Abstract
Before their disappearance from the fossil record approximately 40,000 years ago, Neanderthals, the ancient hominin lineage most closely related to modern humans, interbred with ancestors of present-day humans. The legacy of this gene flow persists through Neanderthal-derived variants that survive in modern human DNA; however, the neural implications of this inheritance are uncertain. Here, using MRI in a large cohort of healthy individuals of European-descent, we show that the amount of Neanderthal-originating polymorphism carried in living humans is related to cranial and brain morphology. First, as a validation of our approach, we demonstrate that a greater load of Neanderthal-derived genetic variants (higher “NeanderScore”) is associated with skull shapes resembling those of known Neanderthal cranial remains, particularly in occipital and parietal bones. Next, we demonstrate convergent NeanderScore-related findings in the brain (measured by gray- and white-matter volume, sulcal depth, and gyrification index) that localize to the visual cortex and intraparietal sulcus. This work provides insights into ancestral human neurobiology and suggests that Neanderthal-derived genetic variation is neurologically functional in the contemporary population.
Similar content being viewed by others
Introduction
The most recent evolutionary relative of H. sapiens, H. neanderthalensis, represents a lineage associated with archeological samples (e.g., tools, pigments) suggesting substantial cognitive achievements1, and is typically contrasted with anatomically modern humans by particular cranial features known from fossil remains2. The initial sequencing of the Neanderthal genome summarily unearthed an unprecedented ingress to Neanderthal biology, resulting not only in a catalogue of shared ancestral alleles and divergent variation relative to H. sapiens but also evidence for gene flow from Neanderthals to modern humans3, 4, which has since been well-supported5,6,7,8. This introduction of Neanderthal alleles most likely occurred in Eurasia, as modern humans ventured out of Africa, between 47,000 and 65,000 years ago7 and accounts for about 2% of the entire non-African human genome8. The functional implications of this observation, however, are not fully understood.
If these alleles are biologically meaningful for their modern bearers5, and by extension, for their extinct originators, individuals endowed with greater proportions of Neanderthal-derived sequence differences might be expected to harbor Neanderthal-like phenotypes. To test this hypothesis, rather than using an additive model in a genetic association analysis, we performed linear regressions of polygenic measures of Neanderthal-derived common variant data with MRI imaging data. Previous reports3, 5 have described methods for testing whether single nucleotide polymorhpisms (SNPs) are derived from admixture of modern humans with Neanderthals. Here, after computing an enhanced D statistic9, which measures the percentage of these alleles present in each individual (subsequently termed “NeanderScore”), we sought to identify whether variation was associated with regional changes in skull and brain shape. Because cranial morphology is a pivotal physical element distinguishing Neanderthals from other human lineages in fossil records, it provides an ideal phenotype to test our approach and the impact of our Neanderthal-derived genetic characteristics. We anticipated that some of the Neanderthal genetic variants responsible for skull development might be represented in our sample and, when sufficiently aggregated in higher NeanderScore individuals, that these variants would be associated with hallmark Neanderthal-like morphological biases, such as increased skull length10, 11 and posterolateral broadening of the cranium2.
As the distinctive cranial morphology of H. neanderthalensis is hypothesized to reflect underlying brain anatomy12,13,14,15,16, in addition to characterizing the relationship between NeanderScore and modern human cranial shape, we also sought to define contributions of Neanderthal-derived genetic variation to brain structure, as measured by gray- and white-matter volume, regional sulcal depth, and gyrification index. We further hypothesized that the morphometry of brain regions underlying the identified NeanderScore-associated cranial regions would also be associated with Neanderthal genetic load.
Results
Calculation of NeanderScore
We first examined common variant genotype data acquired from 223 healthy individuals of self-reported European-descent who also underwent magnetic resonance imaging (MRI) of the head. To ensure all participants were of similar genetic ancestry, multidimensional scaling of ancestry-informative genetic markers17 was performed on the entire sample. Two participants were found to be significantly different from the rest of the group and were excluded, while the remaining 221 individuals (average age 33.7 ± 10.6 years, range 18–60, 116 females) were retained for further analyses examining each individual’s percentage of Neanderthal-derived genetic material. Following previously reported methods3, 5, we identified 100,156 single nucleotide polymorphisms (SNPs) in our sample that are putatively derived from Neanderthals and then calculated an enhanced D statistic9, which determined the percentage that were present in each of the 221 individuals (NeanderScore). Consistent with previous reports3, 5, 8, an expectedly small but normally distributed percentage of Neanderthal sequence variation was present in our participants, with individuals showing an average NeanderScore of 5.4% (standard deviation = 0.38%, range = 3.9–6.5%, coefficient of variation = 0.070). It is important to note that the NeanderScore measure is a related but different measure than prior reports of Neanderthal admixture (as discussed below), and thus has a different mean and standard deviation than those in previous reports; however, our coefficient of variation is identical to those previously reported and indicates sufficient variability to pursue further quantitative trait analyses with the MRI data. Table 1 shows demographic information of the sample. As the high coverage Altai Neanderthal draft sequence was used as the Neanderthal genotypes for each SNP, we examined the subset of 715 SNPs overlapping with the exome data publically available and previously reported for the Vindija and El Sidron Neanderthal samples18, as well as for a Denisovan sample, to determine the degree these SNPs are specific to Neanderthals and maintained in other archaic samples (data acquired from http://cdna.eva.mpg.de/neandertal/exomes/). We found a high degree of concordance of the Altai sequence with the other Neanderthal samples (94.9% for Vindija and 93.8% for El Sidron). In contrast, the Denisovan sample showed a much lower concordance rate of 26.4%, suggesting both sensitivity and specificity of our measure to Neanderthal admixture. As more high coverage archaic samples become available, future work could further describe and investigate the implications of genetic variability in these samples. Additionally, to ensure that the NeanderScore variation observed was not related to possible subtle differences in the microarray chips used, we tested for differences in NeanderScore across these chips and found none (F(2,218) = 0.023, p = 0.98).
Relationships between NeanderScore and Skull Shape
For the subset of our cohort with high-quality, full cranium acquisitions (N = 146, Table 1), we created a 3D skull model for each individual using a previously validated algorithm19,20,21, as well as a 3D template skull model created from the group average. After conducting a nine-parameter alignment between each individual’s model and the template model, we calculated the distance between each node of the template and the nearest node in each individual skull22. We then assessed correlations between these local shape values and NeanderScore.
In line with the contention that Neanderthal genetic load is biologically functional in modern-day humans, we first examined normalized anterior-posterior cranial length, one of the earliest reported parameters differentiating modern human and Neanderthal skulls11, and found this parameter to significantly correlate with NeanderScore (R = 0.240, p < 0.003). Examining morphometry of the entire skull in a data driven fashion, we also discovered that higher NeanderScore was positively and selectively associated with the regional size of a broad posterolateral area of the skull extending from the occipital and inferior parietal bones to bilateral temporal locales (Tmax = 3.5, p < 0.0001, family-wise error (FWE)-corrected; Fig. 1, left and middle). The area of maximal change localized to the right lambdoid suture area, involving both the occipital and parietal bones, a finding that corresponds to published shape differences between modern humans and Neanderthals as documented in fossil samples (Fig. 1, right), thus establishing the validity of our approach. To ensure the results were not driven by the population structure of the cohort, we repeated this analysis while also controlling for the first 4 dimensions from a multidimensional scaling analysis and found that the pattern of association between NeanderScore and skull shape did not significantly change from the original analysis.
NeanderScore related skull shape changes. Associations between skull morphology and percentage of Neanderthal-derived SNPs (NeanderScore) in modern humans (Left), compared to differences between Neanderthal and modern human skulls (Right). Left panel shows regions with morphological changes, measured in vivo, related to NeanderScore and overlaid on the average skull of all 146 participants. Warm colors represent areas for which NeanderScore correlated with skull expansion (p < 0.05, FWE-corrected). There were no findings in the opposite direction. Right panel depicts previously published shape differences between Neanderthals and modern humans, from archeological samples, overlaid on a modern human skull. Red regions represent skull expansion in Neanderthals relative to modern humans; green represents contraction, adapted by permission from Macmillian Publishers Ltd: Nature2, © 2001. Note the convergence of in vivo and archeological data in occipito-parieto-temporal regions (arrows).
Relationships between NeanderScore and Brain Morphology
As previous work has related changes in skull shape to regional brain measures in European populations23, and having established that high NeanderScore is associated with statistically observable Neanderthal-like trends in modern cranial shape, we next sought to define contributions of Neanderthal-derived genetic variation to brain structure, as measured by regional sulcal depth and gyrification index, which we quantified across the entire cortical ribbon via segmentation and subsequent surface-based processing24, 25 of the full intracranial MRI dataset. We hypothesize that we might also finds similar NeanderScore-related changes in the brain regions underlying the NeanderScore-related occipito-parietal skull associations. The first measure we examined, regional sulcal depth, measures the extent to which each fold of the brain descends into the underlying mantle. We found that NeanderScore positively correlated with an increase in sulcal depth in the right intraparietal sulcus (IPS; Tmax = 4.03, pcluster < 0.001, FWE-corrected), directly beneath the area of maximal change in the prior skull shape analysis (Fig. 2), suggesting a significant but regionally circumscribed impact of Neanderthal genetic load on modern human brain.
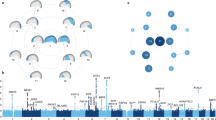
NeanderScore related brain changes in the intraparietal sulcus. Structural variation of the intraparietal sulcus (IPS) related to percentage of Neanderthal-derived SNPs (NeanderScore). Left and middle show lateral and posterior views of the right IPS on the average brain surface, illustrating the anatomical convergence of the associations of NeanderScore with greater sulcal depth (orange; p < 0.05 FWE-corrected), gray matter volume (blue; p < 0.005), and white matter volume (yellow; p < 0.005). Right shows a coronal slice (y = −72 mm) through the IPS where there was a sulcal depth difference of 4 mm between the 30 individuals with highest (blue line) and lowest (red line) NeanderScore. Findings are depicted on the average brain of all 221 individuals included in the study and underlie the maximal skull findings shown in Fig. 1 (arrows).
The other examined neuroanatomic parameter, local gyrification index (LGI), provides a measure of the regional complexity of cortical folding, and is highly modified throughout evolution26,27,28. We found that NeanderScore positively correlated with LGI in early visual regions in the left hemisphere, also directly underlying the NeanderScore-associated skull findings, spanning primary visual cortex, V1 and V2 (Tmax = 4.24, p < 3 × 10−5, FWE-corrected p < 0.0001; Fig. 3), as well as with the right superior temporal sulcus (Tmax = 3.67, p < 3.1 × 10−4, FWE-corrected p < 0.005), such that cortical complexity, and thus cortical surface area, increased with NeanderScore.
NeanderScore related brain changes in the primary visual cortex. Associations between local gyrification index, gray matter volume, and NeanderScore in visual cortex. Medial view of the primary visual cortex showing areas where NeanderScore was significantly associated with local gyrification index (red; p < 0.05 FWE-corrected) and with gray matter volume (yellow; p < 0.005).
The statistical strength and localization of NeanderScore associations with measures of cortical folding raised the possibility of additional structural differences in these brain areas. We therefore conducted follow-up, whole-brain, voxel-based morphometric analyses aimed at quantifying associations between NeanderScore and both gray and white matter volume across the brain. Associations with both gray matter volume (MNIxyz = 28.5, −87, 36; Tmax = 3.87, p < 1.4 × 10−4, uncorrected) and white matter volume (MNIxyz = 18, −82, 28.5; Tmax = 4.32, p < 2.4 × 10−4, uncorrected) were localized to a right parietal region adjacent to the IPS focus described above, such that greater Neanderthal SNP load correlated with more gray and white matter volume (Fig. 2). Similarly, in an area overlapping with the primary visual cortex gyrification finding described above, we observed that gray matter volume, though not white matter volume, was positively associated with NeanderScore (MNIxyz = 0, −99, 10.5; Tmax = 5.07, p < 8.5 × 10−7, uncorrected; Fig. 3). We also found evidence for an inverse correlation between NeanderScore and white matter volume in the orbitofrontal cortex (MNIxyz = 37.5, 39; −7.5, Tmax = −4.44, p < 1.4 × 10−5, uncorrected), a structure that appears to have been laterally restricted in Neanderthals relative to modern H. sapiens 13.
Post-hoc Genetic Analysis of Neanderthal-Derived SNPs
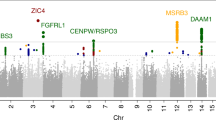
Finally, we conducted an exploratory test of the association of our structural findings to specific variants within the 100,156 SNPs identified as putatively derived from Neanderthals in our participants. The convergence of findings in posterior head regions, across cranial, sulcal depth, gyrification and volumetric variables, led us to hypothesize that common genetic influences would be found in genes preferentially expressed in the human brain. Using the first component from a principal components analysis as a quantitative trait that captures the shared-variance across our structural measures, we tested each of the 100,156 Neanderthal-derived SNPs for association with brain/skull structure using a linear model. SNPs in a 53 kb LD-block of chromosome 10, encompassing the telomeric end of the gene GPR26, a G-protein coupled receptor preferentially expressed in the human brain29, showed strong association with brain/skull structure (t = 5.1, p < 1.04 × 10−6, Bonferroni-corrected p < 0.013, Table 2). Figure 4 shows the manhattan plot of these results and Fig. 5 shows the QQ plot.
Manhattan plot of the shared variance of NeanderScore associated brain and skull changes. Manhattan plot showing the chromosomal position and significance of association between the shared variance of all Neanderthal-associated brain measures for each Neanderthal-derived SNP. Red line indicates Bonferroni-corrected threshold of p < 0.05 for the effective number of SNPs in the analysis. The arrow points to the significant 53-kb LD block on chromosome 10 containing 18 SNPs. See also Table 2.
We further examined this genetic region to assess the plausibility of its Neanderthal introgression. The 53 kb block we identified overlaps a previously predicted Neanderthal-introgressed region – by both Vernot et al.30 and Sankararaman et al.5. Since regions of extended similarity to archaic genomes could arise by incomplete lineage sorting (ILS), we calculated the maximum possible length of such an ILS region given the recombination rate and time since introgression31. Assuming the lowest average recombination rate in the region (which is most conservative), the branch lengths in years8, a generation time of 25 years, and a mutation rate of either 0.5 × 10−9 per site per year or 1 × 10−9 per site per year, we can rule out that any region longer than 1580 bp (0.5 × 10−9) or 3163 bp (1 × 10−9) is the result of ILS. The chance of maintaining a 53 kb block in both lineages is conservatively estimated at p < 9.4 × 10−7, using a gamma distribution with shape parameter 2 and a conservative rate parameter of 1/3163 bp31.
Discussion
In this work, we describe relationships between Neanderthal-derived genetic variation and co-localized cranial and brain morphology in modern humans. The results show that greater NeanderScore is associated with more Neanderthal-like skull shape (corresponding to published shape differences between modern humans and Neanderthals as documented in fossil samples2 and shown in Fig. 1), as well as regional changes in brain morphology underlying these skull changes, specifically in the IPS and visual cortex. This work not only offers an unprecedented window into structure of the Neanderthal brain, but also characterizes the contributions of admixture with H. neanderthalensis to the evolution of the modern human brain.
In examining the associations of NeanderScore with skull shape, it is important to note that the topology of the identified occipito-parieto-temporal patch associated with NeanderScore specifically recapitulates the pattern of expansion in Neanderthal relative to anatomically modern human skulls previously reported from fossil remains (Fig. 1, right)2. This finding provides crucial validation of the NeanderScore measurement and suggests that even in the context of the modern H. sapiens genome, Neanderthal genetic variation is associated with patterns of skull dimensions that mirror known Neanderthal phenotypes.
Additional validation of the NeanderScore metric lies in its comparison with other reports characterizing Neanderthal-derived genetic contributions to modern humans. The coefficient of variation (i.e., the relative standard deviation, independent of the mean and comparable across measures) of the NeanderScore metric is consistent with previous published studies5 that were performed with a different goal: prior studies, unlike ours, sought to test for differences in admixture across populations, and have, therefore, calculated the proportion of the entire genome that was derived from Neanderthals. In those studies, approximately 1.15% of the entire genome of persons of European decent was found to be derived from Neanderthals (standard deviation = 0.08)5. Inherently, that approach and the present one capture related though different characteristics of Neanderthal-derived genetic information and the means (and therefore the standard deviations as well) of these differently derived measures are not the same. Importantly, however, the coefficient of variation in previous studies was also 0.070,5 identical to the coefficient of variation reported here, providing further validity to the NeanderScore measure.
In determining NeanderScore related changes in brain morphology, we found two significant cortical regions, the IPS and primary visual cortex, which both directly underlie the region of skull shape associations. The IPS, though present throughout highly gyrified modern primates28, has been theorized to have undergone substantial evolutionary expansion in hominids, with cross-species functional neuroimaging demonstrating unique visuospatial processing characteristics in modern humans relative to rhesus monkeys32. Additionally, cranial vault analyses of fossil skulls have suggested differences in the parietal lobes of Neanderthals14, 15, and the intraparietal sulcal region in particular has been hypothesized to be a focus for some of these differences33. Moreover, the fact that the IPS is particularly critical for tool manipulation in modern humans34 makes this finding even more intriguing in view of continued debate over the nature and development of Neanderthal tool use35.
The other brain region revealed to have significant associations with NeanderScore was primary visual cortex. This cortical region is responsible for the first steps in processing of visual information in the mammalian cortex and feeds into later brain regions in the ventral and dorsal visual processing streams (which differentially subserve object recognition and visuospatial object location, respectively)36, with the IPS playing a prominent role in the latter. Though the functioning of the primary visual system is relatively conserved in primates, the size of primary visual cortex in modern humans is smaller than would be expected from brain volume37. Our data not only suggest that this may be less the case in Neanderthals, but also are consistent with cranial remains showing more prominent visual systems in Neanderthals than in modern humans15.
It should be noted that we did not find associations of NeanderScore with smaller frontotemporal volumes38 or shortened anterior extension of the temporal lobes13, as might have been hypothesized from previous cranial analyses of H. neanderthalensis, suggesting either that these particular phenotypes, if accurate, are not driven by the allelic variation captured by the NeanderScore measurement, or that such effects are more directly influenced by any of the myriad factors that establish the genetic and biological context of this modern cohort. Additionally, some of these phenotypes may be only partially modulated by genotypic factors inherited from Neanderthals, with an effect too small to be observed in our sample. In contrast, the effect sizes we observed here for the associations of NeanderScore with the skull and brain measures were moderate (average Cohen’s d = 0.58) but appropriate for the sample sizes used.
The analyses reported here were restricted to a sample of individuals of European descent. It is known that the degree of admixture is variable in different modern populations. For example, East Asian populations have been found to have a larger portion of the genome derived from Neanderthals than European populations (up to 20% more), though the admixed regions of the genome are not necessarily overlapping5. This raises the possibility that the findings reported here may not translate to other populations, where Neanderthal introgression may involve other genomic regions that may be functional in different ways. As large neuroimaging and genetic data from different populations become available, future work could investigate this possibility by performing similar analyses in different populations, including using African populations with minimal Neanderthal admixture as potential “null hypothesis” groups.
Finally, our analyses of the relationship of our findings to specific Neanderthal-derived gene variants, revealed a single 53 kb LD block that was significantly associated with the shared variance of the identified Neanderthal-derived brain and skull changes and that encodes for the gene GPR26. In line with our primary hypothesis for this analysis, that such genetic influences would be found in genes preferentially expressed in the human brain, GPR26 in fact encodes a G-protein coupled receptor subtype that is preferentially expressed in the brain29. Interestingly, in human post-mortem brains samples, expression of GPR26 peaks perinatally39, when the visual system is first challenged, indicating that it may play a role in development of the human visual system. Mouse models also show this gene to impact both affective and energy homeostatic functions40, 41. Additionally, this G-protein-coupled receptor has been shown to form oligomers with the 5-HT1a receptor42, perhaps providing a putative mechanism underlying the Neanderthal-related brain changes found here. Although the nature of the influence of this region on modern and archaic human nervous systems is uncertain, a possible link between brain energy regulation, neurodevelopment and mature structure may merit further investigation.
Because the NeanderScore measure employed here is, itself, polygenic, it is likely that the genetic contributions to skull and brain morphology we observed involve a number of different genetic loci. Nonetheless, our exploratory post-hoc genome-wide analysis of the shared variance of these findings identified only a single significant region. It is unlikely that this single locus, the LD block on chromosome 10, fully explains the brain and skull findings, and in fact, the Manhattan plot in Fig. 4 suggests that multiple other regions that do not meet strict Bonferroni corrections may represent some degree of true signal. Our modest sample size may have been underpowered to identify these additional signals. As larger datasets containing both genotype and neuroimaging data of brain and skull become available, future work will likely uncover additional genetic regions contributing to these findings.
Taken together, the associations between Neanderthal sequence variation and co-localized skull and brain morphology in modern humans engender an enduring, living footprint of H. neanderthalensis – a residual echo of shared, intimate history with a fallen lineage close to our own. To the extent that characterization of Neanderthal variation in present-day people can provide insights into archaic human phenotypes, this work can form the basis of future studies aimed at a more thorough understanding of Neanderthal biology. By the same token, we suggest that Neanderthal gene flow into modern humans is not only of evolutionary interest, but may also be functional in the living H. sapiens brain, revealing novel genetic influences on neurodevelopment of the visuospatial system upon which a fuller account of molecular mechanisms of IPS-driven normative mental functions, such as visuospatial integration and tool manipulation, can be built. This, in turn, may inform models of IPS-associated cognitive disability as seen in select developmental and neurological disorders43,44,45,46.
Methods
Subjects
Two hundred twenty-three participants were identified from a pool of healthy control volunteers recruited as part of the NIMH Sibling Study – a multidisciplinary initiative aimed at examining the neurobiology of schizophrenia risk – who had completed both neuroimaging with 3-Tesla MRI and genome-wide SNP genotyping. Participants included in the current work were all adults of self-reported European-descent and were screened to be healthy and free of any neurologic or psychiatric diseases based on clinician-obtained history and physical examination, semi-structured diagnostic interview (Structured Clinical Interview for DSM-IV), clinical MRI of the brain, and routine laboratory testing, including urine toxicology. Table 1 provides the subject demographics. All procedures were carried out in accordance with NIH guidelines and were approved by the National Institutes of Health CNS Institutional Review Board, and all participants provided written informed consent for the study procedures.
Genetic Analysis
Genotyping was carried out on DNA extracted from lymphoblast cell lines derived from each individual. As data from this study has been collected over time, genotyping was performed in four steps with an increasing number of SNPs genotyped at each step. All genotyping was done on Illumina QUAD SNP chips (ranging from 550K-2.5 M SNPs). The 275,659 common SNPs from all chip types were combined and Illumina genotype QC and imputation procedures were performed following the previously reported methods47. After QC procedures, data was pre-phased with SHAPEIT, imputation was done with IMPUTE2, using 1000 genome phase 1 data as a reference panel. Ambiguous SNPs and duplicated SNPs were removed prior to imputation.
To ensure participants were of similar genetic ancestry, clustering and multidimensional scaling was performed in Plink (http://pngu.mgh.harvard.edu/~purcell/plink), using a set of 125 ancestry informative markers17. Two individuals were noted to be of genetically different ancestry, based on the pairwise identity-by-state distance information of all participants and appeared to cluster separately from the rest of the group on plots of the first two principle components of the multidimensional scaling, and were removed from further analysis.
Calculation of NeanderScore
Previous studies aimed at examining Neanderthal ancestry have identified the proportion of the genome derived from Neanderthals, predominantly to examine differences in admixture across populations5. Here, instead of examining for population effects, we sought to quantify the proportion of potential Neanderthal admixture in each individual by calculating an enhanced D statistic. Enhanced D statistics have been shown to be linearly related to the amount of Neanderthal ancestry in an individual48 and are termed “enhanced” because they improve the signal to noise ratio of basic D statistics by restricting computation to sites where an isolated population carries the ancestral allele9. This approach could be interpreted as a polygenic score for Neanderthal inheritance, similar to other scores used to study the genetics of disease49. To accomplish this, we sought to identify single nucleotide polymorphisms (SNPs) derived from Neanderthals via admixture. Admixture may have occurred at a SNP location if (1) the derived (non-ancestral) allele is present in both humans and Neanderthals, (2) the genotyped individual carries the derived allele, and (3) that derived allele is absent in a human population devoid of Neanderthal admixture5. In our case, we specifically identified SNPs in the genome at which Neanderthal genotyping indicates the presence of a derived allele that is not present in a population without admixture, based on publically available data that has previously been used to estimate Neanderthal ancestry5. As in that previous work, we used the high coverage Altai Neanderthal genome8 as the Neanderthal sequence, the 1000 genomes project Yoruba population50 as the ancestral human population devoid of admixture with Neanderthals, and a 6-primate consensus sequence51 to represent the ancestral species.
We determined a “NeanderScore” for each genotyped individual, which is defined as:
where ∑(n ABBA ) is the count of SNP locations at which the genotyped individual shares an allele with the Neanderthal sequence but differs from all Yoruba and primate sequences, and ∑(n AABA ) is the count of locations at which the genotyped individual shares an allele with Yoruba and primates sequences but differs from the Neanderthal sequence. Importantly, the denominator in this equation represents all SNPs where the Neanderthal sequence differs from (1) all individuals in the Yoruba population and (2) the consensus primate sequence (i.e., all SNPs that could be tested for contribution from Neanderthals). The numerator is the number of those sites in which Neanderthal contribution was found in a particular individual. The ratio, or NeanderScore, represents the percentage of potential Neanderthal-derived SNPs in that individual, whereas prior reports have estimated the percentage of the entire non-African genome derived from Neanderthals3, 5, 8. Figure 6 depicts this graphically in a phylogenic tree where genotype ‘A’ represents the ancestral allele, present in both Yoruba and the primate consensus, and genotype ‘B’ represents the derived allele, assumed to have passed from the Neanderthals to the genotyped individual.
Image Acquisition and Preprocessing
Three-dimensional structural MRI scans were acquired on a 3-Tesla GE scanner (GE Medical Systems, Milwaukee, WI) using a magnetization prepared rapid gradient echo (MPRAGE) sequence (repetition time 7.28 ms, echo time 2.74 ms, 120–136 slices, resolution 0.859 × 0.859 × 1.2 mm). Preprocessing included intensity nonuniformity normalization52 and rigid alignment to MNI space.
Skull Surface Reconstruction
All images were visually inspected to ensure that the entire head was represented. Of the 221 participants’ scans, 146 were deemed usable for skull reconstruction. We applied previously published methods19,20,21 to our participants’ MRI data after using SPM8 (http://fil.ion.ucl.ac.uk/spm) to perform segmentation of individual scans into tissue probability maps, representing voxel-wise probabilities of gray matter, white matter, cerebrospinal fluid (CSF), soft tissue, bone or “other.” After identifying head and intracranial masks for each participant, skull voxels were identified as those that (1) were within the head mask, (2) were outside of the intracranial mask, and (3) had a combination of high bone-tissue probability and low probability of gray matter, white matter and CSF. An example of these boundaries is shown in Fig. 7 (Left). Individual volumes representing skull voxels were used to produce 3D surface representations (meshes) of participants’ skulls, using SUMA tools (http://afni.nimh.nih.gov/afni/suma/). An example skull surface is also shown in Fig. 7 (Right).
Skull surface creation from the T1-weighted MRI scan of a single participant. Left shows the segmentation procedure performed in the volume where voxels labeled blue represent scalp, orange voxels represent skull, and gray voxels represent the intracranial area. Right shows the 3D skull surface derived from voxels labeled as skull (orange) in the segmentation.
Skull Surface Analysis
A group-average template of all 146 MRI scans used in the skull analysis was created using ANTs software (http://stnava.github.io/ANTs). The skull surface of this group average template was created as described above for the individual scans. Next, each individual scan was aligned to the template using nine-degrees of freedom (accounting for shifts, rotations and scaling about the x-, y- and z- axes) to normalize for skull size without altering shape. Using SUMA tools, the signed Euclidian distance between each node of the template and the nearest node on the surface of each individual skull was calculated. Areas for which skull shape co-varied with NeanderScore, controlling for age and sex, were determined based on previously reported methods22. As neuroimaging data are highly dimensional and statistical analyses may be biased towards type-I error, a surface-based family-wise error correction for multiple comparisons was calculated using permutation testing on 10,000 Monte-Carlo simulations of synthesized Gaussian white noise with the structure and smoothness of the underlying data53, 54. Resulting statistical maps were thresholded at p < 0.05, corrected for multiple comparisons using this family-wise error (FWE) correction. Additionally, the anterior to posterior distance for each individual skull was calculated using SUMA tools and was correlated with NeanderScore using IBM SPSS Statistics for Windows, version 21.0 (SPSS, IBM Corp, Armonk, NY) after controlling for effects due to age and sex.
Cortical Surface-based brain analyses
The 221 individual brain scans were processed with the FreeSurfer version 5.3 pipeline (http://surfer.nmr.mgh.harvard.edu) to produce surface-based maps of sulcal depth and local gyrification index for each individual. The surface-based maps were then aligned to a 198,812 node standard mesh55. Statistics were computed for each node on a participant’s surface, correlating sulcal depth and local gyrification measures with NeanderScore while controlling for age and sex. A surface-based correction for multiple comparisons was determined using the methods described above for skull shape analysis, with resulting maps thresholded at p < 0.05, corrected for multiple comparisons using FWE correction53, 54.
Voxel-based brain analyses
After the image preprocessing described above, voxel-wise tissue density maps, including gray and white matter maps, were computed using SPM8’s segmentation functions. SPM8’s DARTEL tools were used to create a group-average template. Voxel-based morphometry was performed, based on tissue density maps modulated by the jacobian determinant of participants’ deformation maps to create voxel-wise volumetric maps of each tissue class56. Maps of gray matter volume and white matter volume were spatially smoothed using a 6 mm FWHM Gaussian kernel. Linear regression was performed on the resulting maps on a voxel-wise basis, using NeanderScore as a regressor of interest, while controlling for age and sex effects. Resulting statistical maps were thresholded at a significance level of p < 0.005, uncorrected.
Post-hoc genetic analyses
Post-hoc analyses were conducted to determine whether individual Neanderthal-derived SNPs were of particular importance in influencing converging skull/brain variation found within the intraparietal area and the primary visual cortex in modern humans. We pre-processed the peak results from the skull, IPS sulcal depth, primary visual local gyrification index, IPS/primary visual gray matter volume and IPS white matter volume analyses by removing effects due to age and sex from each result individually using linear regression. We then conducted a principal components analysis using SPSS on the residualized variables, using the first principal component to form a composite score representing the primary inter-individual variance shared across the skull/brain measures. This composite served as a quantitative phenotype in a linear genetic model using Plink (http://pngu.mgh.harvard.edu/~purcell/plink) to examine each Neanderthal-derived SNP. GEC (http://statgenpro.psychiatry.hku.hk/gec) was used to determine the number of effective SNPs in this analysis (13378 SNPs)57, yielding a Bonferroni correction level of pcorrected < 0.05 occurring when puncorrected < 3.7 × 10−6.
References
Lind, J., Lindenfors, P., Ghirlanda, S., Liden, K. & Enquist, M. Dating human cultural capacity using phylogenetic principles. Sci Rep 3, 1785, doi:10.1038/srep01785 (2013).
Ponce de Leon, M. S. & Zollikofer, C. P. Neanderthal cranial ontogeny and its implications for late hominid diversity. Nature 412, 534–538, doi:10.1038/35087573 (2001).
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722, doi:10.1126/science.1188021 (2010).
Burbano, H. A. et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science 328, 723–725, doi:10.1126/science.1188046 (2010).
Sankararaman, S. et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357, doi:10.1038/nature12961 (2014).
Gibbons, A. Human evolution. Neandertals and moderns made imperfect mates. Science 343, 471–472, doi:10.1126/science.343.6170.471 (2014).
Sankararaman, S., Patterson, N., Li, H., Paabo, S. & Reich, D. The date of interbreeding between Neandertals and modern humans. PLoS Genet 8, e1002947, doi:10.1371/journal.pgen.1002947 (2012).
Prufer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49, doi:10.1038/nature12886 (2014).
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226, doi:10.1126/science.1224344 (2012).
Bruner, E., De La Cuetara, J. M. & Holloway, R. A bivariate approach to the variation of the parietal curvature in the genus homo. Anatomical record 294, 1548–1556, doi:10.1002/ar.21450 (2011).
Morant, G. M. Studies of palaeolithic man. Annals of Eugenics 2, 318–381, doi:10.1111/j.1469-1809.1927.tb02056.x (1927).
Balzeau, A., Holloway, R. L. & Grimaud-Herve, D. Variations and asymmetries in regional brain surface in the genus Homo. Journal of human evolution 62, 696–706, doi:10.1016/j.jhevol.2012.03.007 (2012).
Bastir, M. et al. Evolution of the base of the brain in highly encephalized human species. Nat Commun 2, 588, doi:10.1038/ncomms1593 (2011).
Bruner, E., Manzi, G. & Arsuaga, J. L. Encephalization and allometric trajectories in the genus Homo: evidence from the Neandertal and modern lineages. Proc Natl Acad Sci USA 100, 15335–15340, doi:10.1073/pnas.2536671100 (2003).
Pearce, E., Stringer, C. & Dunbar, R. I. New insights into differences in brain organization between Neanderthals and anatomically modern humans. Proc Biol Sci 280, 20130168, doi:10.1098/rspb.2013.0168 (2013).
Ponce de Leon, M. S., Bienvenu, T., Akazawa, T. & Zollikofer, C. P. Brain development is similar in Neanderthals and modern humans. Curr Biol 26, R665–666, doi:10.1016/j.cub.2016.06.022 (2016).
Kosoy, R. et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Human mutation 30, 69–78, doi:10.1002/humu.20822 (2009).
Castellano, S. et al. Patterns of coding variation in the complete exomes of three Neandertals. Proc Natl Acad Sci USA 111, 6666–6671, doi:10.1073/pnas.1405138111 (2014).
Perdue, K. L. & Diamond, S. G. T1 magnetic resonance imaging head segmentation for diffuse optical tomography and electroencephalography. J Biomed Opt 19, 026011, doi:10.1117/1.JBO.19.2.026011 (2014).
Shi, L. et al. Morphometric analysis for pathological abnormality detection in the skull vaults of adolescent idiopathic scoliosis girls. Med Image Comput Comput Assist Interv 9, 175–182 (2006).
Dogdas, B., Shattuck, D. W. & Leahy, R. M. Segmentation of skull and scalp in 3-D human MRI using mathematical morphology. Hum Brain Mapp 26, 273–285, doi:10.1002/hbm.20159 (2005).
Patenaude, B., Smith, S. M., Kennedy, D. N. & Jenkinson, M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage 56, 907–922, doi:10.1016/j.neuroimage.2011.02.046 (2011).
Bakken, T. E., Dale, A. M., Schork, N. J. & Initi, As. D. N. A Geographic Cline of Skull and Brain Morphology among Individuals of European Ancestry. Hum Hered 72, 35–44, doi:10.1159/000330168 (2011).
Dale, A. M., Fischl, B. & Sereno, M. I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage 9, 179–194, doi:10.1006/nimg.1998.0395 (1999).
Van Essen, D. C. Cortical cartography and Caret software. NeuroImage 62, 757–764, doi:10.1016/j.neuroimage.2011.10.077 (2012).
Rogers, J. et al. On the genetic architecture of cortical folding and brain volume in primates. NeuroImage 53, 1103–1108, doi:10.1016/j.neuroimage.2010.02.020 (2010).
Ventura-Antunes, L., Mota, B. & Herculano-Houzel, S. Different scaling of white matter volume, cortical connectivity, and gyrification across rodent and primate brains. Frontiers in neuroanatomy 7, 3, doi:10.3389/fnana.2013.00003 (2013).
Zilles, K., Palomero-Gallagher, N. & Amunts, K. Development of cortical folding during evolution and ontogeny. Trends in neurosciences 36, 275–284, doi:10.1016/j.tins.2013.01.006 (2013).
Jones, P. G. et al. Tissue distribution and functional analyses of the constitutively active orphan G protein coupled receptors, GPR26 and GPR78. Biochim Biophys Acta 1770, 890–901, doi:10.1016/j.bbagen.2007.01.013 (2007).
Vernot, B. & Akey, J. M. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021, doi:10.1126/science.1245938 (2014).
Huerta-Sanchez, E. et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197, doi:10.1038/nature13408 (2014).
Vanduffel, W. et al. Extracting 3D from motion: differences in human and monkey intraparietal cortex. Science 298, 413–415, doi:10.1126/science.1073574 (2002).
Bruner, E. Morphological Differences in the Parietal Lobes within the Human Genus: A Neurofunctional Perspective. Current Anthropology 51, S77–S88, doi:10.1086/650729 (2010).
Vingerhoets, G. Contribution of the posterior parietal cortex in reaching, grasping, and using objects and tools. Front Psychol 5, 151, doi:10.3389/fpsyg.2014.00151 (2014).
Soressi, M. et al. Neandertals made the first specialized bone tools in Europe. Proc Natl Acad Sci USA 110, 14186–14190, doi:10.1073/pnas.1302730110 (2013).
Ungerleider, L. G. & Mishkin, M. In Analysis of visual behavior (eds Ingle, D. J., Goodale, M. A. & Mansfield, R. J. W.) 549–586 (MIT Press, 1982).
de Sousa, A. A. et al. Hominoid visual brain structure volumes and the position of the lunate sulcus. Journal of human evolution 58, 281–292, doi:10.1016/j.jhevol.2009.11.011 (2010).
Lieberman, D. E., McBratney, B. M. & Krovitz, G. The evolution and development of cranial form in Homosapiens. Proc Natl Acad Sci USA 99, 1134–1139, doi:10.1073/pnas.022440799 (2002).
Colantuoni, C. et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523, doi:10.1038/nature10524 (2011).
Chen, D., Liu, X., Zhang, W. & Shi, Y. Targeted inactivation of GPR26 leads to hyperphagia and adiposity by activating AMPK in the hypothalamus. PLoS One 7, e40764, doi:10.1371/journal.pone.0040764 (2012).
Zhang, L. L. et al. GPR26-deficient mice display increased anxiety- and depression-like behaviors accompanied by reduced phosphorylated cyclic AMP responsive element-binding protein level in central amygdala. Neuroscience 196, 203–214, doi:10.1016/j.neuroscience.2011.08.069 (2011).
Salim, K. et al. Oligomerization of G-protein-coupled receptors shown by selective co-immunoprecipitation. The Journal of biological chemistry 277, 15482–15485, doi:10.1074/jbc.M201539200 (2002).
Kippenhan, J. S. et al. Genetic contributions to human gyrification: sulcal morphometry in Williams syndrome. J Neurosci 25, 7840–7846 (2005).
Meyer-Lindenberg, A. et al. Neural basis of genetically determined visuospatial construction deficit in Williams syndrome. Neuron 43, 623–631, doi:10.1016/j.neuron.2004.08.014 (2004).
Molko, N. et al. Functional and structural alterations of the intraparietal sulcus in a developmental dyscalculia of genetic origin. Neuron 40, 847–858 (2003).
Price, G. R., Holloway, I., Rasanen, P., Vesterinen, M. & Ansari, D. Impaired parietal magnitude processing in developmental dyscalculia. Curr Biol 17, R1042–1043, doi:10.1016/j.cub.2007.10.013 (2007).
Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427, doi:10.1038/nature13595 (2014).
Fu, Q. et al. The genetic history of Ice Age Europe. Nature 534, 200–205, doi:10.1038/nature17993 (2016).
Power, R. A. et al. Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nature neuroscience 18, 953–955, doi:10.1038/nn.4040 (2015).
Genomes Project, C. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65, doi:10.1038/nature11632 (2012).
Paten, B., Herrero, J., Beal, K., Fitzgerald, S. & Birney, E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome research 18, 1814–1828, doi:10.1101/gr.076554.108 (2008).
Sled, J. G., Zijdenbos, A. P. & Evans, A. C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17, 87–97, doi:10.1109/42.668698 (1998).
Nichols, T. & Hayasaka, S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Stat Methods Med Res 12, 419–446 (2003).
Hagler, D. J. Jr., Saygin, A. P. & Sereno, M. I. Smoothing and cluster thresholding for cortical surface-based group analysis of fMRI data. NeuroImage 33, 1093–1103, doi:10.1016/j.neuroimage.2006.07.036 (2006).
Saad, Z. S. & Reynolds, R. C. Suma. NeuroImage 62, 768–773, doi:10.1016/j.neuroimage.2011.09.016 (2012).
Ashburner, J. & Friston, K. J. Unified segmentation. NeuroImage 26, 839–851, doi:10.1016/j.neuroimage.2005.02.018 (2005).
Li, M. X., Yeung, J. M., Cherny, S. S. & Sham, P. C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Human genetics 131, 747–756, doi:10.1007/s00439-011-1118-2 (2012).
Acknowledgements
This work was supported by the NIMH Intramural Research Program (ZIAMH002942), clinical protocols 00-M-0085/NCT00004571 and 95M-0150/NCT00001486. Some of the analyses performed utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov). The authors would like to thank Svante Pääbo for his ideas and guidance on this work and Janet Kelso for her assistance with genetic analyses and insightful comments.
Author information
Authors and Affiliations
Contributions
M.D.G. and K.F.B. conceived of the analyses specific to this report; M.D.G. and Z.S. designed novel methodological approaches; M.D.G., J.S.K., P.D.K., D.D., and Q.C. performed the analyses. M.D.G., V.S.M., D.R.W. and K.F.B. were involved in data collection; D.D., V.S.M., D.R.W. and K.F.B. were involved in the overall study design; D.R.W. and K.F.B. oversaw the study as principal investigators; all authors contributed to data interpretation. M.D.G., D.P.E. and K.F.B. wrote the paper, with input from all authors.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gregory, M.D., Kippenhan, J.S., Eisenberg, D.P. et al. Neanderthal-Derived Genetic Variation Shapes Modern Human Cranium and Brain. Sci Rep 7, 6308 (2017). https://doi.org/10.1038/s41598-017-06587-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-06587-0
This article is cited by
-
Echoes of ancient DNA in living modern humans affect risk for neuropsychiatric disease and brain structure and function of networks subserving higher-order cognition
Neuropsychopharmacology (2023)
-
Merging morphological and genetic evidence to assess hybridization in Western Eurasian late Pleistocene hominins
Nature Ecology & Evolution (2022)
-
The nature of Neanderthal introgression revealed by 27,566 Icelandic genomes
Nature (2020)
-
Structural connectivity and functional properties of the macaque superior parietal lobule
Brain Structure and Function (2020)
-
Neanderthal clues to brain evolution in humans
Nature (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.