at the genome level. By comparative analysis in all selected plants, here, we inferred an evolutionary history of trihelix family in the plant kingdom: from Bryophyta to Angiosperm, all five clades exist from Bryophyta; then, GTγ was absent in Lycophyta while existed in angiosperm plants like other four groups (Fig. 10). All the data obtained are compatible with trihelix genes emerging very early in eukaryotic evolution and being transmitting both vertically and horizontally.
Evolutionary history of the trihelix family in plants. (a) GT-2, GT-1, SH4, SIP1 (indicated by different colors) exist in the course of evolution from Bryophyta to Angiosperm. (b) GTγ (indicated by green line) exists in the course of evolution from Bryophyta to Angiosperm except Lycophyta (dummy line indicates inexistence).
In addition to analyses of the evolutionary history of trihelix genes, based on the complete genome sequences and sequence similarities, we attempted to predict their functions in various species46, 47. Due to the similar intron and exon numbers, the homolog genes may have similar gene structures. Their highly conserved sequences were further proved by analyzing the proteins with MEME. In addition, both the duplicated genes in the neofunctionalization or subfunctionalization models and the expansions of the large gene family were associated with the processes of tissue-expression divergence48,49,50. In this study, the tissue-specific expression patterns of trihelix genes were also examined: most trihelix genes were highly expressed in all the five tissues or several at least. Meanwhile, a few of genes showed tissue-specific expression and some trihelix genes of different clades had similar expression patterns, indicating their common importance in plant development. The genes expressed in specific tissues might have acquired new functions related to plant development. The divergences in expression profiles between homologs revealed that some of them may acquire new functions after duplication in the evolutionary process.
In summary, it seems reasonable that repeated WGD events facilitated the increase in trihelix genes network complexity, such as in A. thaliana and B. rapa (Supplementary Fig. 8). By integrating phylogenetic, molecular evolution, gene structure and expression pattern analyses and conducting a comparative analysis with the currently available genome information in the selected plants (Figs 1 and 5), our study provides a deep understanding of the evolutionary history of trihelix gene family in plants. The evolution and origin of the trihelix genes in the plant kingdom were analyzed, and the evolutionary pattern of the trihelix genes was determined (Figs 5d and 10). Due to visible tissue-specific expression patterns, the expansion of trihelix genes seems to be correlated with the evolution of increasingly complex organs in plants. This finding will lead to novel insight into functional divergence and conservation in this gene family.
Materials and Methods
Identification of the Trihelix Genes in Multiple Species
All the B. rapa genome sequence data were downloaded from the Brassica database (BRAD; http://brassicadb.org/brad/)27. The protein sequences of A. thaliana trihelix were obtained from the Arabidopsis Information Resource database (http://www.arabidopsis.org/; Supplementary Table S1). The gene information of Amborella. trichopoda was obtained from the Amborella Genome Database (http://www.amborella.org/)32. The gene information of Ca. papaya, V. vinifera, Po. trichocarpa, Ph.patens and S. moellendorffii were downloaded from Phytozome v9.1 (http://www.phytozome.net/;)51. To identify putative trihelix family members, the Hidden Markov Model (HMM) profiles of trihelix (PF13837) were retained from the Pfam database (http://pfam.xfam.org/) and were used to identify the putative trihelix proteins with the best domain e-value cutoffs of <1 × 10−4. With a cutoff e-value of <10−10, the Arabidopsis trihelix sequences were used as the query to perform a BLASTP search. The SMART tool(http://smart.embl-heidelberg.de/) and the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/) were used to analyze these potential sequences to validate the HMM and BLAST search52.
Synteny Analysis
The Multiple Collinearity Scan toolkit (MCScanX) was used for the synteny analysis between the A. thaliana and B. rapa genomes according to previous reports. (http://chibba.pgml.uga.edu/mcscan2/; match_score: 50, match_size: 5, gap_score: −3, E_value: 1E–05)53,54,55. An all-against-all BLASTP comparison provided the pairwise gene information and P values for primary clustering. The whole-genome protein sequences from B. rapa and A. thaliana, were searched against themselves using BLASTP (E < 1e-10, identity >75%). Further, MCScanX was also used to identify WGD/segmental, tandem, proximal and dispersed duplication events in the BraTH family.
Ks Analysis
The protein sequences of trihelix from B. rapa were aligned with their syntenic genes in A. thaliana using MUSCLE56. To estimate the divergence of the duplicated trihelix genes, the sequences of the duplicated pairs of trihelix genes were aligned using ClustalW2. We calculated the synonymous rate (Ks), non-synonymous rate (Ka), and evolutionary constraint (Ka/Ks).An in-house Perl script based on ParaAT54 was used to translate the protein alignments into coding sequence alignments and based on that we calculated the Ka (nonsynonymous substitution rate) and Ks (synonymous substitution rate) values using the method of Nei and Gojobori implementing in KaKs_calculator57. The Ks values were then used in the density and boxplot through the R3.3.0 program58. The formula T = Ks/2r was used to calculate the divergence time in which the r was taken to be 1.5 × 10−8 synonymous substitutions per site per year, representing the rate of divergence59.
Evolution Analysis of trihelix Gene Family
The MUSCLE program was used to align the full-length trihelix proteins sequences with the default parameters56. The maximum-likelihood method was used to construct the phylogenetic relationship in each analysis. MEGA5.2 was used to calculate Bootstrap values with 1,000 replications60. To estimate the nucleotide divergence between sequences, all nucleotide sequences of the trihelix genes were also analyzed with MEGA 5.2 using the Jukes-Cantor model. Bootstrap (1,000 replicates) analyses were also performed for this estimation.
Motif Identification and Exon–Intron Structural Analysis
To identify the conserved motifs of the trihelix genes of B. rapa, the online Multiple Expectation-maximization for Motif Elicitation program version 4.9.061 was employed among the amino acid sequences with the default parameters, except for the following parameters: Maximum number of motifs, 10; optimum motif width 20 and 120. The position information of the trihelix genes, and trihelix domains was obtained from the Pfam database, and the information of gene structure was obtained from the General Feature Format files. We then draw the domain and exon–intron structures positions through the online program GSDS (http://gsds.cbi.pku.edu.cn/)62.
Expression Pattern Analysis for Trihelix Genes in Five Tissues
For expression profiling of the trihelix genes in B. rapa, we analyzed five tissues of B. rapa accession Chiifu-401-42 (root, stem, leaf, flower, and silique). The Illumina RNA-seq data which were previously generated and analyzed by25 were utilized. Gene expression patterns of each tissue were analyzed and fragments per kilobase of exon model per million mapped (FPKM) values were log2 transformed. The gene expression patterns of each tissue were analyzed using Cluster 3.0, and the expression values were log2 transformed. Finally, heat maps of hierarchical clustering were visualized using Tree View (http://jtreeview.sourceforge.net/). The A. thaliana development expression profiling was analyzed using the AtGenExpress Visualization Tool (AVT; http://jsp.weigelworld.org/expviz/expviz.jsp) with mean-normalized values. Heat maps of the gene FPKM values in B. rapa and A. thaliana were visualized using Tree View (http://jtreeview. sourceforge.net/).
Plant Materials
The Chinese cabbage cultivar Chiifu-401-42 was used for this experiment. This cultivar is a typical cultivar for Chinese cabbage research as its whole genome sequencing has been completed. Seeds were surface sterilized in 12% sodium hypochlorite before germinating on 0.5 Murashige and Skoog (MS) agar plates (0.7%) in a growth chamber at 22 °C in the dark for 2 days. The germinated seeds were grown in pots containing a soil: vermiculite mixture (3:1) in the greenhouse of Nanjing Agricultural University, and the controlled environment growth chamber was programed for 75% humidity, light 16 h/25 °C and dark 8 h/20 °C. One month later, the five-leaf stage seedlings were transferred to 4 or 38 °C growth chambers under the same light intensity and day length as the cold and heat treatments. Pots were irrigated with 250 mM NaCl and 15% (w/v) polyethylene glycol (PEG) for 30 min under normal growth conditions as salt and osmotic treatments, respectively. Simultaneously, some plants were grown in 1/2 Hoagland’s solution in plastic containers with the pH at 6.5 for acclimation. 5 days later, plants were grown in the following three treatments: (1) Control; (2) 100 μM GA; (3) 100 μM ABA; We sampled at 1, 6 and 12 h, the young leaf samples were frozen in liquid nitrogen and stored at −70 °C for further analysis.
RNA Isolation and qRT-PCR analyses Analysis
The RNA was isolated from leaves using an RNA kit (RNAsimply total RNA Kit; Tiangen, Beijing, China) according to the manufacturer’s instructions. We used agarose gel electrophoresis to assess the quality and quantity of every RNA sample. The RNA was then reverse transcribed into cDNA using the Prime Script RT reagent Kit (TaKaRa). The Supplementary Table S1 listed the gene-specific primers used for real-time polymerase Chain Reaction (PCR). The reactions were performed using a Step one plus Real-Time PCR System (Applied Biosystems, Carlsbad, CA). The PCR parameters were as follows: 94 °C for 30 s, 40 cycles at 94 °C for 10 s, and 60 °C for 30 s, and then a melting curve (61 cycles at 65 °C for 10 s) was generated to check the specificity of the amplification. Relative fold expression changes were calculated using the comparative Ct value method63.
Pearson Correlation Analyses
Based on log2-transformed quantitative Real-Time (qRT)-PCR data, we calculated Pearson correlation coefficients (PCCs) of transcript levels of trihelix gene pairs by R program. For a gene coregulatory network analysis, we collected the gene pairs whose PCC was more than 0.5 and significant at the 0.05 significance level (P -value). Based on the PCCs of these gene pairs, the co-expression networks were visualized through Cytoscape64. The interaction network associated with Arabidopsis orthologous of trihelix genes in Chinese cabbage was constructed using the Arabidopsis interaction viewer and cytoscape software64.
Nuclear localization assays
The BraTH28 coding region (without the stop codon) was amplified by PCR and inserted into the psmGFP vector65 to produce the BraTH28-GFP fusion construct. Onion epidermal cells were transfected by biolistic bombardment using the PDS-1000/He system (Bio-Rad) according to the manufacturer’s instructions and imaged with a confocal microscope (LSM 510, Zeiss).
References
Zhang, H. et al. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Research 39, 1114–1117 (2011).
JL, R. et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110 (2001).
Nagano, Y., Inaba, T., Furuhashi, H. & Sasaki, Y. Trihelix DNA-binding protein with specificities for two distinct cis-elements: both important for light down-regulated and dark-inducible gene expression in higher plants. Journal of Biological Chemistry 276, 22238–22243 (2001).
Green, P. J., Kay, S. A. & Chua, N. H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. Embo Journal 6, 2543–2549 (1987).
Nagano, Y. Several Features of the GT-Factor Trihelix Domain Resemble Those of the Myb DNA-Binding Domain. Plant Physiology 124, 491–494 (2000).
Ni, M., Dehesh, K., Tepperman, J. M. & Quail, P. H. GT-2: in vivo transcriptional activation activity and definition of novel twin DNA binding domains with reciprocal target sequence selectivity. Plant Cell 8, 1041–1059 (1996).
Gourrierec, J. L., Li, Y. F. & Zhou, D. X. Transcriptional activation by Arabidopsis GT-1 may be through interaction with TFIIA–TBP–TATA complex. Plant Journal 18, 663–668 (1999).
Zhou, D. X. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends in Plant Science 4, 210–214 (1999).
Kaplan-Levy, R. N., Brewer, P. B., Quon, T. & Smyth, D. R. The trihelix family of transcription factors – light, stress and development. Trends in Plant Science 17, 163–171 (2012).
Barr, M. S., Willmann, M. R. & Jenik, P. D. Is there a role for trihelix transcription factors in embryo maturation? Plant Signaling & Behavior 7, 205–209 (2012).
C, B. et al. The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21, 2307–2322 (2009).
Brewer, P. B. et al. PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131, 4035–4045 (2004).
Gao, M. J. et al. Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21, 54–71 (2009).
Li, C., Zhou, A. & Sang, T. Rice domestication by reducing shattering. Science 311, 1936–1939 (2006).
Tzafrir, I. et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiology 135, 1206–1220 (2004).
Jing, X., Qiu, Y., Du, L. & Poovaiah, B. W. Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Science 185-186, 274–280 (2012).
Xie, Z. M. et al. Soybean Trihelix Transcription Factors GmGT-2A and GmGT-2B Improve Plant Tolerance to Abiotic Stresses in Transgenic Arabidopsis. PloS one 4, e6898 (2009).
Rong, W., Hong, G. & Han, B. Transcript abundance of rml 1, encoding a putative GT1-like factor in rice, is up-regulated by Magnaporthe grisea and down-regulated by light. Gene 324, 105–115 (2004).
Perisic, O. & Lam, E. A tobacco DNA binding protein that interacts with a light-responsive box II element. Plant Cell 4, 831–838 (1992).
Ayadi, M., Delaporte, V., LI, Y.-F. & Zhou, D. X. Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis. Febs Letters 562, 147–154 (2004).
Lin, Z. et al. Origin of seed shattering in rice (Oryza sativa L.). Planta 226, 11–20 (2007).
Willmann, M. R., Mehalick, A. J., Packer, R. L. & Jenik, P. D. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiology 155, 1871–1884 (2011).
Yoo, C. Y. et al. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22, 4128–4141 (2010).
Fang, Y., Xie, K., Xin, H., Hu, H. & Xiong, L. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Molecular Genetics & Genomics Mgg 283, 157–169 (2010).
Tong, C. et al. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. Bmc Genomics 14, 1–10 (2013).
Town, C. D. et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18, 1348–1359 (2006).
Wang, X. et al. The Genome of the Mesopolyploid Crop Species Brassica rapa. Nature Genetics 43, 1035–1039 (2011).
Schranz, M. E., Lysak, M. A. & Mitchell-Olds, T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in Plant Science 11, 535–542 (2006).
Lee, T.-H., Tang, H., Wang, X. & Paterson, A. H. PGDD: a database of gene and genome duplication in plants. Nucleic acids research 41, D1152–D1158 (2013).
Albert, V. A. et al. The Amborella genome and the evolution of flowering plants. Science 342, 1241089 (2013).
Jiao, Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011).
Project, A. G. et al. The Amborella genome and the evolution of flowering plants. Science 342, 1241089–1241089 (2013).
Lee, T. H., Tang, H., Wang, X. & Paterson, A. H. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Research 41, 1152–1158 (2013).
Schmid, M. et al. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506 (2005).
Atkinson, R. G., Schröder, R., Hallett, I. C., Cohen, D. & Macrae, E. A. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiology 129, 122–133 (2002).
Fabi, J., Cordenunsi, B., Seymour, G., Lajolo, F. & Do Nascimento, J. Molecular cloning and characterization of a ripening-induced polygalacturonase related to papaya fruit softening. Plant Physiology & Biochemistry 47, 1075–1081 (2009).
Birchler, J. A. & Veitia, R. A. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19, 395–402 (2007).
Lou, P. et al. Preferential Retention of Circadian Clock Genes during Diploidization following Whole Genome Triplication in Brassica rapa. Plant Cell 24, 2415–2426 (2012).
Riaño-Pachón, D. M., Corrêa, L. G., Trejos-Espinosa, R. & Mueller-Roeber, B. Green transcription factors: a Chlamydomonas overview. Genetics 179, 31–39 (2008).
Pérezrodríguez, P. et al. PInTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Research 38 (2010).
Zhang, H. et al. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic acids research 39, D1114–D1117 (2011).
Lang, D. et al. Genome-Wide Phylogenetic Comparative Analysis of Plant Transcriptional Regulation: A Timeline of Loss, Gain, Expansion, and Correlation with Complexity. Genome Biology & Evolution 2, 488–503 (2010).
Leebensmack, J. Polyploidy and angiosperm diversification. American Journal of Botany 96, 336–348 (2009).
Innan, H. & Kondrashov, F. The evolution of gene duplications: classifying and distinguishing between models. Nature Reviews Genetics 11, 97–108 (2010).
Bekaert, M. & Conant, G. C. Two-Phase Resolution of Polyploidy in the Arabidopsis Metabolic Network Gives Rise to Relative and Absolute Dosage Constraints. Plant Cell 23, 1719–1728 (2011).
Huang, S. et al. Draft genome of the kiwifruit Actinidia chinensis. Nature. Communications 4, 8–8 (2013).
Xu, Q. et al. The draft genome of sweet orange (Citrus sinensis). Nature Genetics 45, 59–66 (2013).
Ganko, E. W., Meyers, B. C. & Vision, T. J. Divergence in expression between duplicated genes in Arabidopsis. Molecular Biology & Evolution 24, 2298–2309 (2007).
Li, W. H., Jing, Y. & Xun, G. Expression divergence between duplicate genes. Trends in Genetics 21, 602–607 (2005).
Huerta-Cepas, J., Dopazo, J., Huynen, M. A. & Gabaldón, T. Evidence for short-time divergence and long-time conservation of tissue-specific expression after gene duplication. Briefings in Bioinformatics 12, 442–448 (2011).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186 (2012).
Ludwig-Müller, J. Auxin conjugates: their role for plant development and in the evolution of land plants. Journal of Experimental Botany 62, 1757–1773 (2011).
Wang, Y. et al. Diversification, phylogeny and evolution of auxin response factor (ARF) family: insights gained from analyzing maize ARF genes. Molecular Biology Reports 39, 2401–2415 (2012).
Wang, Y. et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research 40, e49–e49(41) (2012).
Duan, W. et al. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Molecular Genetics & Genomics 290, 239–255 (2015).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 (2004).
Wong, K. S. KaKs_Calculator:Calculating Ka and Ks Through Model Selection and Model Averaging. Genomics Proteomics & Bioinformatics 4, 259–263 (2006).
Ihaka, R. & Gentleman, R. R: A Language for Data Analysis and Graphics. Journal of Computational & Graphical Statistics 5, 299–314 (2012).
Koch, M. A., Haubold, B. & Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Molecular Biology & Evolution 17, 1483–1498 (2000).
Tamura, K. et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology & Evolution 28, 2731–2739 (2011).
TL, B. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208 (2009).
Hu, B. et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296 (2014).
Heid, C. A., Stevens, J., Livak, K. J. & Williams, P. M. Real time quantitative PCR. Genome Research 6, 986–994 (1996).
Shannon, P. et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research 13, 2498–2504 (2003).
Davis, S. J. & Vierstra, R. D. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Molecular Biology 36, 521–528 (1998).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31330067, 31301782), Science & Technology Pillar Program of Jiangsu Province (No.BE2013429), the Agricultural science and technology independent innovation funds of Jiangsu Province [CX(13)2006].
Author information
Authors and Affiliations
Contributions
W.W., P.W., T.L., H.R. and Y.L. conceived the study. W.W., P.W. and H.R. completed the experiments. T.L. and Y.L. contributed to data analysis and manuscript preparation. W.W. and P.W. participated in the planning of experiments and revising the manuscript. All authors had read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, W., Wu, P., Liu, T. et al. Genome-wide Analysis and Expression Divergence of the Trihelix family in Brassica Rapa: Insight into the Evolutionary Patterns in Plants. Sci Rep 7, 6463 (2017). https://doi.org/10.1038/s41598-017-06935-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-06935-0
This article is cited by
-
Genome-wide characterization, evolutionary analysis, and expression pattern analysis of the trihelix transcription factor family and gene expression analysis under MeJA treatment in Panax ginseng
BMC Plant Biology (2023)
-
Analysis of Trihelix Genes and Their Expression in Potato in Response to Abiotic Stresses
Potato Research (2023)
-
Genome-wide identification of trihelix transcription factors in the apple genome in silico
Journal of Applied Genetics (2023)
-
Insights into the trihelix transcription factor responses to salt and other stresses in Osmanthus fragrans
BMC Genomics (2022)
-
Genome-wide identification and expression profile analysis of trihelix transcription factor family genes in response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench]
BMC Genomics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.