Abstract
Mycobacterium avium subsp. hominissuis is associated with infection of immunocompromised individuals as well as patients with chronic lung disease. M. avium infects macrophages and actively interfere with the host killing machinery such as apoptosis and autophagy. Bacteria alter the normal endosomal trafficking, prevent the maturation of phagosomes and modify many signaling pathways inside of the macrophage by secreting effector molecules into the cytoplasm. To investigate whether M. avium needs to attach to the internal surface of the vacuole membrane before releasing efferent molecules, vacuole membrane proteins were purified and binding to the surface molecules present in intracellular bacteria was evaluated. The voltage-dependent anion channels (VDAC) were identified as components of M. avium vacuoles in macrophages. M. avium mmpL4 proteins were found to bind to VDAC-1 protein. The inactivation of VDAC-1 function either by pharmacological means or siRNA lead to significant decrease of M. avium survival. Although, we could not establish a role of VDAC channels in the transport of known secreted M. avium proteins, we demonstrated that the porin channels are associated with the export of bacterial cell wall lipids outside of vacuole. Suppression of the host phagosomal transport systems and the pathogen transporter may serve as therapeutic targets for infectious diseases.
Similar content being viewed by others
Introduction
Mycobacterium avium subsp. hominissuis (M. avium) is the most common pathogen among non-tuberculosis mycobacteria, and of great public health relevance as one of the leading cause of bacterial infection in patients with HIV/AIDS as well as in individuals with chronic lung conditions1, 2. The opportunistic pathogen has the ability to invade and proliferate within a variety of mammalian cells including mucosal epithelium cells and macrophages. Following uptake, the pathogen is contained within a cytoplasmic vacuole, and intracellular survival is dependent on a number of bacterial virulence determinants used to remodel the vacuolar compartment and to resist the host antimicrobial mechanisms3,4,5,6. M. avium can prevent the recruitment of proton-ATPase to the vacuole and, therefore, inhibits the acidification of the phagosome7. The pathogen arrests the maturation of phagosomes in the early endosome phase8 by interfering with trafficking process5, and grow in non-acidified compartments9. M. avium actively survives and resists the most effective cellular killing mechanisms by molecules of reactive oxygen intermediates (ROIs) and nitric oxide (NO)10,11,12. Another characteristic of M. avium is the ability to use apoptosis as a trigger to escape from phagocytes and infect surrounding cells13, 14. The interaction between virulent mycobacteria and host antimicrobial mechanisms is assumed to be an active process controlled only by a viable bacilli, since none of above effects occur following phagocytosis of dead mycobacterium or after inhibition of bacterial protein synthesis15, 16.
The specialized protein secretion systems are one of the main virulence determinants of pathogenic bacteria that efficiently deliver bacterial secreted effectors directly to the cytosol across eukaryotic membranes, either plasma or vacuolar. Many pathogens coordinately deliver/inject virulence factors via Type III, IV and/or VI secretion machineries to the extracellular (tissues or bloodstream) or intracellular (host cells) environment. Mycobacteria lack all of above virulence-associated secretion machineries, and in addition they are encapsulated in an unique lipid-rich mycolate layer. An increasing body of literature indicate that mycobacterium protein export is facilitated in part by the Type VII secretion system (T7SS), which plays a central role in mycobacterial pathogenesis17, 18. Pathogenic mycobacteria species encode up to five copies (ESX1–5) of T7SS, and disruptions of the T7SS systems or their substrates have been shown to diminish bacterial intracellular fitness or decrease in virulence3, 4, 19. The best-characterized ESX-1 locus of RD1 is involved in the secretion of ESAT-6 and CFP-10 of Mycobacterium tuberculosis and Mycobacterium marinum 20, 21 influencing the host cell signaling and cytokine secretion22 and apparently required for the escape of M. tuberculosis from the phagolysosome into the cytosol23. M. avium, that lacks the ESX-1 region, has been demonstrated to use the ESX-5 system for virulence. The ESX-5 locus exports several extracellular proline-glutamic acid proteins, the PPE and PE virulence factors4, 24, found within the mycobacterial cell envelope25 and characterized by the antigenic variation and consequent immune evasion26, 27. Studies have demonstrated that many PE/PPE proteins found in M. avium are secreted and the disruption of PE/PPE family genes is linked to bacterial attenuation3, 4.
Despite the significant progress made in the past decade, it is still unknown how mycobacteria translocate virulence effectors through the membrane-bound phagosome and deliver effector molecules into the cytosol of the host cell. Since intracellular mycobacterium is found juxtaposed to the phagosome membrane, the goal of this study was to identify possible phagosomal proteins that are employed by M. avium to export virulence factors into the cytosol of host cells.
Results
VDAC porins are associated with M. avium phagosomes
M. avium phagosomes were purified using biotin labeling and magnetic purification technique, previously described for mycobacteral phagosomes28. After magnetic separation, the intact phagosomes isolated from infected THP-1 cells were stained with Alexa Fluor 488 conjugated Annexin V (Fig. 1A), Rab5 (Fig. 1B) and Rab7 (Fig. 1C) markers, and examined under fluorescence microscopy. To visually determine Rab5 and Rab7 labeled phagosomes, we evaluated three hundred bacterial cells expressing the tomato red protein and the percentage of positive phagosomes was calculated. In agreement with previous studies29, the most of M. avium-containing phagosomes were positive for Rab5, whereas the co-localization of bacteria with Rab7 was significantly less seen (Fig. 1D). Using a BD accuri C6 flow cytometer, the phagosomes were further assessed for flow cytometry and the Rab5 co-localization with labeled bacteria was observed in 93% of isolated phagosomes (Fig. 1E).
Magnetically labeled M. avium and isolation of phagosomes. The intact phagosomes of biotin labeled tomato red clone of M. avium were separated from the total THP-1 cells lysate using the streptavidin-coated MACS microbeads as described in Materials and Methods. The labeled phagosomes with the Alexa Fluor 488-conjugated Annexin B (A) Rab5 (B) and Rab7 (C) were visualized for purity under the fluorescent microscopy. Scale bar 5μm. M. avium-containing phagosmes were stained with antibodies against Rab5 or Rab7 for 2 h at a dilution of 1:250 in PBS containing 3% BSA. After washing, phagosomes were probed with FITC-conjugated secondary antibody for 1 h and then processed for fluorescence microscopy. (D) The percentage of co-localized tomato red-labeled M. avium and FITC-labeled Rab5 and Rab7 phagosomal markers was determined by evaluating three hundred bacterial cells and express as the mean ± SD for three separate experiments. Significant differences were observed between Rab5 and Rab7 in their co-localization with the M. avium phagosome. **p < 0.001. The dtTomato M. avium-containing phagosomes stained for Rab5 were analyzed by flow cytometry as well (E). To verify the purity of intracellular M. avium sample and rule out the contaminant host proteins, bacteria isolated from human macrophages at 4 h and 24 h post-infection were incubated with the extraction buffer for 2 h with gentle agitation. The resulting supernatants (F) and the host cell total proteins of infected THP-1 cells (used for isolation of the intracellular M. avium) were visualized on a protein gel with the Coomassie staining (G). The magnetically purified M. avium phagosomes were lysed in 20 mM HEPES supplemented with the 1% Tergitol and protease inhibitor cocktail and visualized on the SDS-PAGE (H).
In order to identify the phagosomal proteins interacting with the surface of M. avium in the host environment, the adherence of vacuolar proteins to the intracellular M. avium was assayed. To insure that the isolated intracellular bacterial sample did not contain contaminant non-phagosomal proteins, the intracellular M. avium isolated at 4 h and 24 h time-points were resuspended in the extraction buffer (20 mM Octyl β-D-glucopyranoside and 25 mM EDTA; Sigma) and incubated for 2 h on a rotator at 4 °C. Resulting supernatants (Fig. 1F) as well as the host cell total protein fraction (Fig. 1G) of THP-1 cells were separated by SDS-PAGE and visualized by Coomassie staining. The proteins of phagosomal lysates are presented in the Fig. 1H. The crude phagosomal extract was incubated with M. avium, as described in the material and methods, and the unbound proteins were removed by washing bacterial cells three times in PBS. Elution of bound proteins from the bacterial surface with the extraction buffer yielded 33 proteins (Table 1). The mass spectrometric analysis identified many previously described phagosomal proteins such as ATP synthase, prelamin, prohibitin, anexin A5 and vimentin30. Unexpectedly, there were many mitochondrial proteins identified raising a possibility for the localization of mitochondrial proteins onto the phagosomal membrane. Interestingly, all three members of the eukaryotic mitochondrial porin ion channels or Voltage-Dependent Anion Channels (VDAC-1, VDAC-2 and VDAC-3) were found to be associated with M. avium surface. In order to eliminate the possibility that washing the bacterial surface with light detergent of 20 mM Octyl β-D-glucopyranoside and 25 mM EDTA, previously described in similar studies6, 31, did not result in M. avium cell lysis, the obtained mass spectrometric (MS) data were analyzed against mycobacterial database. Only seven proteins listed in the Table 2 including two mmpL4 (mycobacterial membrane protein large 4) lipoproteins, known to be localized on the surface of M. avium, were identified in the sample. To extract the bacterial surface proteins, more vigorous techniques such as partial trypsinization or selective labelling of surface proteins and affinity purification have to be applied for mycobacteria32. In addition, we performed the control experiment where the pellet of 7H9 Middlebrook broth grown M. avium was washed twice with HBSS and then incubated with the extraction buffer for 2 h. The mass spectrometric analysis of the resulting sample confirms that the incubation with the extraction buffer does not lead in bacterial cell lysis or in striping the bacterial surface (data not shown). This observation raised a possibility that identified M. avium proteins listed in the Table 2 most likely formed complexes with some of phagosomal proteins. This phenomenon was further confirmed in this study.
Inhibition of VDAC results in reduction of bacterial viability in THP-1 cells
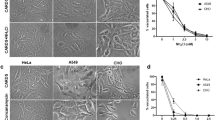
To investigate the relationship between VDAC and M. avium virulence, we inhibited channel proteins by pretreating THP-1 cells with 5 μM Cyclosporine A (CsA), a potent blocker of VDAC complex. Macrophages were treated with CsA 4 hours prior bacterial infection to avoid long incubation with these inhibitors and to prevent adverse effects and triggering functional imbalance in the host cells. While M. avium was able to enter and infect the host cells at the same rate (treated as well as untreated control), the chemical impairment of VDAC function had significant effect on bacterial growth at 1, 2 and 3 days post-infection when compared with untreated group as determined by the number of bacterial CFU (Fig. 2A).
Suppression of M. avium growth in macrophages during VDAC inhibition. (A) THP-1 cells were pretreated with 5 μM CsA and then infected with M. avium up to 3 day; cultures were harvested at indicated time points and bacterial number were determent by CFU counts. Results represent mean ± standard error of three independent experiments. **p < 0.01, the significance of differences between CsA treated and control groups at the corresponding time points. (B) Macrophages were pretreated and cultured in DIDS or control medium. Bacterial number was determined at 4 h, and 1, 2, and 3 days post-infection by plating cell lysates on 7H10 agar plates. The difference in DIDS-treated THP-1 cultures compared with untreated cells was statistically significant at day 1, 2 and 3 (P < 0.01, t test). Data are representative of three experiments. (C) In vitro growth of M. avium in aerated 7H9 medium containing DVAC inhibitors at concentrations used for tissue culture infection studies. (D) THP-1 cells (approximately 105/ml) were seeded and differentiated into macrophages with PMA in 6-well plate. After 24 h, cells were replenished with new medium and allowed to rest additional 48 h. Macrophage monolayers were washed with siRNA transfection medium and replaced with either the VDAC-1 siRNA transfection reagent or the scrabbled sequences of negative control siRNA 24 hours prior M. avium infection. Briefly, cells were lysed in CelLytic™ M lysis buffer supplemented with protease inhibitor cocktail (Sigma) and pre-cleared samples were separated on 12% Tris–HCl gels. Membranes were blocked with 3% BSA for 1 h and incubated with VDAC-1 primary antibody at a 1:250 dilution for 2 h. After, membrane was probed with the corresponding IRDye® secondary antibody (Li-Cor Biosciences, Inc) at a dilution of 1:5,000 for 30 min. THP-1 cells transfected with VDAC-1 siRNA for 72 h demonstrates efficient and specific silencing of VDAC-1 quantified via semi-quantitative western blot on the Odyssey Imager (Li-Cor). The photon emission means were recorded for each band to quantify the signal intensity. Beta-actin was used as a loading control. (E) Intracellular CFU decrease at several times of post-infection of transfected THP-1 cells with VDAC-1 siRNA or siRNA control and infected with M. avium. Data are means ± SD of three independent experiments. **p < 0.01 and *p < 0.05, the significance of differences between VDAC-1 knock down and siRNA control or M. avium infection groups.
Previous studies have suggested that the process of oligomerization of VDAC proteins led to the formation of large pores allowing for release of the proapoptotic factors such as cytochrome C out of mitochondria33. To investigate the possibility that VDAC oligomerization in M. avium phagosome membrane took place and was involved in the translocation of larger molecules into the cytoplasm, we examined the bacterial survival in macrophages that were pretreated with the 4,4′-Diisothiocyano-2,2′-disulfonic acid stilbene (DIDS), a blocker of VDAC oligomerization, 4 hours prior M. avium infection. The CFUs were recorded over 3 days following infection. The results from the inhibition of VDAC by DIDS displayed similar ability of M. avium to invade THP-1 cells as untreated control group. However, M. avium had impaired capability to grow and survive in monolayers that were treated with DIDS, and significant decrease in growth was observed at day 1, 2 and 3 time points when compared with the control group (Fig. 2B).
To rule out toxic effects of CsA or DIDS on M. avium growth in vitro, approximately 1 × 106 bacteria were cultured in 7H9 broth with or without VDAC inhibitors and OD readings were monitored over 3 days. Results indicate that CsA at 5 μM and DIDS at 20 μM had no inhibitory or killing effect on bacterial growth in vitro (Fig. 2C).
Inhibition of the VDAC-1 gene expression is associated with suppression of M. avium growth in THP-1 cells
The mass spectrometric analysis of M. avium phagosomes identified VDAC-1 channels as the most abundant channel present in the sample. Using the small interfering RNA (siRNA), we were able to suppress the expression of VDAC-1. Western blot result shows that VDAC-1 expression was significantly reduced compared to control siRNA and untreated cells (Fig. 2D). To verify the differences in M. avium survival in macrophages with functional VDAC-1 and in cells with impaired gene expression, intracellular bacteria were cultured from phagocytic cells and CFUs were recorded at several time points following infection. As observed in the Fig. 2E, while reduced M. avium growth was observed at 4 h in VDAC-1-inactivated macrophages, differences were even more significant at 1 and 2 days post-infection compared with THP-1 cells transfected with the scrambled siRNA control. M. avium was able to recover on day 2 and 3, however, bacterial growth in VDAC-1-silenced monolayers continued to lag behind when compared with scrambled siRNA controls at the same time points.
M. avium proteins interacting with VDAC-1
We hypothesized that VDAC channels may play a role in the export of bacterial proteins into the cytosol of host phagocytic cells. To examine this hypothesis, we studied interactions between VDAC-1 and selected M. avium secreted effectors (MAV_1177, MAV_2921, MAV_2941 and CipA) using the yeast two-hybrid system. Previous studies identified some of these proteins to be secreted into the cytoplasm of host cells3, 5 while CipA is secreted upon contact with cell surface34. None of these effectors showed to have positive interaction with the channel, as the resulting zygotes of both the bait and pray constructs did not grow in the absence of Ade, His, Leu, and Ttp and presence of 125 ng/ml Aureobasidin and X-a-Gal. An exception was MAV_2921; however, the yeast MAV_2921 clone did not turn blue in the presence of X-a-Gal meaning that the transcription of the α-galactosidase reporter gene MEL-1 did not take place, giving the false interaction result with VDAC-1 (Fig. 3A).
In vitro protein-protein interaction between VDAC-1 and M. avium effectors. The open reading frame of VDAC-1 cDNA encoding a 283 amino acid protein was amplified from the human sequence-verified Clone ID: 6023095 (Dharmacon), which was isolated from the NIH_MGC library. The yeast two-hybrid interaction of VDAC-1 with the bacterial target proteins MAV_1177 (1), MAV_2921 (2), MAV_2941 (3) and CipA (4) showed a negative or false positive interaction. The screening of mmpL4 lipoproteins MAV_0084 (5) and MAV_4996 (6) as well as ATP synthase alpha (7) and beta (8) subunits established positive interaction with VDAC-1. The known interaction between pGBKT7-53 and pGADT7-T served as a positive control (+), whereas pGBKT7-lam and pGADT7-T were used as a control for a negative interaction (−).
We then performed the pull-down assay to expand our search in finding M. avium proteins that might interact with VDAC-1. Only two M. avium proteins, ATP synthase subunit alpha and beta were found to bind VDAC-1 (Table 3). The further investigation through the yeast two-hybrid system, shown in the Fig. 3B, has proved the specificity of the interaction of both subunits of the ATP synthase.
The mmpL4 proteins were identified in the M. avium surface-bound phagosome fraction using mass spectrometric analysis. Due to the fact that mmpL proteins participate in export of mycobacterium cell wall components to the bacterial surface and genetic interferences affect pathogen fitness in vitro and in vivo 35, we examined the yeast two-hybrid interaction between mmpL4 lipoproteins (MAV_0084 and MAV_4996) and VDAC-1, finding it to be positive (Fig. 3B).
Immunostaining reveals co-localization of VDAC-1 with mmpL4
We also performed immunofluorescence staining of VDAC-1 in THP-1 cells that were infected with either a M. avium clone containing the Red Fluorescent Protein (RFP) or a clone overexpressing mmpL4 (MAV_4696) protein in fusion with RFP. While the granular fluorescence of VDAC-1 protein was dispersed in the cytosol of uninfected cells (Fig. 4A and B), M. avium infected cells showed punctate staining on bacterial vacuoles (Fig. 4A). VDAC-1 staining in infected THP-1 cells revels that this channel protein is always localized with bacterial-containing phagosomes. The fact that the phagosome membrane is originated from the host cell plasma membrane during the infection process and VDAC-1 is one of the components of the plasma membrane36, 37, may explain the observation. In addition, the VDAC-1 was stained with a greater intensity on M. avium vacuoles overexpressing the mmpL4 protein (Fig. 4B) than in control macrophages (Fig. 4A), suggesting the host protein co-localization with this bacterial surface protein.
The co-localization of VDAC-1 on phagosomes of M. avium expressing mmpL4 protein. Representative images of constitutively expressing RFP (A) and RFP:mmpL4 (B) proteins in M. avium show subcellular co-localization of VDAC-1 on bacterial vacuoles. The arrows highlight specific regions of interest visualizing the overlapping yellow pixel clusters (co-localization). Images contain uninfected control cells as well. All images were obtained using 100x oil objective of a fluorescent microscope (Leica). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Two images are included for each experimental group. Bar = 10 μm.
The role of VDAC in M. avium cell wall lipid release in macrophages
Mycobacterial mmpL proteins have been well documented to be involved in the biosynthesis and export of cell wall lipid constituents, and play a role in mycobacterium pathogenesis38. In addition, recent studies on VDAC have generated strong evidence on its association/interaction with host lipids39, 40. The ability of VDAC to influence the cholesterol distribution of mitochondrial membrane has been also recently demonstrated41, and cholesterol and ergosterol have been found to form complex with purified VDAC protein42. It also has been established that the oligomerization of VDAC can be significantly influenced by lipids40. In attempts to investigate the possible relation between VDAC, mmpL4 proteins and M. avium surface-associated lipid export into macrophages, we pretreated THP-1 cells with DIDS for 4 hours and then infected cells with Texas red hydrazide-labeled M. avium. The DIDS was kept up to 24 h in the culture medium and lipid release from bacterial surface was analyzed by fluorescent microscopy. THP-1 cells without DIDS treatment served as a control. As previously identified by Beatty et al.15, the extensive release of the Texas red label from mycobacterial surface was observed at 24 h post-infection of THP-1 (Fig. 5A). In contrast, macrophages treated with DIDS had the red fluorescent label markedly contained within M. avium phagosomes, suggesting the involvement of VDAC in bacterial cell wall component translocation. Evaluation of two hundred M. avium-infected THP-1 cells without DIDS treatment confirmed the observation that majority (87%) of the host macrophages permeated the red fluorescence that was released from the Texas Red-labeled bacteria. Conversely, only 19% of the DIDS treated macrophages had a positive staining (Fig. 5B). Results were further confirmed using the flow cytometry (Fig. 5C). To insure that the fluorescent labeling of host cells was not the result of M. avium presence in the cytosol, the percentage of Rab5 positive phagosomes were calculated in THP-1 cells with and without DIDS treatment and the co-localization rate of Rab5 in both groups were observed to be similar (Fig. 5D).
M. avium cell wall lipid release inside of macrophages. (A) THP-1 cells with or without DIDS treatment were infected with Texas Red hydrazyde-labeled M. avium with MOI of 25:1 for 24 h and analyzed by fluorescent microscopy. While significant release of fluorescent label from bacterial phagosomes are observed in wells without DIDS treatment, the export of bacterial cell wall components into the cytosol of macrophages are substantially reduced as observed on micrographs obtained from infected THP-1 cells during VDAC inhibition. Two images are included for each experimental group. Scale bar 10μm. (B) The percentage of the host macrophages permeated the red fluorescence released from the Texas Red hydrazyde-labeled M. avium. Results represent means ± standard error of three independent experiments. *, p < 0.001, the significance of differences between M. avium infected THP1 cells with and without DIDS treatment. (C) M. avium infected THP-1 macrophages with DIDS (blue trace) or without DIDS (red trace) treatment were analyze by flow cytometry to discern lipid transport as described in the materials and methods. The host cells without infection are shaded grey. (D) To visualize and demonstrate the colocalization of Rab5 with the Texas Red hydrazide stained M. avium directly in THP-1 infected cells without DIDS treatment was technically impossible, due to the massive release of lipids within the host cells. Thus, the percentage of M. avium co-localization with Rab5 phagosomal marker was determined by evaluating three hundred M. avium-containing phagosmes, which were isolated from THP-1 cells with and without DIDS treatment at 24 h post-infection as described in materials and methods. Results represent means ± standard error of two independent experiments.
Discussion
M. avium, like many other pathogenic mycobacteria, is highly adapted for survival within phagocytic cells. In the vacuolar compartments, bacteria are isolated from a rich source of nutrients existing in the cytoplasm. The phagosome membrane separates the intravacuolar bacteria from the cytoplasm and therefore is placed between released virulence factors and targets in the host cell cytoplasm. Many studies have demonstrated that M. avium as well as Mycobacterium tuberculosis secrete virulence factors inside the vacuole environment5, 15, 20, 43. Differently from gram-negative bacteria, such as Salmonella, mycobacteria do not have classical type III or type IV secretion systems to inject effector molecules across membranes. Both M. avium and M. tuberculosis are equipped with the type VII secretion system which is a major export mechanism for proteins belonging to the ESX regions3, 17. M. tuberculosis also uses a pili-like structure to secrete two proteins involved in apoptosis inhibition of macrophages44. If other proteins employ the above mentioned secretion apparatus as well, it is currently unknown. Therefore, in the absence of an “injectosome”, such as the types III and IV secretion systems, M. avium depends of an alternative approach to deliver the virulence-associated molecules to the cytoplasm of macrophages.
In this study, we investigated the vacuole membrane transport mechanisms that are used by the pathogen to be able to translocate secreted molecules across the vacuole membrane. We hypothesized that bacterial effectors would recognize a counterpart on the vacuole membrane facilitating the transport of exported products. In fact, our screen identified a large number of host proteins on the phagosome membrane that are recognized by bacterial surface structures. The majority of the models of phagocytosis describe the plasma membrane as the main source of the phagosomal membrane formation. Several proteins identified in our study have been previously demonstrated to be part of the plasma and phagosomal membranes30. The mass spectrometric sequencing, however, also identified several mitochondrial proteins on M. avium phagosomes. The fact is that pathogens such as Salmonella, mycobacteria and Leishmania avoid the degradation into phagolysosomes by inhibiting the fusion of the phagosomes with the endocytic organelles45. The vacuolar compartments of pathogens such as Brucella 46 and Chlamydia 47 receive lipids from the trans-Golgi network and end up having the endoplasmic reticulum or Golgi features on the phagosomes. Previous proteomics studies have also reported the presence of mitochondrial proteins in phagosomal membrane48, 49. Although it is still difficult to define the role of many organelle proteins in the vacuole membrane, evidence suggests that the plasma membrane is probably not the only source of phagosomal membrane and some select class of mitochondrial proteins may also contribute to formation of phagosomes. Among mitochondrial proteins previously shown to be part of the phagosome30, VDAC proteins are strong candidates for translocation of bacterial-derived molecules. VDAC are voltage-channels that initially were described as mitochondria-associated porins responsible for ATP/ADP exchange, and involved in the flux of metabolites across the mitochondrial outer membrane50, 51. More recently, these channels have been identified on the plasma membrane36, 37, and in phagosome membranes of latex beads, M. bovis BCG and Brucella vacuoles30, 52, 53. Studies have shown the ability of VDAC to bind to and transport cholesterol, and influence its distribution between the inner and outer mitochondrial membranes41, 54. The VDAC was found to allow the translocation of DNA sequences across a planar membrane55. In addition, the transport of large molecule such as the cytochrome C across the mitochondrial membrane56 was accomplished following fusion of several VDAC molecules to form a large pore known as an oligomerization process57.
In order to examine if VDAC had a role in the transport of M. avium secreted proteins, first we selected known effector proteins to be exported in the cytosol of macrophages and investigated protein-protein interaction using the yeast two-hybrid system. We also performed the pull-down assay, however, only two M. avium proteins of alpha and beta subunits of ATP synthase (ATPases) were found to bind VDAC-1. These interactions were further confirmed with the yeast two-hybrid system and the binding of the host VDAC-1 molecule to bacterial ATPases were found to be positive. Previous studies have described the association of ATPases with the surface of intracellular M. avium 32 and at the mycobacterial surface during biofilm formation (Rose at). M. tuberculosis ATPases function in the cell envelope58 providing energy for substrate transport59 and driving type VII protein export across the cytoplasmic membrane60. On the other hand, the interaction between host VDAC and ATPases, and regulation of ATP trafficking in and out of the mitochondria has been well documented61. The above facts strongly support our finding that bacterial ATPases can be associated with VDAC and possibly are involved in this channel gating. This hypothesis, however, requires further confirmation in the experimental systems.
We were unsuccessful to demonstrate that bacterial secreted proteins employ the VDAC system as a mechanism of transport. During our investigation, however, it became clear that the function and oligomerization of VDAC are crucial for M. avium growth within phagosomes of the host macrophage. We were able to demonstrate that VDAC-1 protein co-localizes and interacts with M. avium mmpL4 proteins. MmpL family proteins are unique to the mycobacterial core genome, and a growing body of literature indicates that the primary role of most mmpL proteins are dedicated to transport of mycobacterial lipids for incorporation into the cell wall35. Inactivation of many of these genes leads to failure to export the mycolic acid-containing lipids and mycolate ester wax to the bacterial surface. The lipid export function has been described for mmpL3, mmpL7, mmpL8 and mmpL11. The recent study suggests that mmpL3 transport trehalose out of the cell wall, and its inhibition prevents the incorporation of de novo synthesized mycolic acids into the cell wall62. In fact, using the β-lactamase reporter transposon, Dr. Braunstein’s group has mapped the exported protein domains of MmpL463. The location as well as the identity of mmpL4 transporter substrates has not been fully elucidated, however, the functional studies suggest that mmpL4 is involved in the biosynthesis of cell surface polyketides and the glycopeptidolipids64 and most likely is juxtaposed to the cell wall as the majority of the mmpL family proteins.
Beatty and colleagues15 demonstrated that mycobacterial lipids are released from the bacterial phagosome and accumulate in late endosomal/lysosomal compartments of macrophages. Due to the fact that bacterial lipids were also found in extracellular milieu and subsequently internalized by uninfected neighboring macrophages, the authors raised the possibility that mycobacterial exported lipids most likely have an immunomodulatory effect contributing to the control of surrounding uninfected cells. This hypothesis was later confirmed by O’Neil and colleagues65. On the other hand, it has been shown that the presence of specific host lipids can change VDAC conformational equilibrium and regulate the voltage gating of the channel66. VDAC is also capable to bind and transport the host lipids41, 54. In this study, we examined whether blocking the VDAC oligolimerization process had any inhibitory effect on M. avium lipid export. Indeed, we observed the significant decrease in bacterial lipid export in host macrophages during DIDS treatment when compared with the untreated control. Currently, it is unknown whether VDACs are the only channel-forming proteins associated with the translocation of mycobacterial lipids. Previous studies using the morphological and biochemical analysis of phagosomes of isolated latex beads identified the VDAC as one of the component of the phagosome membrane30. The presence of VDAC on phagosomes of Bacille Calmette-Guerin (BCG)53 and Brucella-infected macrophages52 raises the possibility that the transport mechanism may be common among some pathogens. All these observations, including our study, suggest that the VDAC proteins previously identified in other cellular compartments are representative of more than a simple contamination and the VDAC molecules are genuine constituents of phagosomes.
Mycobacteria inside the macrophage vacuole appear to use host cell transport system to translocate virulence factors into the cytoplasm. Our finding is in agreement with the observation by de Chastellier and colleagues67 who discovered that the contact between bacteria and phagosome membrane is required for M. avium survival in macrophages. Our data suggests that at least in some cases, the export of bacterial constituents begins with the recognition of a transport system in the vacuole membrane by a M. avium mmpL4 proteins. Recent report indicated that treatment of M. tuberculosis-infected macrophages with cyclosporin A protects mitochondria from the mitochondrial permeability transition68. This process blocks the host cell necrosis induced by this pathogen and shifts to apoptotic death enhancing antimycobacterial activity of macrophages and killing of intracellular M. tuberculosis. While it may be the only explanation, we also want to highlight that our observation raises another possibility. In the M. avium model, the inhibition of apoptosis and induction of necrosis do not occur, and therefore bacterial attenuation in the macrophage is unlikely to be explained by the cell necrosis. Furthermore, the use of siRNA and the absence of observation of necrosis in monolayers exposed to the inhibitor and control monolayers, ruled out the possibility.
In the current study, we demonstrate that the VDAC transport system interacts with mmpL4 proteins on the vacuole membrane of M. avium, and functional channels are required for the pathogen survival in macrophages. The underlying mechanism of interaction between bacterial ATPases and VDAC molecules is still unknown, but based on the existing research literature there is a possibly that ATPases may regulate the channel function. In this work, we can conclude that M. avium alters the VDAC function in a pathogen-directed manner. The pathogen translocates bacterial lipids via VDAC system and inhibition of the oligomerization process of the VDAC channel contributes to the dynamic changes of mycobacterial intraphagosome and, thus, M. avium survival within the phagocyte. Understanding the molecular basis of phagosome channels, its regulation and activation mechanism most likely will have a crucial importance for designing new therapeutic tools against mycobacterial diseases.
Materials and Methods
Bacterial strain and hydrazide labeling
Mycobacterium avium strain 104 was originally isolated from the blood of AIDS patients with disseminated infection. Bacteria were maintained in Middlebrook 7H9 broth (BD Biosciences) supplemented with 10% (vol/vol) olei acid-albumin-dextrose-catalase (OADC; Hardy Diagonstics) at 37 °C for 6 days. The tomato red clone of M. avium 104 strain was created suing pJDC60 mycobacterial plasmid expressing the tdTomato gene under L5 promoter provided by Dr. Jeffrey Cirillo at Texas A&M University System Health Science Center, College Station, TX. This clone was maintained in the Middlebrook culture medium supplemented with 400 μg/ml kanamycin. Mycobacterium surface-exposed terminal oxidizable carbohydrates were labeled with hydrazide according the protocol published by Beatty et al.15. Before labeling, bacterial cells were washed twice with PBS containing 0.05% Tween 80, and resuspended in 0.1 M sodium acetate and 1 mM sodium periodate (Sigma) solution at pH 5.5. M. avium was gently rotated for 20-min at 4 °C and then the reaction was stopped by adding 0.1 mM glycerol. Bacterial cell suspension was washed three times with PBS supplemented with 0.05% Tween 80 followed by 2 h incubation in PBS/Tween containing 1 mM Texas Red hydrazide (Molecular Probes) at room temperature. The culture was washed twice and, prior infection, the bacterial viability was determined by colony forming units (CFU) on Middlebrook 7H10 agar.
Cell culture maintenance and infection
The THP-1 human monocyte cell line was purchased from the American Type Culture Collection (ATCC) and maintained in Roswell Park Memorial Institute medium (RPMI; Corning) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gemini) in 75 cm3 flasks. Prior infection, cells were differentiated by adding 5ng/ml of phorbol 12-myristate 13-acetate (PMA, Sigma Aldrich) to culture medium and, depending on experiment performed, were seeded in range of 60–95% confluence into 6-, 24-well plates, two-chamber glass slides or T-200 tissue culture flasks. Following 24 h incubation at 37 °C in an atmosphere of 5% CO2, cell culture medium was replenished with fresh new medium and incubated for additional 48–72 h for cell differentiation. Macrophages were infected with mid-log phase grown M. avium and after two hours post-infection, wells were extensively washed with the Hank’s Balanced Salt Solution (HBSS, Life technologies), and the total number of viable bacteria in the inoculum as well as cell-associated bacteria over time were determined by CFU counts. In all experiments, except infections with the hydrazide-labeled bacteria, the multiplicity of infection (MOI) was adjusted to ~10 bacteria per macrophage.
Magnetic isolation of intact phagosomes
The mid-log phase grown M. avium 104 in Middlebrook 7H9 broth were pelleted, washed twice with HBSS and passed 10 times through a 27-gauge needle to ensure a single cell suspension. M. avium was incubated at room temperature with 1 mg/ml EZ-Link sulfo-NHS- LC biotin (Thermo Fisher Scientific) in PBS for 30 minutes. The reaction was stopped by washing bacterial pellet with PBS supplemented with 0.1 M glycine at pH 7.2, and then the pellet was resuspended in PBS with 0.05% Tween-80 to remove unbound biotin. Biotinylated M. avium was incubated under gentle agitation with streptavidin-coated microbeads (Miltenyi Biotech) for 20 minutes at room temperature. Macrophages were seeded up to 95% confluence in T-200 flasks and infected with labeled M. avium at MOI of 10:1. After 4 h and 24 h incubation at 37 °C 5% CO2, macrophages were scraped and resuspended in homogenization buffer (1 M HEPES with 1 M sucrose; Life Technologies) containing protease inhibitors cocktail (Sigma). THP-1 cells were then mechanically lysed by multiple passages through a 27-gauge needle. Intact phagosomes were selected through a MiniMACS column on a magnetic selector obtained from Miltenyi Biotech and the bound phagosomes were eluted in PBS. Isolated phagosomes were incubated with Alexa Fluor 488- conjugated Annexin B (Thermo Fisher Scientific) at a dilution of 1:1,000 and visualized on a Leica DM4000B micriscope. In addition, Rab5 and Rab7 phagosome markers were immunostained using anti-Rab5 and anti-Rab7 mouse monoclonal antibodies (Santa Cruz Biotechnology) at a dilution of 1:500 followed by visualization with corresponding FITC conjugated secondary antibody (1:1,000). Three hundred bacterial cells expressing the tomato red protein were evaluated to calculate the percentage of positive phagosomes for either Rab5 or Rab7. Purified phagosomes were further processed for protein purification as follows: phagosomes were resuspended in 1% Tergitol (Sigma) in 20 mM HEPES (Sigma) supplemented with the protease inhibitor cocktail (Sigma) and lysed overnight. Twenty-four hours later, the suspension was centrifuged to remove bacteria and microbeads, and protein sample was processed for electrophoresis and Coomassie staining.
Isolation of M. avium surface bound phagosomal proteins
The intracellular, non-biotin labeled, M. avium 104 was extracted from THP-1 cells at 4 h and 24 h time-points of infection as previously described69. To assess if samples had any host cell protein contaminants, isolated bacteria were washed twice in cold HBSS, and then incubated in the extraction buffer (20 mM Octyl β-D-glucopyranoside and 25 mM EDTA; Sigma) for 2 h on a rotator at 4 °C. Resulting supernatants were separated by SDS-PAGE and visualized by Coomassie staining.
Isolated phagosomal proteins were combined with the intracellular M. avium and incubated at 4 °C. After 24 h, bacterial pellet was centrifuged at 3,500 rpm for 20 min, washed three times with PBS and resuspended in the extraction buffer to elute any phagosomal protein that was bound to the surface of the intracellular M. avium. The bacteria were pelleted down and collected supernatant was processed for the buffer exchange procedure using 3 kDa filters and the 25 mM ammonium bicarbonate as the exchange buffer. Eluted phagosomal proteins were trypsin digested in solution at 37 °C for 5 h and sequenced by electrospray ionization mass spectrometry (ESI-MS/MS) at the Oregon Health Science University (OHSU) proteome facility.
Construction of mmpL4 overexpression M. avium clone
To demonstrate binding of bacterial mmpL4 protein to VDAC-1, the full length MAV_4696 gene was cloned into HindIII site of pMV261–5HRFP3 as C-terminal fusions to a monomeric RFP moiety with an N-terminus 6X-His tag. Vector construction and gene cloning confirmation were performed in E. coli. Vectors with and without mmpL4 gene were transformed into M. avium and selected on Middlebrook 7H10 agar containing kanamycin 400 μg/ml. Resulting red colonies were selected for immunostaining experiments. Following infection of THP-1 cells, we analyzed bacterial and host protein co-localization with fluorescent microscopy.
Immunofluorescent microscopy
Approximately, 1 × 105 THP-1 cells were seeded in 2-chamber slides and infected with hydrazide-labeled bacteria with MOI of 25 bacteria to 1 cell. Following 4 h and 24 h infection, cells were fixed in 4% formaldehyde for 30 min. VDAC-1 antibody was purchased from the Santa Cruz Biotechnology, Inc and used at 1:100 dilution followed by visualization with corresponding FITC conjugated secondary antibody (1:1,000). Slides were mounted and observed under a Leica DM4000B fluorescent microscope (Leica).
Effect of VDAC inhibitors, cyclosporine A and 4,4′-Diisothiocyano-2,2′-disulfonic acid stilbene, on M. avium growth
Two widely used inhibitors for VDAC channels: cyclosporine A (CsA; Novartis), an inhibitor of the CA2+- dependent VDAC pore (Lobatón et al., 2004; Yuqi et al., 2009), and 4,4′-Diisothiocyano-2,2′-disulfonic acid stilbene (DIDS), a blocker of VDAC oligomerization, were selected to impair the channel function. Prior macrophage inhibition assays, we tested effects of CsA (5 μM) and DIDS concentrations (20–200 μM), used in tissue culture studies, on M. avium viability. Bacteria were incubated with 5 μM CsA and 20–200μM-concentration range of DIDS and CFUs were recorded at 4 h, 1d, 2d, and 3d post-infection. Five micromole CsA and 20 μM of DIDS were used for further studies due to the fact that the 100–200 μM concentration range of DIDS led to significant reduction of bacterial number in culture (Data not shown). There was no inhibitory effect in range of 20–50 μM.
Inhibition of VDAC-1 channel
Approximately, 1 × 105 THP-1 macrophage-like cells were seeded in 24-well plates and pre-treated with either 5 μM CsA or 20 μM DIDS for 4 h. Cells were then infected with M. avium 104 for 2 h at MOI of 10:1, washed 3 times with HBSS and replenished with new RPMI medium supplemented with 10% FBS but without CsA or DIDS. Macrophages were lysed with 0.1% Triton X-100 at 4 h, day1, 2 and 3 post-infection, plated and CFUs were determined.
Inactivation of VDAC-1 by siRNA
THP-1 cells were seeded at 60% confluence in 6-well plates and, 24 hours prior infection, transfected with control (scrabbled sequences) as well as experimental (VDAC-1) siRNAs purchased from Santa Cruz Biotechnology. Briefly, siRNAs were diluted in DMEM without serum at a final concentration of 25 nM and 3μl of ContinuumTM transfection reagent (Gemini) was added into diluted siRNA. The transfection mixture was added drop-wise to monolayers and then incubated at 37 °C in presence of 0.5% CO2 for 24 h. Next day, cells were infected with M. avium for 4 h, 1d, 2d, and 3d and CFUs were recorded on Middlebrook 7H10 agar plates. The VDAC-1 and β-actin protein levels from control and experimental wells were analyzed by semi-quantitative Western blotting on the Odyssey Imager (Li-Cor).
Western Blot
Samples were mixed with an equal volume of 2X Laemmli sample buffer (Bio-Rad), resolved onto SDS-PAGE gel (Bio-Rad) and transferred to a nitrocellulose membrane (Bio-Rad). Membrane was blocked with 3% bovine serum albumin (BSA) in phosphate buffered saline (PBS) overnight. After, the membrane was incubated with primary antibody at a dilution of 1:250 for 2 h. Membrane was washed three times with PBS and then probed with corresponding IRDye secondary antibody (Li-Cor Biosciences, Inc) at a dilution of 1:5000 for 1 h. Proteins were visualized using Odyssey Imager (Li-Cor).
The Yeast Two-Hybrid interaction
The VDAC-1 gene was fused in frame with the GAL4 DNA binding domain by inserting the PCR-generated fragment into the EcoRI and BamHI sites of pGBKT7 (Clontech). The resultant bait vector pGBKT7:VDAC-1 was transformed into Saccharomyces cerevisiae strain Y2HGold using Yeastmaker Yeast Transformation System 2, according to the manufacturer’s instructions (Clontech). The following M. avium genes: MAV_1177, MAV_2921, MAV_2941 and CipA (MAV_4671) encoding secreted proteins identified in previous studies3, 5, 34 were fused with the GAL4 activation domain of pGADT7 and transformed into the yeast strain Y187 (Clontech). In addition, protein interaction experiments were extended for mmpL4 lipoproteins MAV_0084 and MAV_4996 as well as ATP synthase subunits of alpha (MAV_1525) and beta (MAV_1527) following the MS analysis of bacterial proteins from Table 2 and pull-down assay (Table 3). Plasmids pGBKT7–53, pGBKT7-lam, and pGADT7-T were used as positive and negative controls (Clontech). One ml of bait strain was combined with the one ml of prey strain and was grown in 2xYPDA liquid medium containing 50 μg/ml kanamycin at 30 °C for 24 h. The yeast zygotes that grew of blue color on Quadruple Dropout agar plates in absence of tryptophan (Trp), leucine (Leu), histidine (His), adenine, (Ade) (SD–Ade/–His/–Leu/–Trp) that contained 20 mg/ml X-a-Galactosidase and 125 ng/ml Aureobasidin were identified as positive clone.
Immunoprecipitation of VDAC-1 and pull-down assay
THP-1 cells were lysed and proteins were precleared as previously described44. Ten microgram of anti-VDAC-1 mouse monoclonal antibody (Santa Cruz Biotechnology) conjugated to agarose beads was added to approximately 500μg of total cellular protein and incubated at 4 °C for 4 h. The sample was centrifuged at 2,500 rpm for 1 min and washed 3 times with PBS. The captured VDAC-1 was then added to M. avium total cell proteins and incubated at 4 °C for overnight. Twenty-four hours later, sample was washed three times with PBS and processed for tryptic Digestion (Thermo Fisher Scientific). Protein sequencing was performed at the Oregon Health and Science University proteomics facility by electrospray ionization mass spectrometry (ESI-MS/MS).
Statistical analysis
All data are presented as ± SD and comparison of variables is performed using the unpaired Student’s t test. Statistical significance was set at P < 0.05.
References
Armstrong, D. et al. Treatment of infections in patients with the acquired immunodeficiency syndrome. Ann Intern Med 103, 738–743 (1985).
Griffith, D. E. et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175, 367–416, doi:10.1164/rccm.200604-571ST (2007).
McNamara, M., Danelishvili, L. & Bermudez, L. E. The Mycobacterium avium ESX-5 PPE protein, PPE25-MAV, interacts with an ESAT-6 family Protein, MAV_2921, and localizes to the bacterial surface. Microb Pathog 52, 227–238, doi:10.1016/j.micpath.2012.01.004 (2012).
Li, Y., Miltner, E., Wu, M., Petrofsky, M. & Bermudez, L. E. A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell Microbiol 7, 539–548, doi:10.1111/j.1462-5822.2004.00484.x (2005).
Danelishvili, L. & Bermudez, L. E. Mycobacterium avium MAV_2941 mimics phosphoinositol-3-kinase to interfere with macrophage phagosome maturation. Microbes Infect. doi:10.1016/j.micinf.2015.05.005 (2015).
Danelishvili, L. et al. Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc Natl Acad Sci USA 104, 11038–11043, doi:10.1073/pnas.0610746104 (2007).
Sturgill-Koszycki, S. et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263, 678–681 (1994).
Frehel, C., de Chastellier, C., Lang, T. & Rastogi, N. Evidence for inhibition of fusion of lysosomal and prelysosomal compartments with phagosomes in macrophages infected with pathogenic Mycobacterium avium. Infect Immun 52, 252–262 (1986).
Crowle, A. J., Dahl, R., Ross, E. & May, M. H. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect Immun 59, 1823–1831 (1991).
Appelberg, R. & Orme, I. M. Effector mechanisms involved in cytokine-mediated bacteriostasis of Mycobacterium avium infections in murine macrophages. Immunology 80, 352–359 (1993).
Bermudez, L. E. & Young, L. S. Oxidative and non-oxidative intracellular killing of Mycobacterium avium complex. Microb Pathog 7, 289–298 (1989).
Cooper, A. M., Pearl, J. E., Brooks, J. V., Ehlers, S. & Orme, I. M. Expression of the nitric oxide synthase 2 gene is not essential for early control of Mycobacterium tuberculosis in the murine lung. Infect Immun 68, 6879–6882 (2000).
Bermudez, L. E., Parker, A. & Petrofsky, M. Apoptosis of Mycobacterium avium-infected macrophages is mediated by both tumour necrosis factor (TNF) and Fas, and involves the activation of caspases. Clin Exp Immunol 116, 94–99 (1999).
Early, J., Fischer, K. & Bermudez, L. E. Mycobacterium avium uses apoptotic macrophages as tools for spreading. Microb Pathog 50, 132–139, doi:10.1016/j.micpath.2010.12.004 (2011).
Beatty, W. L. et al. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic 1, 235–247 (2000).
Danelishvili, L., McGarvey, J., Li, Y. J. & Bermudez, L. E. Mycobacterium tuberculosis infection causes different levels of apoptosis and necrosis in human macrophages and alveolar epithelial cells. Cell Microbiol 5, 649–660 (2003).
Abdallah, A. M. et al. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol 5, 883–891, doi:10.1038/nrmicro1773 (2007).
Stanley, S. A., Raghavan, S., Hwang, W. W. & Cox, J. S. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci USA 100, 13001–13006, doi:10.1073/pnas.2235593100 (2003).
Lewis, K. N. et al. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis 187, 117–123, doi:10.1086/345862 (2003).
Guinn, K. M. et al. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol 51, 359–370, doi:10.1046/j.1365-2958.2003.03844.x (2004).
Tan, T., Lee, W. L., Alexander, D. C., Grinstein, S. & Liu, J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol 8, 1417–1429, doi:10.1111/j.1462-5822.2006.00721.x (2006).
Wang, X. et al. ESAT-6 inhibits production of IFN-gamma by Mycobacterium tuberculosis-responsive human T cells. J Immunol 182, 3668–3677, doi:10.4049/jimmunol.0803579 (2009).
Simeone, R. et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog 8, e1002507, doi:10.1371/journal.ppat.1002507 (2012).
Ramakrishnan, L., Federspiel, N. A. & Falkow, S. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288, 1436–1439 (2000).
Abdallah, A. M. et al. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol 73, 329–340, doi:10.1111/j.1365-2958.2009.06783.x (2009).
Banu, S. et al. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol 44, 9–19 (2002).
Delogu, G. & Brennan, M. J. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect Immun 69, 5606–5611 (2001).
Grotzke, J. E. et al. The Mycobacterium tuberculosis phagosome is a HLA-I processing competent organelle. PLoS Pathog 5, e1000374, doi:10.1371/journal.ppat.1000374 (2009).
Kelley, V. A. & Schorey, J. S. Mycobacterium’s arrest of phagosome maturation in macrophages requires Rab5 activity and accessibility to iron. Mol Biol Cell 14, 3366–3377, doi:10.1091/mbc.E02-12-0780 (2003).
Desjardins, M. et al. Molecular characterization of phagosomes. The Journal of biological chemistry 269, 32194–32200 (1994).
Rao, S. P., Ogata, K. & Catanzaro, A. Mycobacterium avium-M. intracellulare binds to the integrin receptor alpha v beta 3 on human monocytes and monocyte-derived macrophages. Infect Immun 61, 663–670 (1993).
McNamara, M., Tzeng, S. C., Maier, C., Zhang, L. & Bermudez, L. E. Surface proteome of “Mycobacterium avium subsp. hominissuis” during the early stages of macrophage infection. Infect Immun 80, 1868–1880, doi:10.1128/IAI.06151-11 (2012).
Shoshan-Barmatz, V., Keinan, N., Abu-Hamad, S., Tyomkin, D. & Aram, L. Apoptosis is regulated by the VDAC1 N-terminal region and by VDAC oligomerization: release of cytochrome c, AIF and Smac/Diablo. Biochim Biophys Acta 1797, 1281–1291, doi:10.1016/j.bbabio.2010.03.003 (2010).
Harriff, M. J. et al. Mycobacterium avium genes MAV_5138 and MAV_3679 are transcriptional regulators that play a role in invasion of epithelial cells, in part by their regulation of CipA, a putative surface protein interacting with host cell signaling pathways. Journal of bacteriology 191, 1132–1142, doi:10.1128/JB.01359-07 (2009).
Pacheco, S. A., Hsu, F. F., Powers, K. M. & Purdy, G. E. MmpL11 protein transports mycolic acid-containing lipids to the mycobacterial cell wall and contributes to biofilm formation in Mycobacterium smegmatis. The Journal of biological chemistry 288, 24213–24222, doi:10.1074/jbc.M113.473371 (2013).
Bathori, G. et al. Porin is present in the plasma membrane where it is concentrated in caveolae and caveolae-related domains. The Journal of biological chemistry 274, 29607–29612 (1999).
Bahamonde, M. I., Fernandez-Fernandez, J. M., Guix, F. X., Vazquez, E. & Valverde, M. A. Plasma membrane voltage-dependent anion channel mediates antiestrogen-activated maxi Cl- currents in C1300 neuroblastoma cells. The Journal of biological chemistry 278, 33284–33289, doi:10.1074/jbc.M302814200 (2003).
Lamichhane, G., Tyagi, S. & Bishai, W. R. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun 73, 2533–2540, doi:10.1128/IAI.73.4.2533-2540.2005 (2005).
Martel, C., Wang, Z. & Brenner, C. VDAC phosphorylation, a lipid sensor influencing the cell fate. Mitochondrion 19 Pt A, 69–77, doi:10.1016/j.mito.2014.07.009 (2014).
Betaneli, V., Petrov, E. P. & Schwille, P. The role of lipids in VDAC oligomerization. Biophys J 102, 523–531, doi:10.1016/j.bpj.2011.12.049 (2012).
Campbell, A. M. & Chan, S. H. The voltage dependent anion channel affects mitochondrial cholesterol distribution and function. Arch Biochem Biophys 466, 203–210, doi:10.1016/j.abb.2007.06.012 (2007).
De Pinto, V., Benz, R., Caggese, C. & Palmieri, F. Characterization of the mitochondrial porin from Drosophila melanogaster. Biochim Biophys Acta 987, 1–7 (1989).
Beatty, W. L. & Russell, D. G. Identification of mycobacterial surface proteins released into subcellular compartments of infected macrophages. Infect Immun 68, 6997–7002 (2000).
Danelishvili, L., Yamazaki, Y., Selker, J. & Bermudez, L. E. Secreted Mycobacterium tuberculosis Rv3654c and Rv3655c proteins participate in the suppression of macrophage apoptosis. PLoS One 5, e10474, doi:10.1371/journal.pone.0010474 (2010).
Meresse, S. et al. Controlling the maturation of pathogen-containing vacuoles: a matter of life and death. Nat Cell Biol 1, E183–188, doi:10.1038/15620 (1999).
Pizarro-Cerda, J. et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun 66, 5711–5724 (1998).
Heinzen, R. A., Scidmore, M. A., Rockey, D. D. & Hackstadt, T. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun 64, 796–809 (1996).
Garin, J. et al. The phagosome proteome: insight into phagosome functions. J Cell Biol 152, 165–180 (2001).
Rogers, L. D. & Foster, L. J. The dynamic phagosomal proteome and the contribution of the endoplasmic reticulum. Proc Natl Acad Sci USA 104, 18520–18525, doi:10.1073/pnas.0705801104 (2007).
Schein, S. J., Colombini, M. & Finkelstein, A. Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from paramecium mitochondria. J Membr Biol 30, 99–120 (1976).
Lemasters, J. J. & Holmuhamedov, E. Voltage-dependent anion channel (VDAC) as mitochondrial governator–thinking outside the box. Biochim Biophys Acta 1762, 181–190, doi:10.1016/j.bbadis.2005.10.006 (2006).
Fugier, E. et al. The glyceraldehyde-3-phosphate dehydrogenase and the small GTPase Rab 2 are crucial for Brucella replication. PLoS Pathog 5, e1000487, doi:10.1371/journal.ppat.1000487 (2009).
Lee, B. Y. et al. The Mycobacterium bovis bacille Calmette-Guerin phagosome proteome. Mol Cell Proteomics 9, 32–53, doi:10.1074/mcp.M900396-MCP200 (2010).
Campbell, A. M. & Chan, S. H. Mitochondrial membrane cholesterol, the voltage dependent anion channel (VDAC), and the Warburg effect. J Bioenerg Biomembr 40, 193–197, doi:10.1007/s10863-008-9138-x (2008).
Szabo, I. et al. Double-stranded DNA can be translocated across a planar membrane containing purified mitochondrial porin. FASEB J 12, 495–502 (1998).
Zalk, R., Israelson, A., Garty, E. S., Azoulay-Zohar, H. & Shoshan-Barmatz, V. Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem J 386, 73–83, doi:10.1042/BJ20041356 (2005).
Hoogenboom, B. W., Suda, K., Engel, A. & Fotiadis, D. The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J Mol Biol 370, 246–255, doi:10.1016/j.jmb.2007.04.073 (2007).
Zhang, X. L. et al. Core component EccB1 of the Mycobacterium tuberculosis type VII secretion system is a periplasmic ATPase. FASEB J 29, 4804–4814, doi:10.1096/fj.15-270843 (2015).
Ramsdell, T. L., Huppert, L. A., Sysoeva, T. A., Fortune, S. M. & Burton, B. M. Linked domain architectures allow for specialization of function in the FtsK/SpoIIIE ATPases of ESX secretion systems. J Mol Biol 427, 1119–1132, doi:10.1016/j.jmb.2014.06.013 (2015).
Hou, J. M. et al. ATPase activity of Mycobacterium tuberculosis SecA1 and SecA2 proteins and its importance for SecA2 function in macrophages. Journal of bacteriology 190, 4880–4887, doi:10.1128/JB.00412-08 (2008).
Rostovtseva, T. & Colombini, M. ATP flux is controlled by a voltage-gated channel from the mitochondrial outer membrane. The Journal of biological chemistry 271, 28006–28008 (1996).
Tahlan, K. et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56, 1797–1809, doi:10.1128/AAC.05708-11 (2012).
McCann, J. R., McDonough, J. A., Sullivan, J. T., Feltcher, M. E. & Braunstein, M. Genome-wide identification of Mycobacterium tuberculosis exported proteins with roles in intracellular growth. Journal of bacteriology 193, 854–861, doi:10.1128/JB.01271-10 (2011).
Chalut, C. MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis (Edinb) 100, 32–45, doi:10.1016/j.tube.2016.06.004 (2016).
O’Neill, H. C. & Quah, B. J. Exosomes secreted by bacterially infected macrophages are proinflammatory. Sci Signal 1, pe8. doi:10.1126/stke.16pe8 (2008).
Rostovtseva, T. K. & Bezrukov, S. M. VDAC regulation: role of cytosolic proteins and mitochondrial lipids. J Bioenerg Biomembr 40, 163–170, doi:10.1007/s10863-008-9145-y (2008).
de Chastellier, C., Forquet, F., Gordon, A. & Thilo, L. Mycobacterium requires an all-around closely apposing phagosome membrane to maintain the maturation block and this apposition is re-established when it rescues itself from phagolysosomes. Cell Microbiol 11, 1190–1207, doi:10.1111/j.1462-5822.2009.01324.x (2009).
Gan, H. et al. Enhancement of antimycobacterial activity of macrophages by stabilization of inner mitochondrial membrane potential. J Infect Dis 191, 1292–1300, doi:10.1086/428906 (2005).
Jha, S. S. et al. Virulence-related Mycobacterium avium subsp hominissuis MAV_2928 gene is associated with vacuole remodeling in macrophages. BMC Microbiol 10, 100, doi:10.1186/1471-2180-10-100 (2010).
Acknowledgements
We thank Amy Palmer for technical help in performing the flow cytometry. The research was supported by the NIH grant AI-043199. The mass spectrometric analysis was performed by the OHSU proteomics shared resource with partial support from NIH core grants P30EY010572, P30CA069533, S10OD012246, and S10RR025571.
Author information
Authors and Affiliations
Contributions
L.D. designed studies, performed experiments, analyzed data, wrote the manuscript and prepared Figs 1–5. J.J.J.C., T.P. and R.G. performed experiments. L.E.B. Original idea, design studies, analyzed data, wrote the manuscript, senior investigator.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Danelishvili, L., Chinison, J.J.J., Pham, T. et al. The Voltage-Dependent Anion Channels (VDAC) of Mycobacterium avium phagosome are associated with bacterial survival and lipid export in macrophages. Sci Rep 7, 7007 (2017). https://doi.org/10.1038/s41598-017-06700-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-06700-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.