Abstract
Malaria is caused by mosquito-borne Plasmodium spp. parasites that must infect and survive within mosquito salivary glands (SGs) prior to host transmission. Recent advances in transcriptomics and the complete genome sequencing of mosquito vectors have increased our knowledge of the SG genes and proteins involved in pathogen infection and transmission. Membrane solute carriers are key proteins involved in drug transport and are useful in the development of new interventions for transmission blocking. Herein, we applied transcriptomics analysis to compare SGs mRNA levels in Anopheles stephensi fed on non-infected and P. berghei-infected mice. The A. stephensi solute carriers prestinA and NDAE1 were up-regulated in response to infection. These molecules are predicted to interact with each other, and are reportedly involved in the maintenance of cell homeostasis. To further evaluate their functions in mosquito survival and parasite infection, these genes were knocked down by RNA interference. Knockdown of prestinA and NDAE1 resulted in reduction of the number of sporozoites in mosquito SGs. Moreover, NDAE1 knockdown strongly impacted mosquito survival, resulting in the death of half of the treated mosquitoes. Overall, our findings indicate the importance of prestinA and NDAE1 in interactions between mosquito SGs and Plasmodium, and suggest the need for further research.
Similar content being viewed by others
Introduction
Over the last 15 years, massive prevention measures and new treatment tools have greatly decreased the global malaria burden. However, despite these advances, about 214 million new malaria cases and 438,000 associated deaths were registered in 2015 alone1. Malaria is caused by mosquito-borne Plasmodium parasites. In particular, adult female mosquitoes of the genus Anopheles are efficient Plasmodium vectors in many diverse ecosystems2, 3. Anopheles stephensi is a competent vector for both Plasmodium falciparum and P. vivax, the most virulent malaria-associated species3, as well as for Plasmodium species that infect rodents and, thus, this mosquito species is widely used as a laboratory model. A. stephensi is found throughout the Indian subcontinent—with its territory extending from the Arabian Peninsula, Iran, and Iraq, to Bangladesh, southern China, Myanmar, and Thailand4.
Malaria transmission from mosquitoes to vertebrate hosts occurs through the mosquito’s salivary glands (SGs), and for this reason many studies have focused on this organ to develop new malaria control measures5,6,7,8,9. Sporozoite invasion of mosquito SGs is receptor-mediated, and involves several parasite and SG proteins10, 11. Characterization of the salivary components involved in parasite infection and transmission would greatly enhance our understanding of the mosquito–host–pathogen interface, and potentially reveal candidate targets for malaria prevention and control.
Sialotranscriptomic and functional genomic studies of Anopheles spp. after feeding or infection have created transcriptome catalogues that reveals the influence of Plasmodium infection process in certain genes5, 12. Recently, a role of a membrane glucose transporter gene in Anopheles gambiae was reported in malaria transmission using RNA interference studies5. Membrane proteins are responsible for the transport of a high diversity of substrates through the SGs, maintaining basic cell physiological functions and preserving optimal conditions for completion of the parasite life cycle5, 13, 14.
Solute carrier (SLC) families include membrane proteins that transport solutes, such as ions and metabolites maintain ionic homeostasis using ion or electrochemical gradients15. Over 300 human SLCs have been identified and studied with regards to their pharmacological applications as key proteins for drug transport16. Thus, research focusing the ion transporters in mosquito SGs will be useful not only for increasing our knowledge of these biological processes, but also for the development of new interventions for transmission blocking17.
In the present study, SGs were isolated from mosquitoes fed on P. berghei-infected and on non-infected mice, and RNA sequencing (RNA-seq) was performed. Gene expression levels were determined and compared between the infected and non-infected sialomes. The membrane ion transporters prestinA and NDAE1 were found to be up-regulated in response to infection. Based on the gene expression during Plasmodium infection, putative co-interaction and transmembrane function to maintain cell homeostasis, these genes were selected to perform silencing experiments. Further gene silencing of prestinA and NDAE1 resulted in significantly reduced vector survival and decreased sporozoite numbers in the SG. This work completes the analysis of the first Illumina-derived mRNAs assembly from P. berghei-infected A. stephensi SGs, constituting a valuable tool for future research18. Our present results suggest that membrane ion transporters in A. stephensi SGs affect Plasmodium infection, potentially representing new targets for development of measures to block malaria transmission.
Results and Discussion
Expression profiling and functional annotation of A. stephensi sialotranscriptome during P. berghei infection
Recently‚ it was demonstrated that, according to the system under study, transcriptomic changes are highly correlated with protein levels19, 20. Thus, the analysis of the transcriptomic response of A. stephensi SG to P. berghei infection can predict, at some extend, proteome regulation. Previous studies have investigated the Anopheles sp. sialotranscriptome12, 21,22,23,24, but concerning A. stephensi, there is only a previous work reporting an immune related response to P. berghei infection18. Herein, RNA-seq analysis enabled the production of two catalogs of transcripts corresponding to the P. berghei infected and non-infected SGs, which constitute a body of fundamental information to help with the selection of potential key targets for malaria control.
From the RNA sequencing data, we obtained a total of 26,357 and 36,393 transcripts for control and infected samples, respectively. Among these transcripts, 2536 showed differential expression in response to P. berghei infection (P < 0.05, Biological Coefficient Variation value = 0.2), of which 1996 (79%) were upregulated and 540 (21%) were downregulated. These findings suggest a vast network of interactions between the vector and parasite, with certain molecules likely being essential for parasite maintenance in the mosquito and/or for part of the mosquito’s response to infection. Other studies have shown comparable results25, 26.
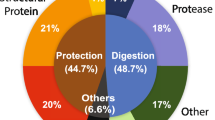
The differentially expressed A. stephensi transcripts were functionally annotated using Gene Ontology (GO) terms, and were grouped into 14 classes: cell function, chemosensory, detoxification, diverse, immunity, metabolism, proteolysis, recognition, regulatory, replication-transcription-translation (RTT), signaling, structural, transport, and unknown function. Figure 1 presents the number of transcripts in each functional class, and their expression. Due to the importance of the innate immune response of anophelines to Plasmodium infection, the data concerning immunity class was discussed elsewhere18.
Differentially expressed genes in Anopheles stephensi salivary glands in response to Plasmodium berghei infection. (A) Distribution of differentially expressed transcripts by functional classes. (B and C) Distribution of upregulated (B) and downregulated (C) transcripts by functional classes. For annotation, gene identifiers were obtained from VectorBase (www.vectorbase.org) and compared to the corresponding sequences in A. gambiae.
The assembly of differentially expressed A. stephensi SG transcripts revealed that the more commonly represented functional classes in this data set included metabolism, RTT, cell function, and transport (539, 363, 303, and 238, respectively), while less common functional classes included recognition, detoxification, and chemosensory (16, 20, and 33, respectively) (Fig. 1A). The most over-expressed and under-expressed transcripts were found in metabolism, RTT, cell function, and transport. Among the metabolism class transcripts, 429 were upregulated while 110 transcripts were downregulated. Of the transcripts of the RTT and cell function classes, respectively, 305 and 257 transcripts were upregulated while 58 and 46 were downregulated. Regarding the 238 transport class transcripts, 190 were upregulated and 48 downregulated. As SLCs play a role in Plasmodium–SGs interactions hence here main preference was given to this class that includes 33 upregulated transcripts and 5 downregulated transcripts.
Validation of RNA-seq results
For validation of the RNA-seq data, qPCR was conducted. Fourteen genes that were differentially expressed in response to Plasmodium infection, including 8 that were upregulated (fold-change of 5.67 to 1.49) and 6 that were downregulated (fold-change of −3.16 to −1.39), were analyzed (see Supplementary Table S1). There was a strong correlation between the mRNA levels obtained by both RNA-seq and qPCR methods (Pearson’s correlation coefficient = 0.667, P = 0.009) (see Supplementary Fig. S1).
Metabolism genes
Changes in metabolism-related transcripts may be associated with increases of metabolites favoring sporozoite development within SGs before transmission to a vertebrate host22, 27. Such alterations could also be related to SG maintenance and repair after Plasmodium invasion. Metabolism was the most enriched functional class, accounting for 21.49% of upregulated genes and 20.37% of downregulated genes. Metabolic processes can be induced by parasites, for example, de novo biosynthesis of pyrimidines is required by P. falciparum 28. Our present data included a fold-change of 4.17 in the transcript AGAP004172, which is described as showing orotate phosphoribosyltransferase (OPRT) activity mediating this metabolic pathway28. Another transcript, AGAP003184 (fold-change of 2.67), reportedly plays a role in the heme-biosynthetic pathway favoring Plasmodium survival29, 30.
Previous studies show that chondroitin glycosaminoglycan presence in the apical midgut microvilli of A. gambiae enables parasite adhesion31, 32, suggesting its involvement in the metabolic process. Herein, the chondroitin 4-sulfotransferase represented by AGAP005721 was upregulated in P. berghei-infected SGs, supporting the possibility that this molecule may be related to parasite adhesion to SGs.
RTT genes
Translational control of gene expression may result from cell stress and could allow cells to regulate protein levels in relation to infection processes. A total of 363 transcripts from the RTT functional class were differentially expressed, including 15.28% of upregulated and 10.74% of downregulated transcripts. Increased translational regulation has also been observed in P. falciparum-infected A. gambiae 33 and in the SGs of P. berghei-infected A. coluzzii 5. Moreover, differential transcription has been detected in hemocytes of P. berghei-infected A. coluzzii 34, and a high number of upregulated genes related to DNA replication were found in dengue virus-infected A. aegypti 35. It is possible that the Plasmodium parasite may interfere with or manipulate the expressions of genes required for invasion and development within the SGs.
Immunity genes
The results concerning the immune response were discussed in a previous study conducted by the authors18. Since the innate immune response is activated when pathogens are recognized, it is expected that this class will be strongly represented in studies focusing on infection.
Transport genes
Membrane transport proteins mediate the passage of specific molecules and/or ions across the membrane bilayer of the SGs, controlling nutrient uptake, removal of metabolic wastes and xenobiotics, and the maintenance of transmembrane electrochemical gradients36. This activity also ensures that SGs meet the requirements for Plasmodium survival15, 37.
A SAGE analysis revealed that P. berghei infection impacts the expression of transport-related transcripts, suggesting that the parasite may exploit A. gambiae cellular mechanisms. Moreover, a microarray-based experiment revealed the upregulation of 326 transport-related genes among a total of 4978 transcripts from A. gambiae SGs after P. berghei infection23.
Our present results showed that transport-related transcripts were among the most differentially expressed (238 transcripts of the total of 2536), with 9.52% upregulated and 8.89% downregulated. This is in accordance with a previous report in A. coluzzii 5. Within this category, 50 transcripts were identified as ion transmembrane transporters. These proteins mediate the passage of specific molecules and/or ions through SGs membrane, and are associated with a diverse range of physiological roles, including the maintenance of transmembrane electrochemical gradients36. In particular, we identified transcripts related to the solute carrier family (SLC), including AGAP007054 (SLC13), AGAP007743 (SLC16), AGAP005405 (SLC39), AGAP000097 (SLC25), AGAP005537 (SLC35), AGAP010725 (SLC26), and NDAE1 (SLC4). Based on the influence and shared function of transporters in mosquito SGs during infection, and the high expression levels of such genes in our data, we selected the solute carriers SLC26 and NDAE1 as targets for further studies. These molecules are described below in greater detail.
Other functional classes
Some genes that were identified in this study belonged to functional classes other than those mentioned above, and have been previously reported as associated with Plasmodium infection. The transcript AGAP010035 from the cell function class was upregulated (fold-change of 2.34), and is related to the TOR nutritional hormone signaling pathway, which is affected by parasite infection in the midgut38. Additionally, we detected downregulation of two salivary gland surface (SGS) proteins, SGS4 (fold-change of −1.51) and SGS5 (fold-change of and −2.75), which are considered highly immunogenic and related to both blood feeding and parasite infection39, 40, suggesting parasite expression regulation.
Proteins from the proteolysis class are involved in several proteolytic events during blood feeding, including digestion of blood, sugar, and extracellular matrix components23, 41. The proteolysis gene AGAP009212 (fold-change of 1.49)—known as SRPN6, a malaria parasite invasion marker in mosquitoes—suggests that serpins are associated with the immune response against Plasmodium infection in A. stephensi. In fact, mosquito serpins control melanization42,43,44,45 and host homeostasis during blood feeding44. Previous reports showed that serine proteases are also related to dengue virus transmission in Aedes mosquitoes46.
Apoptosis, i.e., programmed cell death, is a crucial cellular mechanism for tissue homeostasis and normal immune system functioning, which is highly regulated by a variety of caspases, Bcl-2 proteins, and inhibitors of apoptosis (IAPs)47. Studies using P. berghei reveal the induction of cell death and apoptosis during infection48, 49. Our present data showed upregulation of the initiator caspase CASPL2 (fold-change of 0.95), which is reportedly related to apoptosis during parasite infection. Ramphul et al.and collaborators (2015) silenced initiator caspases, such as CASP-L1, CASP-L2 and IAP1, and reported no significant changes in Plasmodium infection intensity or infection prevalence among mosquitoes50. Downstream substrates of caspases, such as Bcl proteins, have been found in the A. stephensi sialome. Here we detected upregulation of Anob-1 (AGAP011552) (fold-change of 1.44), which is a protein from the Bcl-2 family (DEBCL) that acts as an apoptosis regulator and is related to autophagy51.
PrestinA and NDAE1 interactions
The prestinA and NDAE1 genes, respectively, were annotated as encoding a protein with sulfate transmembrane transporter activity (Gene Ontology annotation: http://www.ebi.ac.uk/QuickGO/GTerm?id = GO:0015116) and a protein with anion transmembrane transporter activity (Gene Ontology annotation: http://www.ebi.ac.uk/QuickGO/GTerm?id = GO:0008509). PrestinA is an anion exchanger belonging to the SLC26 family, while NDAE1 participates in anion transport and belongs to SLC4 family15. The transmembrane localizations of prestinA and NDAE1 were confirmed using location predictor software.
Prestin transporters have been identified in Drosophila melanogaster (dPrestin) and A. gambiae (AgPrestinA and AgPrestinB) as homologues of mammalian SLC26a5. AgPrestinA and AgPrestinB are highly expressed in the SGs and midgut, respectively15, 52. In our data, the mosquito orthologue was found over-expressed (RNA-seq: fold-change of 2.19; qPCR: fold-change of 4.70) in Plasmodium-infected SGs. PrestinB is a transmembrane protein that supports the exchange of ions HCO3–, oxalate2−, SO42−, and formate across the midgut membrane15. Uptake of SO4 2− by prestinB is essential for biosynthesis of chondroitin sulfate (ChSO4), an epithelial ligand that allows midgut invasion31. PrestinB also mediates the uptake of bicarbonate produced by carbonic anhydrase or supplied by Na+-driven anion exchanger (NDAE1)53 that was also found to be over-expressed (RNA-seq: fold-change of 3.77; qPCR: fold-change of 1.98) as revealed in our studies. NDAE1 is a solute carrier that plays a central role in bicarbonate, sulfate, and oxalate metabolism, contributing to maintaining pH homeostasis in the cell lumen. Disruption of this gene is reportedly lethal to Drosophila larva54. As expected, both these SLC transcripts were found over represented in the previously reported sialotranscriptome of A. coluzzii 5. Moreover, both SLC are presented in both A. gambiae (NCBI (https://www.ncbi.nlm.nih.gov/) with the Accession no. UP_000007062) and A. stephensi (VectorBase (https://www.vectorbase.org/) with the Accession no. AsteI2.3.) proteomes, which indicates the possibility of these transporters being expressed in the mosquito. We further analyzed the potential interaction between these two solute carriers using STRING software for prediction of protein–protein interaction (Fig. 2).
PrestinA and NDAE1 showed close homology to the A. gambiae genes AGAP010725 and AGAP009736, respectively, with higher bitscores and lower E values. PrestinA (SLC26a5; 641 amino acids) may interact with at least two possible network partners: the proteins NDAE1 and K13811. NDAE1 (AGAP009736) showed the closest relationship, with a high confidence score of 0.729. The protein K13811, a 3′-phosphoadenosine 5′-phosphosulfate synthase (666 aa) encoded by the AGAP001256 gene, showed an interaction score of 0.731 with prestinA. In sulfation reactions, K13811 is considered an obligate cosubstrate for active sulfate55. Remarkably, our data showed that the AGAP001256 transcript was also upregulated (fold-change of 1.86). NDAE1 network predictions indicated the existence of two proteins: prestinA (confidence score of 0.729) and FKBP12 (108 aa; confidence score of 0.753), which was not present in the transcriptomic data.
Furthermore, the evolutionary history of the prestin (Fig. 3) indicates a high probability that the A. stephensi protein is closely related to the majority known proteins from other malaria vectors.
Prestin molecular phylogenetic analysis by Maximum Likelihood method. Using the Maximum Likelihood method based on the best fitted models (LG + G), the tree with the highest log likelihood (−1660.211) is shown. A Gamma distribution (G) was used to model evolutionary rate differences among sites (5 categories ( + G, parameter = 0.79). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 17 amino acid sequences with respective VectorBase accession number: A. albimanus (AALB009835); A. arabiensis (AARA008979); A. atroparvus (AATE014136); A. coluzzii (ACOM027477); A. culicifacies (ACUA008536); A. darlingi (ADAC002173); A. dirus (ADIR005276); A. epiroticus (AEPI009604); A. farauti (AFAF017372); A. funestus (AFUN006954); A. gambiae (AGAP010725); A. maculatus (AMAM007756); A. melas (AMEC018923); A. merus (AMEM004325); A. minimus (AMIN009235); A. stephensi (ASTE009391) and Ixodes scapularis (ISCW019766) (outgroup). All positions containing gaps and missing data were eliminated and bootstrap values for internal branches are shown.
The molecular phylogenetic analysis showed that A. stephensi and A. maculatus NDAE1 proteins are in the same monophyletic group, being closely related (Fig. 4).
NDAE1 molecular phylogenetic analysis by Maximum Likelihood method. Using the Maximum Likelihood method based on the best fitted models (LG + G), the tree with the highest log likelihood (−4016.814) is shown. A Gamma distribution (G) was used to model evolutionary rate differences among sites (5 categories ( + G, parameter = 0.27). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 17 amino acid sequences with respective NCBI accession number: A. albimanus (AALB000104); A. arabiensis (AARA004033); A. atroparvus (AATE011952); A. coluzzii (ACOM026960); A. culicifacies (ACUA004881); A. darlingi (ADAC005721); A. dirus (ADIR005084); A. epiroticus (AEPI001894); A. farauti (AFAF017853); A. funestus (AFUN000817); A. gambiae (AGAP009736); A. maculatus (AMAM011081); A. melas (AMEC001882); A. merus (AMEM008317); A. minimus (AMIN009479); A. stephensi (ASTE003102) and Ixodes scapularis (ISCW000340) (outgroup). All positions containing gaps and missing data were eliminated and bootstrap values for internal branches are shown.
This high degree of conservation of prestin between anophelines emphasized the potential of this target to block malaria transmission in different malaria vector species.
Considering this information, we further used RNAi to assess the co-participation of these two SLCs, which share the same physiological processes and functions, in parasite invasion and transmission52, 54 in mosquito SGs.
Knockdown of prestinA and NDAE1: Solute carrier pathways as a potential target for blocking malaria transmission
Membrane transporter proteins of mosquito SGs are considered potential candidates for the development of malaria control measures. For example, saglin is essential for Plasmodium sporozoites to invade the SGs56 and AGAP007752 is up-regulated in the A. gambiae sialotranscriptome after P. berghei invasion5. As mentioned above, we conducted further studies focusing on two SLCs based on their important roles as transmembrane transporters and co-interaction with Plasmodium and mosquito-cell survival. RNA interference-mediated gene silencing was used to evaluate the potential effects of prestinA and NDAE1 on A. stephensi mosquitoes upon infection with P. berghei.
For dsRNA synthesis, both fragments of interest were first amplified and further sequenced using the T7 promoter. The obtained sequences were compared to available databases using the Basic Local Alignment Search Tool (BLAST) from VectorBase. The products showed high similarity to the prestinA sequence (99.8% reverse primer and 96.3% forward primer) and the NDAE1 sequence (99% reverse and forward primer). Synthetized dsRNA was checked for integrity, and ~276 ng of dsRNA was injected into each mosquito. The dsRNA was injected fourteen days post-infection because at this time point the ruptured oocyts release sporozoites which can then migrate to the salivary glands. After silencing assays, we evaluated the gene expression levels (Fig. 5A–C), percentage of gene silencing (Fig. 5D), survival rate (Fig. 6A–D), and numbers of sporozoites on SGs (Fig. 6E).
Results of prestinA and NDAE1 gene knockdown in Anopheles stephensi salivary glands. (A–C) The relative expressions of prestinA and NDAE1 were evaluated in Anopheles stephensi salivary glands with gene knockdown of prestinA, NDAE1, and in co-silenced groups, respectively. Normalized against RPS7, Cq values were compared between mosquitoes injected with dsRNA for the selected genes and with β2M control dsRNA. The expression of the genes in the prestinA, NDAE1 and both silenced groups is set to 1 for a better interpretation. Statistical analyses were conducted using the Pfaff method. *P < 0.05, **P < 0.01, ***P < 0.001. (D) Summary of silencing assay results. Gene expression levels and gene knockdown percentage of the treated groups.
Knockdown of the prestinA and NDAE1 genes in Anopheles stephensi salivary glands. (A–C) Kaplan-Meier plots of the survival of Anopheles stephensi mosquitoes after silencing of prestinA (A), NDAE1 (B), or both genes together (C). (D) Kaplan-Meier statistics with values from the Log-Rank, Breslow, and Tarone-Ware tests. *P < 0.05, **P < 0.01, ***P < 0.001. (E) Effects of silencing prestinA, NDAE1, or both genes together in the Anopheles stephensi salivary glands with regards to the number of sporozoites. Distributions were compared using the Mann-Whitney test. *P < 0.05, **P < 0.01, ***P < 0.001.
In Fig. 5, the relative expression of prestinA and NDAE1 genes is represented in the prestinA (Fig. 5A), NDAE1 (Fig. 5B), and co-silenced A. stephensi groups (Fig. 5C). Mosquitos injected with prestinA dsRNA showed 87.57% gene knockdown, significantly reducing the prestinA expression level (−8.05; P = 0.005). NDAE1 was knocked down by 92.98%, significantly reducing the gene expression level (−14.23; P = 0.047). PrestinA and NDAE1 expression levels were also evaluated under the effect of knockdown of the other gene. PrestinA silencing was associated with a decline of NDAE1 mRNA levels (−1.13; P = 0.264), and NDAE1 silencing was associated with reduced prestinA expression (−6.54; P = 0.271), suggesting interaction between these two proteins. Double knockdown (silencing 62.02% of prestinA and 55.25% of NDAE1) resulted reduced mRNA expression levels for both genes (prestinA = −2.63, P = 0.018; NDAE1 = −2.23, P = 0.012) compared with the control group. The lower silencing efficiency in this case can be explained by the amount of specific dsRNA used, since in the double knockdown, we inoculated mosquitoes with ~138 ng/injection of each dsRNA.
For each group, the survival rate was recorded daily until the 18th day PBM (Fig. 6A–C), and compared to the β2M-dsRNA-injected controls.
Under the conditions of the present study, our results showed that prestinA silencing seems to not influence the survival of A. stephensi mosquitoes (P > 0.05) (Fig. 6A,D). Since an abnormal mortality in the initial time points, in both control and prestinA silenced groups was observed data was re-tested excluding the 14th and 15th day. Nevertheless, statistical analysis continues to point to a weak effect of prestinA silencing in mosquito survival (see Supplementary Table S4). This suggests that other molecules can compensate for the cellular absence of prestinA. Other studies of different transporters, such as apolipophorin-III (a lipid transporter) and fondue (a component of the hemolymph clot), show that manipulation of their gene expressions in A. gambiae G3 strain mosquitoes resulted in survival patterns similar to that of the control57. However fondue-silenced mosquitoes reportedly influence fungal infections57. Our results showed that double knockdown profoundly affected mosquito mortality, with a 23% population reduction. Moreover, knockdown of NDAE1 alone led to a 51% population reduction (P < 0.05) (Fig. 6B–D). This influence of NDAE1 was also detected in a study of Drosophila larvae, in which NDAE1 was found to be crucial for fly development54. Our present results support this molecule’s vital role in Cl−/HCO3 − exchange as a component of the steady-state cell alkalization in the mosquito58. Based on previous findings that acute intracellular pH acidification can rapidly kill a cell, targeting cellular acidification is being considered as a method of cancer therapy59. Recently, a study showed that the survival rate of transgenic mosquitos lacking SGs was not affected8, which could point to a weak effect of NDAE1 knockdown in mosquito mortality, but NDAE1 is not exclusive to SGs15 and therefore, manipulating its expression in the whole organism had a visible physiological effect (lower survival rate). On the other hand, prestinA is SG exclusive, and thus the disruption of mRNA is tissue specific.
To evaluate infection levels, sporozoites were quantified at four days post-dsRNA injection. All knockdown groups showed statistically significant reduction of sporozoite numbers. Knockdown of prestinA reduced infection by 42.2% (P = 0.003), the NDAE1 silenced group showed an infection reduction of 73.5%, (P = 0.006), and double knockdown resulted in a reduction of 53.2% (P = 0.001) (Fig. 6E).
These results together with previous findings15, 31, support that reducing transporter expression in A. stephensi SGs, and thus lowering sulfate uptake and leading to deficient chondroitin sulfate biosynthesis, could prevent Plasmodium invasion by reducing the ability for parasite adhesion.
Sustained pH homeostasis is essential for viable Plasmodium invasion/multiplication in mosquito tissues60. Manipulation of NDAE1 expression in the whole vector may produce a synergetic effect, resulting in an acute reduction of sporozoites in SGs due to pH alteration. Silencing of both genes may have altered system equilibrium, exposing P. berghei to different/adverse environmental parameters. Figure 7 presents our hypothesis of prestinA and NDAE1 physiological function in A. stephensi-SGs cells and their influence on Plasmodium infection.
Proposed model for prestinA and NDAE1 physiological functions and impact on SG cell and Plasmodium berghei infection. PrestinA is expressed at apical membrane of cells, mediates the uptake of sulfate and excretes bicarbonate which is produced by carbonic anhydrase or supplied by NDAE1. Disruption of prestinA alters the biosynthesis of chondroitin sulfate which interferes with Plasmodium adhesion. A reduction of both prestinA and NDAE1 changes cell pH, affecting Plasmodium and mosquito survival.
Suppression of ion and solute exchangers can disrupt different metabolic pathways; therefore, these transmembrane proteins may be useful in new transmission-blocking approaches. The impact of trehalose and glucose transporters has already been demonstrated in terms of survival of A. gambiae 37 and A. coluzzii, as well as P. berghei maintenance in SGs. Our present findings provide crucial information for the selection of target genes for preventing malaria transmission, including our report of the A. stephensi sialotranscriptome during P. berghei infection and our functional characterization of prestinA and NDAE1. These ion and solute membrane transporters are tightly connected, and were demonstrated to be upregulated after infection. Moreover, RNAi-mediated gene knockdown results suggested that the proteins encoded by these genes may affect Plasmodium viability in the SGs. Further investigation is needed to improve our understanding of basic transport, transport regulation, and how these processes regulate vector–parasite interaction. To eradicate and control malaria disease, it is crucial that we explore new targets for developing control measures and transmission-blocking vaccines.
Methods
Ethics statement
Animals were maintained at the Institute of Hygiene and Tropical Medicine (IHMT), in accordance to Europe Directive 86/609/ EEC and Portuguese law (Decreto-Lei No. 129/92). Animal experiments were conducted with the approval of the Divisão Geral de Alimentação e Veterinária (DGAV), Portugal, under Art° 8, Portaria n°1005/92 from 23rd October (permit number n° 023357) and the Council of Ethics of the IHMT. All experiments were performed in accordance with relevant guidelines and regulations.
Mosquito rearing
The A. stephensi mosquitoes were obtained from the IHMT insectary and reared at 20 °C in 70% humidity, under a 12 h light/dark photoperiod. Adult mosquitoes were fed ad libitum on a 10% glucose solution. For experiments, only mosquitoes aged between 3–5 days were used.
Plasmodium infection
To validate gene expression and gene knockdown efficiency, mosquitoes fed on uninfected and P. berghei -infected mice were obtained. Female CD1 mice were intraperitoneally inoculated with 107 P. berghei ANKA parasitized red blood cells. The levels of parasitaemia were measured from blood smears of the mouse tail using light microscopy after staining with Hemacolor® kit (EMD Millipore, Germany). Mice were used when the parasitaemia reached about 10% and exflagellation was observed (4–6 exflagellations/field).
Three hundred female mosquitoes per group were allowed to feed on anesthetized mice, and only fully engorged females were kept at 19–21 °C and 80% humidity.
RNA extraction
Midguts and SGs were dissected from cold anaesthetized female mosquitoes, under a stereoscopic microscope at 4X magnification (Motic SMZ-171B, China). Detailed methodology of tissue and RNA isolation is described elsewhere5. In order to check Plasmodium infection, midguts were dissected at day 8–9 post-blood-meal (PBM) confirming the presence of oocysts; at day 18 PBM, SGs were extracted and kept either in ice cold PBS for sporozoites quantification or in ice-cold RNA later (Ambion, Austin, TX, USA) for RNA extraction.
RNA was promptly extracted using RiboZol™ (AMRESCO, USA) and stored at −80 °C. The quantification and integrity of the RNA was assessed using a ND-1000 Spectrophotometer (NanoDrop ND1000, Thermo Fisher Scientific, Whaltman, MA) and a Agilent 2100 Bioanalyser® system (Agilent Technologies, USA), respectively.
RNA sequencing
RNA-seq was conducted in SGs of mosquitoes fed on uninfected and P. berghei-infected mice. Individual SGs of from each condition were grouped to obtain the quantity needed to perform high throughput sequencing. Sequencing was performed at Parque Científico de Madrid (FPCM, Spain). From 1 µg of total-RNA, enrich poly (A) mRNA were used to bead with magnetic beads with Oligo (dT) to construct the cDNA library. Next, from purified mRNAs, two different fragmentation conditions were applied so that a “shorter” and a “longer” preparation was made for both control and infected RNA samples. Further, double-stranded cDNAs were synthesized and subjected to end-repair, adenylation, ligation with sequencing adapters and PCR amplification. Proper fragments were purified by size using AMPure XP beads (Beckman Coulter, High Wycombe, UK) and size and quality confirmed using an Agilent High Sensitivity DNA kit (Agilent Technologies, CA, USA). The size range of fragments was around 310 bp for the short insert and 465 bp for the longer insert preparations. Next, the libraries were pooled and titrated using qPCR to get an accurate estimation of concentration. In a Cluster Station (Illumina, CA, USA), clusters were generated and using a GAIIx equipment (Illumina, CA, USA), the libraries were sequenced using a 2 × 100 cycle sequencing run program separated by a paired-end turnaround. TruSeq RNA kit (Illumina, CA, USA) was used to prepare cDNA library as per manufacturer instructions. Based on the results provided by FPCM, Era7 Company performed the transcriptomic analysis. Trimmed sequences were used for de novo assembly using Oases program and functional information obtained from Uniprot (GO annotations, Enzyme Commission (EC) number, Interpro motifs). For each condition, Bowtie and Samtools were used to map and determine the RPKM values of reads. After clustering transcripts, eXpress program was used to obtain the quantification per protein, and each set of transcripts annotated by the same protein was considered as a Unigene provided with functional annotation. Differential expression analysis of Unigenes in infected versus uninfected mosquitoes was carried out using EdgeR61, 62. For this data, each transcript compiles information of UniProt, RefSeq, GO, VectorBase and NCBI Taxonomy Enzyme databases. After the initial analysis, transcripts were functionally annotated into 14 classes: cell function, chemosensory, detoxification, diverse, immunity, metabolism, proteolysis, recognition, regulatory, RTT, signaling, structural, transport and unknown. Previous studies have investigated the sialotranscriptome of A. stephensi regarding exclusively the immunity class18. The remaining data is analyzed in the present study.
Up or down gene regulation was also considered in this analysis. RNA-seq data can be found in the database: ArrayExpress (www.ebi.ac.uk/arrayexpress) with the Accession no. E-MTAB-3964.
Validation of transcriptomics data
Fourteen transcripts identified by RNA-seq of A. stephensi SGs with differential regulation and belonging to different functional classes with a potential interference in the parasite development, were chosen for RNA-seq validation through relative qPCR. Using three biological replicates of each condition (infected and non-infected), total RNA (100 ng/µL of each sample) were used to synthesize cDNA using the iScript™ cDNA Synthesis Kit (Bio-Rad, CA, USA). qPCR reactions of 20 μl were performed in triplicate using IQ™ SYBR® Green Supermix kit (Biorad, CA,USA) in a CFX96 Touch Real-time PCR (Bio-Rad, CA, USA) thermocycler. The following conditions were used: an initial cycle of denaturation at 95 °C for 10 min; followed by 45 cycles of 95°C for 15 seconds and temperature of each primer set for 45 seconds. Fluorescence readings were taken at 62 °C after each cycle and a melting curve (60–95 °C) was performed. The sequences of the primers were design using Primer3 platform (http://bioinfo.ut.ee/primer3–0.4.0/) and their conditions are listed in Supplementary Table S2.
Standard curves were constructed with 10-fold serial dilutions of cDNA synthetized from total RNA from a pool of eight females kept exclusively on a sugar diet, in order to determine the reaction efficiency. To ensure that only one amplicon was formed and the amplicons denatured at the same temperature, a dissociation curve was run at the end of the reactions. Relative expression levels were normalized using the ribosomal protein S7 (RPS7) (Vectorbase: ASTE004816) as reference gene (see Supplementary Table S2) and the Pfaff method for analysis63. Normalized Cq values were compared between infected and control samples by Mann-Whitney test (P < 0.05). Pearson’s correlation was used to compare the expression values between RNA-seq and qPCR methods for the 14 selected genes (see Supplementary Fig. S1).
Network and localization analysis
Genes selected for functional analysis were further characterized using the software STRING 10.0 (Search Tool for the Retrieval of Interacting Genes/Proteins available at http://string-db.org) in order to identify known/novel protein-protein interactions. Since STRING software until date only accepts A. gambiae data, orthologues from A. stephensi obtained sequences were used7, 64. Briefly, the program by single protein name or amino acid sequence generates the network images based on a spring model. To predict transmembrane domains/protein localization, TMHMM, based on a hidden Markov model65, 66, WoLF PSORT67 and MultiLoc268 were applied to amino acid sequences.
Sequence analysis
From VectorBase database (https://www.vectorbase.org/), the amino acid sequence of the selected SLC proteins were used to search for the orthologues (https://www.vectorbase.org/blast). The sequences of the orthologues and main vectors of malaria were obtained. Ixodes scapularis-related sequence was used as outgroup. All sequences were aligned with MAFFT (v7) (Katoh & Standley 2016) and all positions containing gaps were exclude with GUIDANCE2 (Sela et al. 2015). Molecular Evolutionary Genetics Analysis (MEGA, version 6) software was used to obtain the best model to build69 a Maximum Likelihood tree based on a bootstrapping method with 1000 replicates. Generated trees were visualized and edited using FigTree v1.4.370.
RNAi
RNAi-mediated gene-silencing assays were performed to evaluate the effect of the knockdown of the genes prestinA (ASTE009391) and NDAE1 (ASTE003102) on P. berghei-infected A. stephensi mosquitoes. Previously obtained mosquito RNA was used to synthesize cDNA using the iScript™ cDNA Synthesis Kit (Bio-Rad, CA, USA) and cDNA was used to amplify fragments of interest, using iProof™ High-Fidelity DNA Polymerase (BioRad, CA, USA) and specific primers containing T7 promoter sequences at the 5′-end (see Supplementary Table S3). PCR reactions of 50 µL included: 1× iProof HF Buffer, 10 mM of dNTP mix, 0.5 µM of each primer, 1 mM of MgCl2, 0.02 U/µL Proof DNA Polymerase and 50–500 ng of template cDNA. The conditions were: initial denaturation performed at 98 °C for 3 minutes; following for denaturation at 98 °C for 10 seconds, annealing at specific temperature (see Supplementary Table S3) for 30 seconds and extension at 72 °C for 15 seconds cycling for 35 cycles; and then a final extension at 72 °C for 10 minutes. Amplification results were analyzed on a 0.5× TBE, 1.2% (w/v) agarose gel, purified using Zigmoclean™ Gel DNA Recovery Kit (Zymo Research, USA) and sequenced. dsRNA was synthetized using the MEGAscript T7 kit (Ambion, Austin, TX, USA) according to manufacturer’s instructions and analyzed by spectrometry and agarose gel.
For dsRNA injection three independent assays per group (target gene) were conducted one week apart. For each group, 100 P. berghei-infected female were used to knockdown prestinA, NDAE1 and both genes simultaneously. Similarly, the control groups were injected with an exogenous gene, mouse beta-2-microglobulin dsRNA (dsβ2 M) (GenBank: NM_009735). Fourteen days PBM, cold anesthetized female mosquitoes were injected intrathoraxically with 69 nl (4 mg/ml) of each dsRNA using a nano-injector (Nanoject, Drummond Scientific, Broomall, PA, USA). For double knockdown a mixture of equal parts of dsRNA was used. At this time point, the majority of oocysts ruptured and the sporozoites start to invade salivary glands.
Gene knockdown assessment
After dsRNA injection, the number of viable mosquitoes was evaluated using Log Rank, Breslow and Tarone-Ware tests71, 72. Four days post injection, three biological replicas containing 15 to 30 SGs per treatment were used to perform sporozoite quantification, by light microscopy using a haemocytometer5. Infection levels were compared between control and treated groups using the non-parametric Mann-Whitney test (SPSS v24.0)73. To assess gene knockdown efficiency three biological replicas of 20–40 SGs per group were used to extract total RNA and synthetize cDNA as described previously. qPCR assays were performed under the conditions aforementioned. Gene expression was analysed by the CFX Manager™ Software (Bio-Rad, CA, USA) using Pfaff method63.
References
WHO. World Malaria Report 2015. (2015). Available at: http://apps.who.int/iris/bitstream/10665/200018/1/9789241565158_eng.pdf?ua = 1. (Accessed: 23rd June 2016).
Sokhna, C., Ndiath, M. O. & Rogier, C. The changes in mosquito vector behaviour and the emerging resistance to insecticides will challenge the decline of malaria. Clin. Microbiol. Infect. 19, 902–7 (2013).
Sinka, M. E. et al. The dominant Anopheles vectors of human malaria in the Asia-Pacific region: occurrence data, distribution maps and bionomic précis. Parasit. Vectors 4, 89 (2011).
Sinka, M. E. Anopheles mosquitoes - New insights into malaria vectors. (2013). Available at: http://horizon.documentation.ird.fr/exl-doc/pleins_textes/divers13-11/010060049.pdf. (Accessed: 23rd March 2015).
Pinheiro-Silva, R. et al. Gene expression changes in the salivary glands of Anopheles coluzzii elicited by Plasmodium berghei infection. Parasit. Vectors 8, 485 (2015).
Francischetti, I., Ma, D., Andersen, J. & Ribeiro, J. Evidence for a lectin specific for sulfated glycans in the salivary gland of the malaria vector, Anopheles gambiae. PLoS One 9, e107295 (2014).
Vijay, S., Rawat, M. & Sharma, A. Mass Spectrometry Based Proteomic Analysis of Salivary Glands of Urban Malaria Vector Anopheles stephensi. (2014). Available at: https://www.hindawi.com/journals/bmri/2014/686319/. (Accessed: 19th March 2016).
Yamamoto, D. S., Sumitani, M., Kasashima, K., Sezutsu, H. & Matsuoka, H. Inhibition of Malaria Infection in Transgenic Anopheline Mosquitoes Lacking Salivary Gland Cells. PLoS Pathog 12, e1005872 (2016).
Doucoure, S. et al. Biomarkers of Vector Bites: Arthropod Immunogenic Salivary Proteins in Vector-Borne Diseases Control. Gen. Methods Biomark. Res. their Appl. 1177–1205 (2015).
Mueller, A., Kohlhepp, F. & Hammerschmidt, C. Invasion of mosquito salivary glands by malaria parasites: Prerequisites and defense strategies. Int. J. Parasitol. 40, 1229–1235 (2010).
Wells, M. & Andrew, D. Salivary gland cellular architecture in the Asian malaria vector mosquito Anopheles stephensi. Parasit. Vectors 8, 1 (2015).
Francischetti, I. & Valenzuela, J. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J. Exp. Biol. 205, 2429–2451 (2002).
Brennan, J. D., Kent, M., Dhar, R., Fujioka, H. & Kumar, N. Anopheles gambiae salivary gland proteins as putative targets for blocking transmission of malaria parasites. Proc. Natl. Acad. Sci. USA. 97, 13859–64 (2000).
Rosenberg, R. Inability of Plasmodium knowlesi sporozoites to invade Anopheles freeborni salivary glands. Am. J. Trop. Med. Hyg. 34, 687–691 (1985).
Hirata, T., Czapar, A., Brin, L. & Haritonova, A. Ion and solute transport by Prestin in Drosophila and Anopheles. J. Insect Physiol. 58, 563–569 (2012).
Schlessinger, A., Khuri, N., Giacomini, K. M. & Sali, A. Molecular modeling and ligand docking for solute carrier (SLC) transporters. Curr. Top. Med. Chem. 13, 843–56 (2013).
Pullikuth, A., Filippov, V. & Gill, S. Phylogeny and cloning of ion transporters in mosquitoes. J. Exp. Biol. 206, 3857–3868 (2003).
Couto, J., Ferrolho, J. & de la Fuente, J. Anopheles gambiae and A. stephensi Immune Response during Plasmodium berghei Infection Uncovered by Sialotranscriptomic Analysis. Int. Trends Immun. 4, 58–72 (2016).
Li, J. & Biggin, M. Statistics requantitates the central dogma. Science (80-.). 347, 1066–1067 (2015).
Rokyta, D., Margres, M. & Calvin, K. Post-transcriptional mechanisms contribute little to phenotypic variation in snake venoms. G3 Genes| Genomes| Genet. 5, 2375–2382 (2015).
Oviedo, M. N. et al. The salivary transcriptome of Anopheles gambiae (Diptera: Culicidae) larvae: A microarray-based analysis. Insect Biochem. Mol. Biol. 39, 382–94 (2009).
Rosinski-Chupin, I. et al. SAGE analysis of mosquito salivary gland transcriptomes during Plasmodium invasion. Cell. Microbiol. 9, 708–24 (2007).
Das, S., Radtke, A. & Choi, Y. Transcriptomic and functional analysis of the Anopheles gambiae salivary gland in relation to blood feeding. BMC Genomics 11, 1 (2010).
Valenzuela, J. G., Francischetti, I. M. B., Pham, V. M., Garfield, M. K. & Ribeiro, J. M. C. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. Biol. 33, 717–32 (2003).
Xu, X. C., Qu, F. Y., Song, G. H. & Xu, J. N. Enrichment and screening of up-regulated genes of the mosquito Anopheles stephensi in response to malaria parasite. Chinese J. Parasitol. Parasit. Dis. 19, 325–329 (2001).
Oduol, F., Xu, J., Niaré, O. & Natarajan, R. Genes identified by an expression screen of the vector mosquito Anopheles gambiae display differential molecular immune response to malaria parasites and bacteria. Proc. Natl. Acad. Sci. 97, 11397–11402 (2000).
Peterson, T. M. L., Gow, A. J. & Luckhart, S. Nitric oxide metabolites induced in Anopheles stephensi control malaria parasite infection. Free Radic. Biol. Med. 42, 132–42 (2007).
Zhang, Y., Evans, G., Clinch, K. & Crump, D. Transition state analogues of Plasmodium falciparum and human orotate phosphoribosyltransferases. J. Biol. Chem. 288, 34746–34754 (2013).
Nagaraj, V., Sundaram, B. & Varadarajan, N. Malaria parasite-synthesized heme is essential in the mosquito and liver stages and complements host heme in the blood stages of infection. PLoS Pathog 9, e1003522 (2013).
Ke, H., Sigala, P., Miura, K. & Morrisey, J. The heme biosynthesis pathway is essential for Plasmodium falciparum development in mosquito stage but not in blood stages. J. Biol. Chem. 289, 34827–34837 (2014).
Dinglasan, R., Alaganan, A. & Ghosh, A. Plasmodium falciparum ookinetes require mosquito midgut chondroitin sulfate proteoglycans for cell invasion. Proc. Natl. Acad. Sci. 104, 15882–15887 (2007).
Sinnis, P., Coppi, A., Toida, T. & Toyoda, H. Mosquito heparan sulfate and its potential role in malaria infection and transmission. J. Biol. Chem. 282, 25376–25384 (2007).
Mead, E., Li, M., Tu, Z. & Zhu, J. Translational regulation of Anopheles gambiae mRNAs in the midgut during Plasmodium falciparum infection. BMC Genomics 13, 366 (2012).
Baton, L., Robertson, A. & Warr, E. Genome-wide transcriptomic profiling of Anopheles gambiae hemocytes reveals pathogen-specific signatures upon bacterial challenge and Plasmodium. BMC Genomics 10, 1 (2009).
Behura, S., Gomez-Machorro, C. & Harker, B. Global cross-talk of genes of the mosquito Aedes aegypti in response to dengue virus infection. PLoS Negl Trop Dis 5, e1385 (2011).
Kirk, K., Martin, R. E., Bröer, S., Howitt, S. M. & Saliba, K. J. Plasmodium permeomics: membrane transport proteins in the malaria parasite. in Current topics in microbiology and immunology 295, 325–56 (Springer Berlin Heidelberg, 2005).
Liu, K., Dong, Y., Huang, Y., Rasgon, J. L. & Agre, P. Impact of trehalose transporter knockdown on Anopheles gambiae stress adaptation and susceptibility to Plasmodium falciparum infection. Proc. Natl. Acad. Sci. USA. 110, 17504–9 (2013).
Hansen, I., Attardo, G. & Park, J. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc. Natl. Acad. Sci. USA. 101, 10626–10631 (2004).
King, J., Vernick, K. & Hillyer, J. Members of the salivary gland surface protein (SGS) family are major immunogenic components of mosquito saliva. J. Biol. Chem. 286, 40824–40834 (2011).
Drame, P., Poinsignon, A., Besnard, P. & Cornelie, S. Human antibody responses to the Anopheles salivary gSG6-P1 peptide: a novel tool for evaluating the efficacy of ITNs in malaria vector control. PLoS One 5, e15596 (2010).
Baker, D. A. et al. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics 12, 296 (2011).
Michel, K., Budd, A., Pinto, S. & Gibson, T. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 6, 891–897 (2005).
Michel, K. & Suwanchaichinda, C. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc. Natl. Acad. Sci. 103, 16858–16863 (2006).
An, C., Budd, A., Kanost, M. & Michel, K. Characterization of a regulatory unit that controls melanization and affects longevity of mosquitoes. Cell. Mol. Life Sci. 68, 1929–1939 (2011).
Zou, Z., Shin, S., Alvarez, K., Kokoza, V. & Raikhel, A. Distinct melanization pathways in the mosquito Aedes aegypti. Immunity 32, 41–53 (2010).
Conway, M., Watson, A. & Colpitts, T. Mosquito saliva serine protease enhances dissemination of dengue virus into the mammalian host. J. Virol. 88, 164–175 (2014).
Puglise, J., Estep, A. & Becnel, J. Expression Profiles and RNAi Silencing of Inhibitor of Apoptosis Transcripts in Aedes, Anopheles, and Culex Mosquitoes (Diptera: Culicidae). J. Med. Entomol. 53, 304–314 (2016).
Han, Y. & Barillas-Mury, C. Implications of Time Bomb model of ookinete invasion of midgut cells. Insect Biochem. Mol. Biol. 32, 1311–1316 (2002).
Kumar, S. Caspase function in programmed cell death. Cell Death Differ. 14, 32–43 (2007).
Ramphul, U. & Garver, L. Plasmodium falciparum evades mosquito immunity by disrupting JNK-mediated apoptosis of invaded midgut cells. Proc. Natl. Acad. Sci. 112, 1273–1280 (2015).
Colussi, P., Quinn, L. & Huang, D. Debcl, a proapoptotic Bcl-2 homologue, is a component of the Drosophila melanogaster cell death machinery. J. Cell Biol. 148, 703–714 (2000).
Brin, L., Hirata, T., Scileppi, E., Holmes, H. & Linser, P. Characterization of Anopheles gambiae Slc26a5 and potential role in malaria. FASEB J. 24, 1002–25 (2010).
Sciortino, C. & Romero, M. Cation and voltage dependence of rat kidney electrogenic Na+ -cotransporter, rkNBC, expressed in oocytes. Am. J. Physiol. Physiol. 277, F611–F623 (1999).
Romero, M., Henry, D., Nelson, S. & Harte, P. Cloning and Characterization of a Na+ -driven Anion Exchanger (NDAE1) A NEW BICARBONATE TRANSPORTER. J. Biol. Chem. 275, 24552–24559 (2000).
Klaassen, C. D. & Boles, J. W. Sulfation and sulfotransferases 5: the importance of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 11, 404–18 (1997).
Ghosh, A., Devenport, M., Jethwaney, D. & Kalume, D. Malaria parasite invasion of the mosquito salivary gland requires interaction between the Plasmodium TRAP and the Anopheles saglin proteins. PLoS Pathog 5, e1000265 (2009).
Kamareddine, L., Nakhleh, J. & Osta, M. Functional Interaction between Apolipophorins and Complement Regulate the Mosquito Immune Response to Systemic Infections. J. Innate Immun. 8, 314–326 (2016).
Boudko, D., Moroz, L. & Harvey, W. Alkalinization by chloride/bicarbonate pathway in larval mosquito midgut. Proc. Natl. Acad. Sci. 98, 15354–15359 (2001).
Parks, S., Chiche, J. & Pouysségur, J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat. Rev. Cancer 13, 611–623 (2013).
Mohring, F. et al. Determination of glutathione redox potential and pH value in subcellular compartments of malaria parasites. Free Radic. Biol. Med. 104, 104–117 (2017).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 (2010).
Chen, Y. & McCarthy, D. edgeR: differential expression analysis of digital gene expression data User’s Guide. Available at: http://bioconductor.riken.jp/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf. (Accessed: 29th February 2016) (2014).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Reid, A. J. & Berriman, M. Genes involved in host-parasite interactions can be revealed by their correlated expression. Nucleic Acids Res. 41, 1508–18 (2013).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–80 (2001).
Sonnhammer, E. L., von Heijne, G. & Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–82 (1998).
Horton, P., Park, K., Obayashi, T. & Fujita, N. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587 (2007).
Blum, T. & Briesemeister, S. MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics 10, 1 (2009).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Rambaut, A. FigTree. Available at: http://tree.bio.ed.ac.uk/software/figtree. (Accessed: 9th February 2017) (2006).
Kaplan, E. & Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53, 457–481 (1958).
Christiansen-Jucht, C. & Parham, P. Temperature during larval development and adult maintenance influences the survival of Anopheles gambiae ss. Parasit. Vectors 7, 1 (2014).
IBM Corporation. IBM SPSS Statistics for Windows. (2016)
Acknowledgements
This work was partially supported by Fundação para a Ciência e a Tecnologia - Global Health and Tropical Medicine (FCT-GHTM) [UID/Multi/04413/2013].
Author information
Authors and Affiliations
Contributions
J.F., V.R., and A.D. designed the study; J.C., S.A., and R.P.-S. performed the mosquito experiments and molecular work; and J.C., S.A., J.F., and A.D. performed data analysis and wrote the manuscript. All authors edited and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Couto, J., Antunes, S., Pinheiro-Silva, R. et al. Solute carriers affect Anopheles stephensi survival and Plasmodium berghei infection in the salivary glands. Sci Rep 7, 6141 (2017). https://doi.org/10.1038/s41598-017-06317-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-06317-6
This article is cited by
-
Glucose transporter GLUT1 influences Plasmodium berghei infection in Anopheles stephensi
Parasites & Vectors (2020)
-
Unraveling the Plasmodium vivax sporozoite transcriptional journey from mosquito vector to human host
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.