Abstract
Nicotinamide mononucleotide (NMN), a precursor of nicotinamide adenine dinucleotide (NAD), is known to act as a functional molecule in animals, whereas its function in plants is largely unknown. In this study, we found that NMN accumulated in barley cultivars resistant to phytopathogenic fungal Fusarium species. Although NMN does not possess antifungal activity, pretreatment with NMN and related metabolites enhanced disease resistance to Fusarium graminearum in Arabidopsis leaves and flowers and in barley spikes. The NMN-induced Fusarium resistance was accompanied by activation of the salicylic acid-mediated signalling pathway and repression of the jasmonic acid/ethylene-dependent signalling pathways in Arabidopsis. Since NMN-induced disease resistance was also observed in the SA-deficient sid2 mutant, an SA-independent signalling pathway also regulated the enhanced resistance induced by NMN. Compared with NMN, NAD and NADP, nicotinamide pretreatment had minor effects on resistance to F. graminearum. Constitutive expression of the NMNAT gene, which encodes a rate-limiting enzyme for NAD biosynthesis, resulted in enhanced disease resistance in Arabidopsis. Thus, modifying the content of NAD-related metabolites can be used to optimize the defence signalling pathways activated in response to F. graminearum and facilitates the control of disease injury and mycotoxin accumulation in plants.
Similar content being viewed by others
Introduction
Plants protect themselves from pathogen attack by activating a variety of defence responses, including the production of antimicrobial compounds and proteins1. Phytoalexins and phytoanticipins are antimicrobial compounds that accumulate in plant tissues with and without phytopathogen attack, respectively. Plants secrete antimicrobial proteins such as thionin to prevent the entry of pathogens into cells2. These antimicrobial substances directly inhibit the growth of pathogens on plant surfaces, whereas plant activators induce the innate immune response rather than possessing antimicrobial properties themselves. SA analogues such as 2,6-dichloroisonicotinic acid (INA) and benzo (1,2,3) thiadiazole-7-carbothionic acid S-methyl ester (BTH) hyperactivate the SA-dependent immune response. Vernooij et al.3 showed that INA acts via the systemic acquired resistance (SAR) signal transduction pathway. Similarly, BTH has been shown to enhance disease resistance against Pseudomonas syringae pv. tomato strain DC3000 (PstDC3000) and induce the expression of SAR genes4, 5. These compounds do not possess antimicrobial activities and are referred to as plant defence activators. In addition, BTH and SA positively regulate disease resistance against fungal pathogens in Arabidopsis6 and wheat7.
Some plant-derived metabolites are also known to act as plant activators8. These natural compounds are therefore potential safe agents that could be used to control disease injury caused by a broad range of phytopathogens. It has been previously reported that the extracellular nicotinamide adenine dinucleotide (NAD) and nicotinamide adenine dinucleotide phosphate (NADP) caused an accumulation of SA and the induction of pathogenesis-related (PR) genes via the Ca2+-dependent signalling pathway, enhancing resistance to Pseudomonas syringae pv. maculicola ES 4326 (Psm ES4326)9. NAD and NADP are known to act as cofactors in various redox reactions as well as to be involved in biotic and abiotic stress responses10, 11. In addition, manipulation of certain NAD biosynthesis genes in plants has been shown to affect redox signalling through the production of reactive oxygen species (ROS) and pyridine alkaloids, resulting in activation of disease resistance to phytopathogens including PstDC3000 via the SA-dependent signalling pathway12, 13. In animals, NMN, a precursor of NAD biosynthesis, is known to exert anti-ageing effects via sirtuin (SIRT) genes encoding NAD-dependent protein deacetylases14. However, the biological function of NMN in plants is largely unknown.
Fusarium species such as F. graminearum, F. culmorum, and F. asiaticum are hemibiotrophic or necrotrophic fungal phytopathogens and causal agents of head blight in wheat and barley15. These fungi often produce trichothecene mycotoxins, which are known to act as translation inhibitors in eukaryotic cells16. DON (deoxynivalenol)-producing F. graminearum exists worldwide, whereas NIV (nivalenol)-producing F. asiaticum is found only in Asia16. Under high humidity conditions, these Fusarium species can easily infect the flowers (spikes) of wheat and barley15. Contamination of grains with trichothecenes is often found worldwide, threatening human and animal health17. However, commercial cereal cultivars showing strong resistance to Fusarium diseases are not yet available17. Therefore, wheat and barley plants are treated at the flowering stage with fungicides to decrease yield and quality losses. However, fungicide residues on crops are not beneficial to human or animal health. In addition, fungicide-resistant Fusarium strains are frequently reported18. Safe and effective agents are therefore required for the control of Fusarium disease in cereal crops.
We previously identified many barley cultivars showing a Fusarium head blight (FHB) resistance phenotype using cut-spike methods19, 20. Among these cultivars, Maja and Sirius O.525 exhibited high FHB resistance. We therefore performed a comparative analysis of the metabolite profiles of these resistant cultivars vs. FHB-susceptible cultivars (Turkey 45 and H.E.S.4). Some metabolites, including NMN, accumulated in the FHB-resistant lines. NAD is known to play roles in plant defence responses against bacterial pathogens13. To estimate the effect of NMN on disease resistance to F. graminearum, we used Arabidopsis model plants, which are known to be susceptible to this pathogen. Exogenous application of NMN activated defence signalling and resulted in enhanced disease resistance against F. graminearum in Arabidopsis. Furthermore, exogenous application of NMN also significantly decreased disease development and DON production by F. graminearum in an FHB-susceptible barley cultivar. Thus, NMN could be useful in controlling disease injury and mycotoxin contamination by Fusarium species in various plant species.
Results and Discussion
NMN accumulation in FHB-resistant barley cultivars
We previously identified many barley cultivars that exhibited an FHB-resistant phenotype19, 20. Two 2-row cultivars (U389; Maja and U121; Sirius O-525) showed high FHB resistance compared with susceptible cultivars (Fig. S1a,b). We also performed a comparative analysis of the metabolite profiles of these FHB-resistant and FHB-susceptible (T615; Turkey 45) 2-row cultivars. NMN was found to be enriched in the uninoculated barley spikes of the two resistant cultivars compared with the susceptible cultivar (Fig. S1c). At 2 days post inoculation, the NMN contents had not significantly changed in any of the three cultivars (data not shown). NMN is a precursor of NAD, which functions as a cofactor for many enzymes involved in the catalysis of various metabolic reactions21. A similar accumulation pattern was not observed for other NAD-related metabolites (NaMN: nicotinic acid mononucleotide, NA: nicotinate, NIC: nicotinamide; data not shown). Unfortunately, NAD(H) and NADP(H) were not quantified in this system. Zhang & Mou9 reported that extracellular NAD induced PR gene expression and resistance against the bacterial pathogen P. syringae in Arabidopsis thaliana. These results suggest that NMN is also involved in plant disease resistance against Fusarium species. It has also been reported that the amount of NAD significantly increased in barley leaves inoculated with the biotrophic fungal pathogen Erysiphe graminis, the causal organism of powdery mildew22. These findings suggest that NAD biosynthesis is likely involved in disease resistance against a broad range of phytopathogens in barley. The NAD(H) and NADP(H) pools function as coenzymes for the various redox reactions. NMN is a precursor of not only NAD(H) and NADP(H) but also antimicrobial pyridine alkaloids23. These facts imply that NMN controls plant disease more effectively than NAD or NADP.
In addition, we examined the antifungal activity of NMN against F. graminearum (Fig. S1d). For this purpose, the inhibitory effect on mycelial growth was investigated. We used the 3-(4,5-di-methylthiazol-2-yl)-2,5-diphenyltetrazollium bromide (MTT) method to analyse the growth of F. graminearum 2, 24. We found that F. graminearum mycelium growth was not inhibited by NMN (Fig. S1d). These results indicate that NMN does not have antifungal activity against F. graminearum.
As stated above, a high concentration (>1 mM) of extracellular NAD activates defence signalling events, including the induction of pathogenesis-related 1 (PR1) expression9. To examine whether NMN can activate defence signalling in Arabidopsis plants, we investigated the expression of the SA-responsive PR1 gene, the SA biosynthesis gene ICS1, and the JA/ET-responsive plant defensin 1.2a (PDF1.2a) gene by RT-qPCR (Fig. S2a, S2b, S2c). Plants were treated with NMN at a concentration of 0.3 mM, which was sprayed onto the surface of rosette leaves. The leaves were then harvested at different time points (0, 6, 24, and 48 h) after spraying. The mRNA levels of the PR1 gene increased at 6 h after NMN treatment and then decreased (Fig. S2a). Correspondingly, ICS1 gene expression was also transiently increased by NMN treatment (Fig. S2b). On the other hand, NMN treatment did not induce expression of the PDF1.2a gene (Fig. S2c). These results suggest that NMN transiently activates the SA-dependent signalling pathway in Arabidopsis leaves. It has previously been reported that SA-dependent signalling positively regulates plant disease resistance against F. graminearum 6, 7.
NMN activates disease resistance against F. graminearum in Arabidopsis leaves and flowers
We examined the effect of NMN pretreatment on disease resistance against F. graminearum in Arabidopsis plants. Six hours after spraying NMN at various concentrations onto leaf surfaces, conidia solutions of F. graminearum were infiltrated into the leaves using a needleless syringe (Fig. S3). When water-pretreated Arabidopsis leaves were inoculated with F. graminearum conidia, severe disease symptoms and extended hyphae were observed (Fig. 1a,c). Pretreatment with 0.3 mM NMN alleviated the disease symptoms of leaves inoculated with F. graminearum (Fig. 1b,c, Fig. S3). However, similar effects were not observed in 3 mM NMN-pretreated Arabidopsis leaves (Fig. S3). We also quantified the F. graminearum genomic DNA in inoculated leaves by qPCR (Fig. 1d). The amount of fungal genomic DNA in NMN-pretreated leaves was significantly decreased compared with that in water-pretreated leaves (Fig. 1d). To examine the effects of NMN on Fusarium resistance in Arabidopsis flowers, an NMN solution was sprayed onto flowers, and 6 h after the NMN treatment, the flowers were inoculated with F. graminearum conidia via spraying. The fungal genomic DNA derived from F. graminearum was also decreased in NMN-treated flowers (Fig. 1e). The inoculated flower samples used for gDNA isolation contained uninfected tissues such as inflorescence stems. Therefore, the difference between water- and NMN-pretreated flowers was relatively small compared with that of leaves. Thus, pretreatment with NMN enhanced disease resistance against F. graminearum in Arabidopsis leaves and flowers.
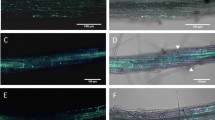
NMN induced disease resistance against F. graminearum in Arabidopsis. (a,b) Water or NMN (0.3 mM) was sprayed onto the surface of Arabidopsis rosette leaves and flower buds prior to incubation for 6 h. Conidia solutions (1 × 105 conidia/ml) of F. graminearum were then injected into leaves and sprayed onto flowers. Plants inoculated with F. graminearum were kept under high humidity conditions. Representative photographs of F. graminearum-inoculated leaves with water (a) or NMN pretreatment (b). Scale bars (a,b): 1 cm. (c) Disease severity of F. graminearum-inoculated leaves with and without NMN pretreatment. Disease severity was evaluated by observations of symptoms on inoculated leaves 3 days after inoculation (n = 18). Open box: normal, cross-hatched box: colour change, dot box: partial aerial mycelium, closed box: expanded aerial mycelium. (d) F. graminearum gDNA was measured by qPCR in inoculated leaves. Error bars represent the standard deviation (n = 3) (Student’s t-test *P < 0.05). (e) F. graminearum gDNA was measured by qPCR in inoculated flower buds. Error bars represent the standard deviation (n = 3) (Student’s t-test *P < 0.05).
Similarly, it has been reported that exogenous application of NAD and NADP enhanced disease resistance against a bacterial pathogen, Psm ES43269. Pétriacq et al.25 reported that higher NAD contents in nadC-overexpressing plants with the addition of quinolinate activated the plant immune response and resulted in disease resistance against a virulent bacterial pathogen, Pst-AvrRpm1. The nadC gene derived from E. coli catalyses the conversion of quinolinate to NaMN25. Therefore, the accumulation of NAD and related metabolites induces disease resistance against bacterial phytopathogens. However, a higher concentration (3 mM) of NMN did not induce disease resistance to F. graminearum (Fig. S3). The optimal concentration of NAD-related metabolites may differ depending on the type of phytopathogen. In addition, different signalling events may occur upon treatment with a lower concentration (0.3 mM) of NMN.
As stated above, NMN application transiently induced expression of the PR1 gene in Arabidopsis leaves. Figure 2a shows that expression of the PR1 gene was up-regulated 48 h after inoculation in NMN-pretreated leaves compared with water-pretreated leaves. By contrast, induction of the PDF1.2a gene by F. graminearum inoculation was significantly suppressed in NMN-pretreated leaves (Fig. 2b). In addition, the SA biosynthesis gene ICS1 was also up-regulated by NMN pretreatment in F. graminearum-inoculated leaves (Fig. S4). These results suggest that pretreatment with NMN enhanced the SA-dependent signalling pathway but completely suppressed the JA/ET signalling pathway in leaves inoculated with F. graminearum. Correspondingly, it has previously been reported that resistance to F. graminearum was positively and negatively regulated by SA and JA/ET signalling, respectively6, 26. The accumulation of SA in F. graminearum-inoculated leaves increased with NMN pretreatment (Fig. 2c). It is likely that this enhanced accumulation of SA mediated by NMN pretreatment led to enhanced resistance to F. graminearum. We next analysed NMN-induced resistance against F. graminearum in the SA-deficient salicylic acid induction-deficient 2 (sid2) mutant. Although the sid2 mutant exhibited a more susceptible phenotype than the wild type (WT), NMN-induced resistance to F. graminearum was also observed in the sid2 mutant (Fig. 2d,e). These results suggest that the enhanced disease resistance caused by NMN is also regulated by an SA-independent signalling pathway. As stated above, NMN-derived antifungal metabolites may be involved in disease resistance to F. graminearum,
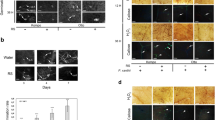
Involvement of the SA signalling pathway in F. graminearum-infiltrated Arabidopsis leaves with and without NMN pretreatment. (a,b) RT-qPCR analysis of PR1 and PDF1.2a mRNA expression in F. graminearum-inoculated WT plants with and without NMN pretreatment. Six hours after NMN pretreatment (time point 0), leaves were injected with F. graminearum (n = 18). Plants were incubated under high humidity conditions and harvested at 0, 6, 24, 48 and 72 h. ACTIN2/8 (Act2/8) was used as the reference gene. The data represent the average of all samples, and error bars represent the standard deviation (n = 3). ACTIN2/8 (Act2/8) was used as the reference gene. Each value is shown as fold change (each sample vs 0 h of water treatment). Error bars represent the standard deviation (n = 3). (c) Accumulation of salicylic acid (SA). (Student’s t-test *P < 0.05) (d,e) NMN induces disease resistance against F. graminearum via the salicylic acid (SA)-independent signalling pathway. NMN (0.3 mM) was sprayed onto the surface of sid2-2 rosette leaves prior to incubation for 6 h. Six hours after NMN treatment, conidia solutions (1 × 105 conidia/ml) of F. graminearum were injected into leaves. (d) The disease severity was evaluated by the disease symptoms of inoculated leaves 3 days after inoculation (n = 18). Open box: normal, cross-hatched box: colour change, dot box: partial aerial mycelium, closed box: expanded aerial mycelium. (e) F. graminearum DNA was measured by qPCR. Error bars represent the standard deviation (n = 3) (Student’s t-test **P < 0.01).
Furthermore, we performed a transcriptome analysis using an Agilent Arabidopsis 3 44k Microarray (Palo Alto, CA, USA) to profile NMN-induced disease resistance. We identified 196 up-regulated and 92 down-regulated genes (Fold Change > 2, P < 0.05) in NMN-treated leaves compared to water-treated leaves without inoculation. In addition, we also identified 688 up-regulated and 786 down-regulated genes (Fold Change > 2, P < 0.05) in NMN-pretreated leaves inoculated with F. graminearum compared to water-pretreated leaves inoculated with F. graminearum. We then performed a gene ontology (GO) enrichment analysis. Table S1 shows the top 30 GO terms enriched in genes induced by NMN without inoculation. These GO terms include response of SA stimulus, SAR, immune response, and negative regulation of cell death. These genes were up-regulated prior to inoculation with F. graminearum. These data support the hypothesis that SA signalling pathway activation and cell death repression are involved in NMN-induced disease resistance against F. graminearum (Fig. 3). Table S2 shows the top 30 GO terms enriched in genes induced in plants that were pretreated with NMN and collected 3 days post inoculation (dpi) with F. graminearum. Table S2 suggests that SA-dependent signalling, the immune response, and systemic acquired resistance were also activated by NMN pretreatment in leaves inoculated with F. graminearum. The number of genes in each GO term was apparently increased compared with those in Table S1. Table S3 suggests that both the JA and ET signalling pathways are suppressed by NMN pretreatment in inoculated leaves. These data also indicate that suppression of JA and ET signalling contributed to NMN-induced disease resistance against F. graminearum. Table S3 also suggests that the ABA signalling pathway and abiotic stress response were repressed by NMN pretreatment. Thus, NMN pretreatment may optimize the defence signalling pathways, leading to enhanced resistance to F. graminearum.
Hierarchical clustering of NMN-regulated genes with or without inoculation of F. graminearum in SA (a), JA (b), and ET (c) categories. SA-, JA- and ET-related genes were selected from NMN-induced or NMN-suppressed genes (FC > 2, P < 0.05) with or without inoculation, based on their GO term names. Then, hierarchical clustering analysis was performed using GeneSpring GX ver.12.5. The level of the bar colour indicates the magnitude of higher expression (red colour) or lower expression (blue colour) of each gene after normalization. F.g.; inoculated with Fusarium graminearum. Asterisk: Benjamini and Hochberg FDR was greater than 0.05.
A hierarchical clustering analysis of NMN-regulated genes containing SA-, JA-, and ET-related GO terms was performed (Fig. 3). NMN-induced SA-related genes (Fig. 3a) in F. graminearum-inoculated leaves included cysteine-rich receptor-like protein kinase 45 (CRK45)27, ACCELERATED CELL DEATH 6 (ACD6)28, WRKYs 29, calcium-dependent protein kinase 31 (CPK31)30, and calmodulin-binding protein 25 (CAMBP25)31. The CRK45 and WRKY53 genes have been reported to positively regulate disease resistance against PstDC300027, 32. In addition, ACD6 is required for SA-dependent disease resistance against virulent bacterial pathogens28. These results suggest that these genes are also involved in disease resistance to F. graminearum. Since calcium signalling is also known to be involved in the defence signalling activated by extracellular pyridine nucleotides13, the CPK31 and CAMBP25 genes may function in NMN-induced disease resistance30, 31. Interestingly, the expression of azelaic acid induced 1 (AZI1), which is involved in the systemic immunity triggered by pathogen infection and azelaic acid26, 33, was up-regulated by NMN treatment with or without inoculation. In addition to the ICS1 gene, expression of the UGT74F1 gene, which catalyses the formation of SA-glucoside (SAG) and the glucose ester of SA (SGE), was significantly reduced by NMN treatment in inoculated leaves34. The down-regulation of the UGT74F1 gene likely contributed to the accumulation of free SA in NMN-treated leaves inoculated with F. graminearum. The DOX1 gene, which is involved in protection against oxidative stress, is reportedly up-regulated by high NAD content25. Similarly, the expression of the DOX1 gene was up-regulated by NMN treatment in uninoculated leaves. However, the induction of DOX1 expression in response to F. graminearum inoculation was down-regulated by pretreatment with NMN. We examined hydrogen peroxide accumulation in water- or NMN-pretreated leaves inoculated with F. graminearum by DAB staining (Fig. S5). These results indicated that NMN pretreatment suppressed the ROS (reactive oxygen species) generation induced by F. graminearum inoculation in Arabidopsis leaves. The suppression of ROS accumulation by NMN was likely involved in the down-regulation of the DOX1 gene by NMN in infected leaves. Therefore, ROS accumulation may negatively affect disease resistance against F. graminearum in Arabidopsis leaves, since host cell death caused by ROS likely contributes to the virulence of F. graminearum necrotrophic growth. Figure 3b shows that JA-inducible (JAZs and VSP1) and JA-biosynthetic genes (DGL and AOC2) were down-regulated by NMN treatment in inoculated leaves35, 36. Although the expression of the WRKY50 and WRKY51 genes was induced by NMN pretreatment in inoculated leaves, these genes control the repression of JA-dependent signalling in low oleic acid (18:1)-containing Arabidopsis plants37. These data indicated that the JA-dependent signalling pathway was suppressed by NMN pretreatment in inoculated leaves, resulting in enhanced disease resistance against F. graminearum 6.
For the ET-related genes, the expression of 7 ERFs (ethylene-responsive transcription factors) was down-regulated by NMN pretreatment in inoculated leaves (Fig. 3c). Five of these genes belong to the ERF subfamily38, 39. In particular, the expression of ERF-1 and ERF96 was induced by ET treatment and activated the transcription of the PDF1.2a gene through a GCC box40, 41. Therefore, the down-regulation of these genes is likely involved in the suppression of the ET response in leaves inoculated with F. graminearum. Figure 3c also shows 7 ERFs (lower parts of the cluster) that are induced by NMN after F. graminearum inoculation. These genes belong to the DREB subfamily38, 39. Among them, RAP2.1 and DEAR2 have EAR repression domains and act as transcriptional repressors42, 43. Thus, suppression of ET-dependent signalling by NMN pretreatment in inoculated leaves likely contributes to enhanced disease resistance against F. graminearum 26.
We next quantified the amount of NMN and NA in NMN-treated Arabidopsis leaves with or without F. graminearum inoculation (Fig. S6). NMN and NA were apparently increased 6 h after NMN spraying (Fig. S6b). In the inoculated leaves, NMN and NA accumulation was also enhanced by NMN pretreatment (Fig. S6c).
Furthermore, we quantified both NAD(H) and NADP(H) levels in leaves with and without NMN treatment (Fig. S7). Extracellular NMN induced NAD and NADP accumulation after 6 h (0 h of inoculation). By contrast, the contents of the reduced forms NADH and NADPH were not significantly altered by NMN treatment. The contents of NAD and NADP were also increased by the inoculation of F. graminearum in Arabidopsis leaves (Fig. S7). Therefore, changes in the NAD+/NADH balance may affect defence signalling. Further increases in the NAD and NADP content were observed in inoculated leaves pretreated with NMN (Fig. S7). These results suggested that extracellular NMN induced disease resistance against F. graminearum without inducing host cell death.
We then investigated the effects of NMN pretreatment and F. graminearum inoculation on the expression of the representative NAD biosynthetic genes nicotinamide mononucleotide adenyltransferase (NMNAT)44 and nicotinamidase 2 (NIC2) (Fig. S8). The expression of both genes was induced by F. graminearum inoculation but not by NMN treatment. Since NMNAT is a rate-limiting enzyme for NAD biosynthesis, the induction of NMNAT likely contributes to the accumulation of NAD. In addition, the induction of the NIC2 gene may be involved in the biosynthesis of pyrimidine alkaloids such as trigonellin13 (Fig. S6a, S8b).
As stated above, NMN is a precursor of NAD, a coenzyme and essential redox-active constituent of all living organisms21. We examined whether other pyridine nucleotides in the NAD biosynthesis pathway also affect disease resistance against F. graminearum. Pretreating plants with NAD and NADP by spraying significantly decreased disease symptoms in Arabidopsis leaves compared with water treatment (Fig. 4a,b). Quantification of gDNA derived from F. graminearum also indicated enhanced disease resistance with NAD and NADP treatment (Fig. 4b). These effects were comparable to those of NMN. On the other hand, NIC had a relatively weak effect on disease resistance compared with the other metabolites. Taken together, these findings suggest that NMN, NAD and NADP are effective suppressors of F. graminearum-induced disease development in Arabidopsis.
The effects of pretreatment with pyridine nucleotides and overexpression of the NMNAT gene on disease resistance against F. graminearum in Arabidopsis. Water (mock), NMN, NAD, NADP and NIC (0.3 mM) were sprayed onto the surface of Arabidopsis rosette leaves prior to incubation for 6 h. A conidia solution (1 × 105 conidia/ml) of F. graminearum was then injected into leaves. (a) The disease severity was evaluated by the visible symptoms of inoculated leaves 3 days after inoculation (n = 18). Open box: normal, cross-hatched box: colour change, dot box: partial aerial mycelium, closed box: expanded aerial mycelium. (b) Pretreatment with pyridine nucleotides was carried out as described above. F. graminearum gDNA was measured by qPCR. Error bars represent the standard deviation (n = 3) (Student’s t-test *P < 0.05 **P < 0.01). (c) Transgenic plants (35 S::AtNMNAT) show enhanced disease resistance. Conidia solutions (1 × 105 conidia/ml) of F. graminearum were inoculated into transgenic plant (35 S::AtNMNAT) leaves and flower buds. Photographs of representative leaves from WT and transgenic plants (35 S::AtNMNAT) at 3 days after inoculation. Scale bars: 1 cm. (d) The disease severity was evaluated by the disease symptoms of inoculated leaves 3 days after inoculation (n = 18). Open box: normal, cross-hatched box: colour change, dot box: partial aerial mycelium, closed box: expanded aerial mycelium. (e) F. graminearum gDNA was measured by qPCR in inoculated leaves. Error bars represent the standard deviation (n = 3) (Student’s t-test **P < 0.01). (f) F. graminearum gDNA was measured by qPCR in inoculated flower buds. Error bars represent the standard deviation (n = 3) (Student’s t-test *P < 0.05).
Furthermore, we prepared Arabidopsis transgenic plants to examine the involvement of intracellular NAD biosynthesis in disease resistance against F. graminearum. Based on the effects of NAD-related metabolites on Fusarium resistance, we prepared transgenic plants constitutively expressing NMNAT, which catalyses the conversion of NMN to NAD (salvage pathway) and NaMN to NaAD (common pathway; Fig. S6a 44). NMNAT is a rate-limiting enzyme involved in NAD biosynthesis in plants41. In addition, expression of the NMNAT gene was induced by F. graminearum inoculation, suggesting that NMNAT plays a role in defence signalling in response to F. graminearum (Fig. S8a). NAD contents were increased in both inoculated and uninoculated leaves of two independent 35S::AtNMNAT lines compared with those of WT plants, whereas the NADP content was unchanged (Fig. S7). By contrast, NMN pretreatment increased the metabolic pool of the NAD biosynthetic pathway (Fig. S6, S7). These transgenic plants showed weak disease symptoms following inoculation with F. graminearum compared to the WT plants (Fig. 4c–g). In addition, the amount of F. graminearum genomic DNA in these transgenic plants was also significantly decreased compared with the WT plant (Fig. 4e). Quantification of gDNA derived from F. graminearum also indicated enhanced disease resistance in flowers of the transgenic plants (Fig. 4f). We found that the level of PR1 mRNA increased in transgenic plants without inoculation compared with WT plants (Fig. S9a). However, enhanced accumulation of PR1 mRNA was not observed in the inoculated leaves of 35S::AtNMNAT plants. The constitutive activation of the SA-dependent signalling pathway is likely involved in the enhanced disease resistance to F. graminearum in 35S::AtNMNAT plants. PDF1.2a expression was suppressed in F. graminearum-inoculated transgenic plants (Fig. S9b). Thus, the higher NAD content in the 35S::AtNMNAT plants resulted in enhanced disease resistance against F. graminearum through the activation of SA signalling and the suppression of JA/ET signalling.
NMN pretreatment also activates disease resistance against F. graminearum in barley plants
Since NMN and related metabolites activated disease resistance against F. graminearum in Arabidopsis leaves and flowers, we also investigated whether it could do so in the FHB-susceptible barley cultivar H.E.S.4, which shows very weak resistance to F. graminearum. Disease symptoms such as aerial hyphae were observed in the water-pretreated flowers (spikes) of H.E.S.4, whereas NMN pretreatment significantly alleviated the disease symptoms (Fig. 5a). In addition, genomic DNA of F. graminearum in NMN-pretreated spikes was significantly decreased compared with plants that received the mock pretreatment (Fig. 5b). DON accumulation in NMN-pretreated spikes was also decreased compared with water-pretreated spikes (Fig. 5c). These results indicate that NMN is also effective in controlling F. graminearum-induced disease injury and mycotoxin accumulation in barley.
NMN pretreatment induces disease resistance against F. graminearum in barley. (a) NMN was sprayed on spikes of the FHB-susceptible line H.E.S.4, followed by incubation for 4 h. A conidia solution (1 × 104 conidia/ml) of F. graminearum was then applied by spraying (n = 3). (a) Scale bars: 1 cm. (b) F. graminearum gDNA was measured by qPCR. Error bars represent the standard deviation (n = 3) (Student’s t-test *P < 0.05). (c) DON was measured by QuickScan DON3 (n = 3) (Student’s t-test *P < 0.05).
In this study, we found that NMN, which is a precursor of NAD, is enriched in FHB-resistant barley cultivars compared with susceptible ones. Although NMN does not have antifungal activity, exogenous application of NMN enhanced disease resistance to F. graminearum in Arabidopsis leaves and flowers. This NMN-induced Fusarium resistance is controlled by the activation of the SA-mediated signalling pathway and repression of the JA/ET-dependent signalling pathway in Arabidopsis. NMN pretreatment induced the expression of the ICS1 gene and resulted in increased SA content in Arabidopsis leaves inoculated with F. graminearum. This enhanced SA accumulation likely suppressed JA biosynthesis and JA/ET responses in Arabidopsis leaves. A higher NAD content in plants is sufficient to enhance disease resistance against F. graminearum through the activation of SA signalling and suppression of JA/ET signalling. However, enhanced resistance was also observed in the SA-deficient sid2 mutant (Fig. 2e). In addition, the increase in SA accumulation mediated by NMN in F. graminearum-infected leaves was less than 2-fold (Fig. 2c). Therefore, an SA-independent signalling pathway is also involved in NMN-induced disease resistance. Thus, elucidating both SA-dependent and SA-independent signalling pathways is necessary to understand the effects of NMN and related metabolites on disease resistance against F. graminearum. How do NAD and related metabolites activate the defence signalling pathways leading to enhanced resistance against F. graminearum? As stated above, an imbalance in the NAD+/NADH and NADP/NADPH ratios affects not only various NAD- and NADP-dependent enzymatic reactions but also defence signalling pathways13. NAD-dependent protein deacetylases (SIRTs) are activated by an increased NAD+/NADH ratio14. Furthermore, target proteins such as histones are deacetylated and then poly(ADP-ribosyl)ated by SIRT and PARP (poly(ADP-ribose)polymerase), resulting in the modulation of the expression of many genes14. Since SIRTs and PARPs are also found in plants, similar events may trigger transcriptional activation of defence-related genes such as ICS1. Finally, we revealed that NMN pretreatment induced disease resistance against F. graminearum in an FHB-susceptible barley cultivar (H.E.S.4). Thus, modifying the content of NAD-related metabolites is useful for the control of disease injury and mycotoxin accumulation in plants.
Methods
Plant materials and growth conditions
Arabidopsis thaliana (ecotype Columbia; Col-0) and its derivatives were used in this study. The salicylic acid induction-deficient 2-2 (sid2-2) mutant45 was obtained from the Arabidopsis Biological Resource Center (ABRC). 35S::AtNMNAT (At5g55810) transgenic plants were prepared in this study. Arabidopsis seeds were sown in soil at 4 °C in the dark for 2 days and thereafter grown at 22 °C under a 16/8-h light/dark cycle. The FHB-susceptible cultivars Turkey 45 (2-row) and H.E.S.4 (6-row) and FHB-resistant cultivars Maja and Sirius O-525 were grown in the field of the Institute of Plant Science and Resources, Okayama University, as previously described19.
Fungal material and growth conditions
F. graminearum strain H3 was used in this study46. The fungus was grown at 28 °C on potato dextrose agar (PDA; Nippon Becton Dickinson Company, Ltd., Tokyo, Japan) medium. F. graminearum conidia were prepared using SN liquid medium, which is composed of 0.1% KH2 PO4, 0.1% KNO3, 0.1% MgSO4·7H2O, 0.05% KCl, 0.02% glucose, and 0.02% sucrose47.
Fusarium head blight (FHB) assay (cut-spike test)
The cut-spike test using field-grown barley spikes was performed as previously described19. Spikes were sprayed with an NMN solution with 0.001% silwetL77. After 4 h, an F. graminearum conidia solution (1 × 104 conidia/ml) with 0.001% silwetL77 was sprayed on the spikes. Inoculated spikes were collected at 7 dpi for disease incidence determination (colour change of barley grains), F. graminearum gDNA analysis and quantification of DON.
Pyridine nucleotide treatment
Pyridine nucleotides (NMN, NAD, NADP and NIC; Sigma-Aldrich Japan, Tokyo, Japan) were dissolved in water at a concentration of 300 µM, and the pH was adjusted to 6.0 with 1 N NaOH. The resulting solution plus 0.001% silwetL-77 was sprayed onto Arabidopsis leaves, flowers and barley spikes (H.E.S.4). To analyse disease resistance, sprayed leaves and flowers were inoculated with F. graminearum 6 h after treatment. Sprayed barley spikes were inoculated with F. graminearum 4 h after treatment.
Antifungal activity assay
Conidial suspensions of F. graminearum (1 × 103 conidia/ml) were grown in SN liquid medium for 2 days with or without NMN. The growth of Fusarium was measured by a 3-(4,5-di-methylthiazol-2-yl)-2,5-diphenyltetrazollium bromide (MTT) assay2.
F. graminearum infection
To prepare conidia solutions, F. graminearum was cultured in SN liquid medium at 22 °C for 3 days. The conidia were then collected by centrifugation and washed with phosphate-buffered saline (PBS) at least 3 times. The collected conidia were suspended in PBS, and the number of conidia was counted using a haemocytometer2. Arabidopsis leaves were sprayed with NMN, NAD, NADP, or NIC plus 0.001% silwetL77. After 6 h, conidia solutions were injected into Arabidopsis leaves. For infiltration inoculation, a conidial suspension of F. graminearum (1 × 105 conidia/ml) was injected into the abaxial side of the leaves using a needleless syringe2. Inoculated plants were incubated in a chamber under approximately 100% relative humidity at 22 °C for 3 days. Disease development on leaves was assessed as described previously2. Inoculated leaves were collected at 3 dpi to for further analysis.
Flowering plants were used for the inoculation assay. Flowers were sprayed with an NMN solution with 0.001% silwetL77. After 6 h, F. graminearum conidia solutions (1 × 105 conidia/ml) with 0.001% silwetL77 were sprayed on Arabidopsis flowers as previously described47. The inoculated plants were then kept in a dew chamber at 100% relative humidity for 7 days. Inoculated flowers were collected at 7 dpi for further analysis.
DAB staining
To detect H2O2, inoculated leaves were infiltrated under a gentle vacuum with 1 mg ml−1 DAB containing 0.05% v/v Tween 20 and 10 mM PBS at pH 7.0. The leaves were then fixed by boiling for 15 min in ethanol: acetic acid: glycerol at 3:1:1 and were then observed under a microscope48. We used an OLYMPUS BX-50 microscope (Olympus Optical Co., Ltd., Tokyo, Japan).
DNA isolation and quantification of F. graminearum gDNA
Genomic DNA was isolated using a Nucleon Phytopure Kit (GE Healthcare., Tokyo, Japan). The amount of F. graminearum gDNA in inoculated leaves and flowers was quantified by qPCR. For inoculated flower samples, primary inflorescences of approximately 5 cm were cut from approximately 6-week-old Arabidopsis plants. qPCR was performed an Mx3000P instrument (Stratagene Japan Inc., Tokyo, Japan) with 2X SYBR Premix Ex Taq II (Takara Bio Inc., Shiga, Japan). The Arabidopsis Act2/8 gene, barley GAPDH gene and Fusarium EF1α gene were used for analysis. ACT2/8 was amplified using the primers Act2/8_F-1, GGTAACATTGTGCTCAGTGGTGG and Act2/8_R-1, AACGACCTTAATCTTCATGCTGA, and the EF1α gene was amplified using the primers EF1α_F-1, CCATTCCCTGGGCGCT and EF1α_R-1, CCTATTGACAGGTGGTTAGTGACTGG. HvGAPDH was amplified using the primers HvGAPDH_F-1, TTGACAGGAACCCTGAGGAG and HvGAPDH_R-1, TGTGAATGAGCGAAAACCAG. qPCR analysis was performed using an Mx3000P instrument (Stratagene Japan Inc., Tokyo, Japan). To generate a standard curve, homologous standards were used in all experiments. The initial gDNA quantities were calculated by the standard curve method using MxPro software (Agilent).
Measurements of NAD/NADH and NADP/NADPH
Samples were ground to fine powder in liquid nitrogen. Then, 40 mg of each sample was used for NAD/NADH and NADP/NADPH quantification. NAD/NADH and NADP/NADPH accumulation was measured using a Fluorescent NAD/NADH and NADP/NADPH Detection Kit (Cell Technology Inc., Mountain View, CA) according to the manufacturer’s protocol.
RT-qPCR
Total RNA was isolated using a Plant RNA Isolation Mini Kit (Agilent Technologies., CA, USA). First-strand cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara Bio Inc., Shiga, Japan). RT-qPCR (intercalator method) was performed as described above. The following primers were used for RT-qPCR: PR1 49, PDF1.2a 46. ICS1_F-1, ATGAGATTCAGCCTCGCTGT and ICS1_R-1, TGATGGATCTCCAATCGTCA. NMNAT_F-1, CCAACGAACTTTGACGGTTT and NMNAT_R-1, CGGGAGTGCAGAAAGATAGC. NIC2_F-1, CGAGAACGTACGAGACACGA and NIC2_R-1, TTCCGTCGAGGATTAGATCG. The primers used to amplify the Act2/8 gene are listed above. To generate a standard curve, homologous standards were used in all experiments. The cDNA quantities of target genes were calculated by the standard curve method using MxPro software (Agilent), and all quantifications in Arabidopsis were normalized to the mRNA level of the Actin2/8 reference gene.
Generation of transgenic plants
To create 35S::AtNMNAT Arabidopsis transgenic plants, Arabidopsis NMNAT (At5g55810) cDNA was amplified using the primers AtNMNAT_F-1, CACCATGGATGTCCCGTTACCAGT and AtNMNAT_R-1, TCATGTGAGCTCAGTGTATAGTTGAT, and recombined into the GatewayTM vector pENTRTM/D-TOPO (Invitrogen., Carlsbad., CA, USA). The gene was then sequenced and transferred into the transformation vector pK2GW7.050. This plasmid was then transformed into Agrobacterium tumefaciens strain GV2260. Transgenic Arabidopsis plants were generated according to the Agrobacterium-mediated floral dip method51. Transgenic plants were selected on Murashige and Skoog (MS) medium supplemented with 2% sucrose and 0.8% Bacto agar at pH 5.8 and with 50 µg ml−1 kanamycin.
Quantification of DON
Barley spikes were ground to fine powder in liquid nitrogen. Then, 200 mg of each sample was used for DON quantification. DON accumulation was measured using a QuickToxTM Kit for QuickScan DON3 (ENVIRLogix, Inc., Portland, Maine, USA) according to the manufacturer’s protocol.
Microarray analysis
Total RNA was prepared from 8 samples (2 biological replicates for 4 different treatments) using a Plant RNA Isolation Mini Kit (Agilent Technologies., CA, USA). Four different treatments were analysed: (1) water-treated Arabidopsis leaves, (2) NMN-treated leaves, (3) water-treated, F. graminearum-inoculated leaves, and (4) NMN-treated, F. graminearum-inoculated leaves. For (1) and (2), these leaves ware collected at 6 h after the spraying of water or NMN. For (3) and (4), after water or NMN pretreatment, F. graminearum-inoculated leaves were collected at 3 dpi. RNA quality was assessed using the 2200 TapeStation (Agilent Technologies), and a one-colour microarray experiment was performed using the Agilent Arabidopsis ver. 3 oligomicroarray, according to the manufacturer’s 60-mer oligo microarray processing protocol. Total RNAs (200 ng) were used to prepare Cy3-labeled cRNA, using a Low Input Quick Amp Labeling Kit (Agilent Technologies). The hybridized and washed arrays were scanned at maximum laser intensity in the Cy3 channel using an Agilent microarray scanner (G2565BA; Agilent Technologies). The images were analysed using Feature Extraction Software (Ver. 10.7.3.1; Agilent Technologies). These data were analysed further with GeneSpring GX12.5 software (Agilent Technologies). Normalization was performed as follows: 1, intensity-dependent Lowess normalization; 2, data transformation, with measurements less than 0.01 set to 0.01; 3, per-chip normalization, in which the 75th percentile method was used to normalize each array; and 4, per-gene normalization, in which the data were normalized to the median value of 8 measurements. After normalization, statistically significant gene sets were defined as those exhibiting P values below the cut-off of 0.05. Genes with a fold change (FC) in the expression ratio (water-treated vs NMN-treated) >2, either up-regulated or down-regulated, were identified. Thus, a combination of statistical analyses and FC methods was used. In addition, we calculated false discovery rate (FDR) by Benjamini-Hochberg procedure. GO enrichment analyses were also performed using GeneSpring GX12.5. Using Fisher’s exact test (corrected P < 0.01), we identified the top 30 GO categories significantly enriched in genes that were up-regulated by NMN treatment. For the hierarchical clustering, SA-, JA- and ET-related genes were selected from the NMN-induced or NMN-suppressed genes (FC > 2, P < 0.05) with or without inoculation, based on their GO term names. For the clustering analysis, we eliminated genes that over-lapped among these categories. Then, hierarchical clustering analysis was performed using GeneSpring GX ver.12.5. The microarray data from this publication have been submitted to the GEO database (http://www.ncbi.nlm.nih.gov/geo/) and assigned the identifier GSE93079.
Metabolome analysis
The freeze-dried samples were measured and added to extraction buffer (0.1% (v/v) formic acid in 80% (v/v) MeOH) containing internal standards (positive: 33.6 nM lidocaine, negative: 210 nM 10-camphorsulfonic acid). The extracted solutions of all samples were adjusted to the same concentration (4 mg dry weight/mL). Then, they were diluted 10 times with extraction buffer, and 25 µL of each sample was dried with N2 gas. The dried samples were dissolved in 250 µL of LC-MS grade H2O. After filtration (UNIFILTER 384 well, Whatman), 1 µL of the solution was measured by LC-QqQ-MS52,53,54. Then, the peak area of each metabolite was normalized to the peak area of the internal standard. Using the resulting MS intensity of each metabolite per dried weight, we compared each treatment. The differentially accumulated metabolites were identified by statistical analysis.
References
Ahuja, I., Kissen, R. & Bones, A. M. Phytoalexins in defense against pathogens. Plant Sci. 17, 73–90 (2012).
Asano, T. et al. The secreted Antifungal Protein Thionin 2.4 in Arabidopsis thaliana Suppresses the Toxicity of a Fungal Fruit Body Lectin from Fusarium graminearum. PLoS Pathog. 9, e1003581 (2013).
Vernooij, B. et al. Salicylic Acid Is Not the Translocated Signal Responsible for Inducing Systemic Acquired Resistance but Is Required in Signal Transduction. Plant Cell. 6, 959–965 (1994).
Friedrich, L. et al. A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 10, 61–70 (1996).
Ravichandran, S., Stone, S., Benkel, B. & Prithiviraj, B. Purple Acid Phosphatase5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidosis. BMC Plant Biol. 13, 107 (2013).
Makandar, R. et al. Involvement of Salicylate and Jasmonate signaling Pathways in Arabidopsis Interaction with Fusarium graminearum. Mol. Plant Microbe Interact 7, 861–870 (2010).
Makandar, R. et al. Salicylic Acid Regulates Basal Resistance to Fusarium Head Blight in Wheat. Mol. Plant Microbe Interact 25, 431–439 (2012).
Aranega-Bou, P. et al. Priming of Plant resistance by natural compounds. Hexanoic acid as a model. Front. Plant Sci 5, 488 (2014).
Zhang, X. & Mou, Z. Extracellular pyridine nucleotides induce PR gene expression and disease resistance in Arabidopsis. Plant J 57, 302–312 (2009).
Berger, F., Ramirez-Hernandez, M. H. & Ziegler, M. The new life of a centenarian: signalling functions of NAD(P). Trends Biochem. Sci 29, 111–118 (2004).
Hunt, L. & Gray, J. E. The relationship between pyridine nucleotides and seed dormancy. New Phytologist 181, 62–70 (2008).
Berrin, J. G. et al. Stress induces the expression of AtNADK-1, a gene encoding a NAD(H) Kinase in Arabidopsis thaliana. Mol. Genet. Genomics 273, 10–19 (2005).
Pétriacq, P., Bont, L., Tcherkez, G. & Gakière, B. Not just a pawn on the board of Plant-Pathogen interaction. Plant Signal. Behav 8, e22477 (2013).
Cantó, C., Sauve, A. A. & Bai, P. Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes. Mol Aspects Med 34, 1168–1201 (2013).
Kazan, K., Gardiner, D. M. & Manners, J. M. On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol 13, 399–413 (2012).
Nishiuchi, T. Plant Response to Fusarium Metabolites. In Fusarium: genomics, molecular and cellular biology. (Brown, D.W. and Proctor, R.H. eds). Horizon Scientific Press, UK, pp.165–178 (2013).
McMullen, M. et al. A unified effort to fight an enemy of wheat and barley: fusarium head blight. Plant Dis 96, 1712–1728 (2012).
Yin, Y., Liu, X., Li, B. & Ma, Z. Characterization of Sterol Demethylation Inhibitor-Resistant Isolates of Fusarium asiaticum and F. graminearum Collected from Wheat in China. Phytopathol. 99, 487–497 (2009).
Hori, K., Kobayashi, K., Sato, K. & Takeda, K. QTL analysis of Fusarium head blight resistance using a high-density linkage map in barley. Theor. Appl. Genet 11, 1661–1672 (2005).
Hori, K., Sato, K., Kobayashi, T. & Takeda, K. QTL Analysis of Fusarium Head Blight Severity in Recombinant Inbred Population Derived from a Cross between Two-rowed Barley Varieties. Breeding Science 56, 25–30 (2006).
Noctor, G., Queval, G. & Gakiere, B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot 57, 1603–1620 (2006).
Ryrie, I. J. & ScottK, J. Metabolic Regulation in Diseased Leaves II. Changes in Nicotinamide Nucleotide Coenzymes in Barley Leaves Infected with Powdery Mildew. Plant Physiol 43, 687–692 (1968).
Ashihara, H. et al. Trigonelline and related nicotinic acid metabolites: occurrence, biosynthesis, taxonomic considerations, and their roles in planta and in human health. Phytochem Rev 14, 765–798 (2015).
Meletiadis, J. et al. Comparison of NCCLS and 3-(4,5-dimethyl-2-Thiazyl)-2, 5-diphenyl-2H-tetrazolium bromide (MTT) methods of in vitro susceptibility testing of filamentous fungi and development of a new simplified method. J. Clin. Microbial 38, 2949–2954 (2000).
Pétriacq, P. et al. Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. Plant J 70, 650–665 (2012).
Chen, X., Steed, A., Travella, S., Keller, B. & Nicholson, P. Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol 182, 975–983 (2009).
Zhang, X. et al. Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae. Plant Physiol. Biochem 73, 383–391 (2013).
Lu, H., Rate, D. N., Song, J. T. & Greenberg, J. T. ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signalling in Arabidopsis defense response. Plant Cell 15, 3408–2420 (2003).
Thomas, E. et al. The WRKY superfamily of plant transcription factor. Trends Plant Sci 5, 199–206 (2000).
Hrabak, E. et al. The Arabidopsis CPK-SnRK Superfamily of Protein kinases. Plant Physiol 132, 666–680 (2003).
Perruc, E. et al. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J 38, 410–420 (2004).
Murray, S. L., Ingle, R. A., Petersen, L. N. & Denby, K. J. Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact 20, 1431–1438 (2007).
Jung, H. W. et al. Priming in systemic plant immunity. Science 324, 89–91 (2009).
Dean, J. V. & Delaney, S. P. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol Plant. 132, 417–425 (2008).
Hyun, Y. et al. Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev. Cell 14, 183–192 (2008).
Leon-Reyes, A. et al. Salicylate- mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232, 1423–1432 (2012).
Gao, Q. H., Venugopal, S., Navarre, D. & Kachroo, A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY 51 protein. Plant Physiol 1, 464–476 (2011).
Nakano, T., Suzuki, K., Fujimaru, T. & Shinshi, H. Genome-wide analysis of ERF gene family in Arabidopsis and rice. Plant Physiol 140, 411–432 (2006).
Sakuma, Y. et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs transcription factors involved in dehydration- and cold –inducible gene. Biochem. Biophys. Res. Commun 290, 998–1009 (2002).
Fujimoto, S. Y. et al. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404 (2000).
Catinot, J. et al. ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate-and ethylene-responsive defensce gene. Plant Cell Environ 38, 2721–2734 (2015).
Dong, C. J. & Liu, J. Y. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol 10, 47 (2010).
Tsutsui, T. et al. DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J Plant Res 122, 633–643 (2009).
Hashida, S. et al. NAD+ Accumulation during Pollen Maturation in Arabidopsis Regulating Onset of Germination. Mol. Plant 6, 216–225 (2012).
Wildermuth, M. C., Dewdney, J., Wu, G. & Ausubel, F. M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 29, 562–565 (2001).
Asano, T., Kimura, M. & Nishiuchi, T. The defense response in Arabidopsis thaliana against Fusarium sporotrichioides. Proteome Sci 10, 61 (2012).
Urban, M., Daniels, S., Mott, E. & Hammond, K. Arabidopsis is susceptible to the cereal ear blight fungal pathogens Fusarium graminearum and Fusarium culmorum. Plant J 32, 961–973 (2002).
Bindschedler, L. V. et al. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47, 851–863 (2006).
Asano, T. et al. AtNFXL1, an Arabidopsis homologue of the human transcription factor NF-X1, functions as a negative regulator of the trichothecene phytotoxin-induced defense response. Plant J 53, 450–64 (2008).
Karimi, M., Inzé, D. & Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7, 193–195 (2002).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743 (1998).
Sawada, Y. et al. Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol 50, 37–47 (2009).
Sawada, Y. et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: a plant-specific MS/MS-based data resource and database. Phytochemistry 82, 38–45 (2012).
Sawada, Y. & Hirai, M. Y. Integrated LC-MS/MS system for plant metabolomics. Comput Struct Biotechnolo J 4, e201301011 (2013).
Acknowledgements
This research was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan TRS1005, Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (28007A), and the Joint Research Program implemented at the Institute of Plant Science and Resources, Okayama University in Japan. Barley seed samples were provided by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was also supported in part by Research for Promoting Technological Seeds (no. 07-070) and by the Adaptable and Seamless Technology Transfer Program through Target-driven R&D (AS232Z02753E and AS242Z03390N) from the Japan Science and Technology Agency (JST). This work was partly supported by KAKENHI (no. 23580060, 26450053 and 15H05780) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We thank Muneo Sato, Yutaka Yamada, and Akane Sakata (RIKEN) for technical assistance in metabolomics. We are deeply grateful to Drs. Shigeru Shigeoka and Masahiro Tamoi (Kinki University) and Dr. Maki Kawai-Yamada for their helpful comments.
Author information
Authors and Affiliations
Contributions
A.M., K.S. and T.N. designed the experiment; A.M., Y.S., K.S. and T.N. conducted the experiment; A.M., Y.S., D.T., M.H., M.K., K.S. and T.N. analysed the data; A.M. and T.N. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miwa, A., Sawada, Y., Tamaoki, D. et al. Nicotinamide mononucleotide and related metabolites induce disease resistance against fungal phytopathogens in Arabidopsis and barley. Sci Rep 7, 6389 (2017). https://doi.org/10.1038/s41598-017-06048-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-06048-8
This article is cited by
-
An agroecological structure model of compost—soil—plant interactions for sustainable organic farming
ISME Communications (2023)
-
Combined transcriptome and metabolome analysis of Nerium indicum L. elaborates the key pathways that are activated in response to witches’ broom disease
BMC Plant Biology (2022)
-
Overexpression of nicotinamidase 3 (NIC3) gene and the exogenous application of nicotinic acid (NA) enhance drought tolerance and increase biomass in Arabidopsis
Plant Molecular Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.