Abstract
A novel series of hybrid analogues of monastrol-1,3,5-triazine were designed and developed via one-pot synthesis using Bi(NO3)3 as a catalyst. Entire compounds were evaluated for their anticancer activity against HeLa (cervical cancer), MCF-7 (breast cancer), HL-60 (Human promyelocytic leukemia), HepG2 (Hepatocellular carcinoma) and MCF 12A (normal epithelial breast cell line) using MTT assay, where they showed highest inhibitory activity against MCF-7. The molecules were also found to be non-toxic to MCF 12A cells. These molecules showed considerable inhibitory percentage against Epidermal Growth Factor Receptor tyrosine kinase (EGFR-TK), in in-vitro assay. Molecular docking study was carried out on the analogs and reference compound (Erlotinib) into the ATP binding site of EGFR-TK domain (PDB ID:1M17) to elucidate vital structural residues necessary for bioactivity. The effect of most active compound 7l was also estimated in-vivo in DMBA induced mammary tumor in female Sprague-Dawley rats. The effect of anti-breast cancer effect of 7l was quantified on the basis of tumour incidence, body weight and tumor volume in DMBA-induced rats. Its effect on biochemical parameters, such as antioxidant status (SOD, CAT, GPX and GSH) and lipid peroxidation was also studied. The compound 7l showed inhibition of EGFR downstream signalling in the western blot analysis.
Similar content being viewed by others
Introduction
The search of novel medicinal agent endowed with therapeutic efficacy is always a great concern for the medicinal chemist. At the same time, the potential development of tolerance or resistance to that compound from the time it is first employed seriously compromised its clinical utility. This holds true and creates major hitch for the agents used in the treatment of chronic diseases such as cancer1.According to an estimate, Cancer is a second leading cause of morbidity and mortality after the cardiovascular diseases. It accounts for 12 million deaths across the globe by 2030, as per WHO2. Particularly in the developing countries, where resources are scarce, the impact of cancer on all populations is truly devastating. It became a serious concern for poor, vulnerable and socially disadvantaged people who get sicker and unable to afford expensive cancer medicines and treatments which die sooner as a result of it3. Modern day’s cancer research has been shifted towards the development of selective inhibitors which can able to target deregulated pathways to stop cancer growth in a classical drug–receptor fashion. This makes compounds less toxic to normal cells, and thus improves tolerability.
The epidermal growth factor receptor (EGFR) is a trans-membrane protein belonging to the erbB/HER-family of tyrosine kinase (TK) receptor, which includes four members defined as ErbB-1/EGFR/HER1, ErbB-2/HER2/neu, ErbB-3/HER3 and ErbB-4/HER4. These receptors share the same molecular structure with an extracellular, cysteine-rich ligand-binding domain, a single alpha-helix transmembrane domain, and an intracellular domain with TK activity in the carboxy-terminal tail (excepting the HER3)4. The receptors of EGFR family regulate the transcription of molecules that play a vital role in normal organ development by mediating morphogenesis and differentiation through effects on cell proliferation, differentiation, apoptosis, invasion, and angiogenesis. Unlike normal cells, the EGFR signalling has been aberrantly altered in tumour cells and often dysregulated. This behaviour leads to the proliferation of tumour cells under adverse conditions where they invade surrounding tissues, and thereby increases angiogenesis5, 6. EGFR intracellular signalling is mainly mediated by two interrelated downstream pathways, viz., the Ras-Raf-mitogen activated protein kinases (MAPK, also known as extra cytoplasmatic regulated kinases, ERK1 and ERK2) and the phosphatidylinositol 3-kinase (PI3K)/Akt pathways7. The overexpression of EGFR has been associated with advanced stages of many types of cancers especially breast, colon and bladder cancers8. Particularly in some breast cancer, subtypes The EGFR-dependent pathway appears to be a driver mechanism for malignant carcinogenesis, Fig. 1 9,10,11. Thus, selective inhibition of this target by EGFR inhibitors offers various advantages.
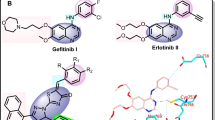
On the basis of hierarchical based virtual screening protocol, Bai et al., has identified number of 1,3,5-triazine derivative as dual-effective inhibitor against both WT and mutant EGFR TKs12. Out of them, compound 1a exhibited most potent activity against WT EGFR (IC50 = 25.9 µM) and mutant EGFRT790M/L858R (IC50 = 6.5 µM), Fig. 2. Moreover, it also exhibit considerable antiproliferative activity against A549, A431 and NCIH1975 cell lines, IC50 = 7.7 ± 1.3, 8.0 ± 1.6 and 10.5 ± 0.9 µM, respectively. It has been found that that, when the fluoro group in the phenyl part was removed along with inclusion of the para-hydroxy group on the another phenyl, the inhibitory potency for WT EGFR has lost and reduced about fivefold for mutant EGFR, compound 1f, Fig. 2. With the help of molecular docking analysis, they have shown that, the selectivity of molecules for WT EGFR and mutant EGFRT790M/L858R would be achieved by exploiting the additional hydrophobic pocket located in the back of the ATP-binding site by inclusion of other groups. This observation was found in agreement with the previous studies carried out by other researches on selective inhibitors against kinases13, 14.
Concerning our endeavour on discovery of novel chemotherapeutic agents from 1,3,5-triazine15,16,17,18,19,20,21,22 and its significance as EGFR TKs inhibitor, present study deals with advancement of novel 1,3,5-triazine derivatives as anticancer agents. The designed targeted molecules were developed via molecular hybridization of 1,3,5-triazine and Monastrol (3,4-dihydro-1H-pyrimidine) in a search of new hit. The later has been selected to get substituted on 1,3,5-triazine because of its anticancer potential and bulky nature, which could be tolerated at the active site of EGFR-TKs due its extended large hydrophobic cavity, Fig. 3. The synthesis of target hybrid compounds were accomplished by the means of one-pot synthesis using Bi(NO3)3 as catalyst.Consequently, the novel target molecules were also tested for anticancer activity.
Results and Discussion
Chemistry
To access heterocyclic compounds in fast and economic way along with high molecular diversity, one-pot synthetic approach has gaining attention. It is a synthetic strategy, whereby reactant/s is subjected to successive chemical reaction in a single reactor. This will improve the chemical reaction without isolating intermediates10, 23,24,25,26. Thus, considering the utility of one-pot reactions, the synthesis of the target compounds has been accomplished by utilizing Bismuth Nitrate as a catalyst.
As depicted in Step 1 (Fig. 4), the synthesis of 1-(4,6-bis(substituted phenylamino)-1,3,5-triazin-2-yl)thiourea derivatives 4 (a–o) were accomplished by reacting di-substituted 1,3,5-traizines 3 (a–o) with thiourea in the presence of the activating base at vigorous condition. While, compounds 3 (a–o) were synthesized via reacting cyanuric chloride (1) with two equivalents of substituted amines 2 (a–o) taking care that reaction mixture does not become acidic, with frequent addition of NaOH.
The next part of the study was aimed at optimising the reaction condition for the Biginelli’s one-pot condensation, Step 2, Fig. 4. In first instance, we tried to optimise the type of catalyst, where, we had taken numerous Lewis acids e.g. AlCl3, FeCl3, and ZnCl2 along with various bismuth salts viz., bismuth chloride, bismuth triflate, bismuth subnitrate, bismuth bromide, bismuth iodide and bismuth nitrate using benzaldehyde, ethyl acetoacetate (6) and 1-(4,6-bis(phenylamino)-1,3,5-triazin-2-yl)thiourea as a model reaction. The results were shown in Table 1.
During the process of optimising the catalyst, we found that 10 mol% of Bi(NO3)3 could effectively catalyse the reaction for the synthesis of desired product with maximum yield (92%) accompanied with shorter reaction time (4 h). The inclusion of other Bismuth salts e.g., BiBr3, Bi5O(OH)9(NO3)3, BiI3, Bi(OTf)3 and BiCl3 as a catalyst were not proved to be efficient enough and the reaction took longer time. It is remarkable to note that, the presence of various Lewis acids could not catalyses the reaction in an efficient way, e.g., FeCl3 take maximum of 9 h with 58% yield. While, in the absence of catalyst, the reaction did not precede smoothly, entry 10. This confirms the utility of the catalyst for the completion of reaction.
To divulge the role of solvent on the reaction and product yield, the next part of the study was aimed to determine the best solvent. Consequently, the reaction has been carried out with different solvents and the results were shown in Table 2. It was inferred that ethanol furnish the product in highest yield with shorter reaction time, entry 5. The time taken to complete the reaction has been augmented two-fold in the case of THF (entry 2) and toluene (entry 3) in comparison to ethanol and further increased in the case of dichloromethane (entry 4). While, the water was proved as ineffective solvent to this type of transformation owing to its high reaction time and less product yield, entry 1.
After optimisation of the catalyst (Bi(NO3)3) and solvent (ethanol), numerous target products containing diverse structural motifs has been synthesized as shown in Fig. 4. The synthetic procedure came out as very straight forward; the target products were isolated and purified by simple filtration and column chromatography.
Anticancer activity
The synthesized compounds were evaluated for in-vitro cytotoxic activity against various cell lines such as HeLa (cervical cancer), MCF-7 (breast cancer), HL-60 (Human promyelocytic leukemia), HepG2 (Hepatocellular carcinoma) and MCF 12A (normal epithelial cell) by the MTT assay method. Cisplatin, one of the most effective anticancer agents was used as a reference drug in this study. The relationship between surviving fraction and drug concentration was plotted to obtain the survival curve of all the cancer cell lines HeLa, MCF-7, HL-60 and HepG2. The response parameter calculated was the IC50 values, which responds to the concentration required for 50% inhibition of cell viability. The in-vitro cytotoxic activity of the synthesized compounds is summarized Table 3.
Results showed that absence of the substituent i.e., intact phenyl rings render the compound inactive against HepG2, while less active against rest of the cell lines, 7a. In the case of compounds 7b and 7c, against entire cell lines, no significant change in activity has been observed. To our surprise, the activity was significantly increased against HepG2 on the introduction of 3-Cl at the phenyl of the triazine along with electron donating group (4-OH), compound 7d. Whereas, on replacing chloro with bromo along with shifting of hydroxyl group to meta, the activity was dropped, 7e. Minor upsurge in activity was reported in the case of compounds 7f and 7g aganist entire cell lines. Presence of non-halogen substituent at the phenyl of triazine along with halogen on the other counterpart significantly lowers the activity against all tested cell lines, compounds 7h and 7i. A momentous increase in activity was reported by the compounds 7j, 7l and 7m having halogen groups except mild activity in the case of compound 7k having NO2 as one the substituent. It is noteworthy to mention that, presence of electron donating substituent significantly diminishes the anticancer potential and makes the compounds inactive against HepG2. All the tested hybrid conjugates exhibited moderate to significant anticancer activity except compound 7a, 7n and 7o for HepG2. Results showed that none of the synthesized compounds exhibited pronounced activity than cisplatin as standard.
It was corroborated that, compound 7l was emerged as most active molecule while compound 7n and 7o as non-active against the entire tested cell lines. Structure-activity relationship studies suggest that presence of halogen electron withdrawing group is necessary for generation and escalation of activity. The presence of non-halogen substituent along with electron donating substituent seriously jeopardise the anticancer potential. Results showed a clear pattern of the effect of substituent on the activity and the same has been discussed in Fig. 5. To determine the selectivity of these molecules between normal and cancerous cells, the designed compounds were also tested against MCF 12A, a normal epithelial breast cell. The results showed that, most of molecules found non-toxic to the normal cell.
EGFR-TKs Inhibitory Activity
The synthesized 1,3,5-triazine-3,4-dihydro-1H-pyrimidine derivatives were evaluated in vitro for EGFR-TK enzyme inhibitory activity (at 10 µM) and the results obtained are reported as percent of inhibition in comparison to Erlotinib as a standard. The results were displayed in Table 4. Against EGFR-TKs, entire set of target compounds showed excellent inhibitory activity, where, compound 7l and 7g revealed as most potent inhibitor with 96.4% and 94.3%, respectively. Further decline in activity was reported by compound 7m (88.5%) and 7f (81.4%). A significant drop in inhibitory potency was reported by compound 7e and 7j which was followed by further reduction against EGFR-TK by 7e and 7d (78.5% and 74.8%, respectively). It was surprising to note that similar pattern of inhibition was disclosed by compound 7h, 7i and 7k near to 69%. The assay revealed that, rest of compound showed inhibitory percentage nearly half to that of Erlotinib, except compound 7o which showed least inhibition, i.e., 45.6%.
Molecular Docking Study
Binding Analysis of inhibitors docked to EGFR-TK domain
Captivated by the exceptional anticancer activity shown by target molecules against MCF-7 (breast cancer) where EGFR enzyme is over-expressed, and potent EGFR-TKs inhibitory activity, we used binding information of our receptor against EGFR-TK inhibitors for validation of our results. Present study was based on genetic algorithm and in-silico approaches. The details of ligand docking scores are given in Table 5.
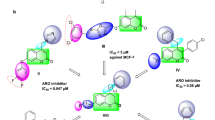
Docking study was performed to understand the interactions of the compounds with the active center of the EGFR-TK receptor by validating the binding mode and interacting amino acids from the active conformation of the co-crystal ligand Erlotinib. We selected PDB ID: 1M17 as our target protein and the protein had co-crystal ligand Erlotinib with literature reference on binding affinities and the protein receptor EGFR-TK was our focused target27. While comparing the docking results of ligands, we observed that erlotinib, 7l, 7m, 7e has the lowest binding energy and predicted inhibition constant values as more negative binding energy score (kcal/mol) corresponds to the binding affinity. On the basis of lower binding free energy, better electrostatic interaction and inhibition constant value of these inhibitors against EGFR-TK, we came to know that 7l and 7m are inhibitors with efficacy for EGFR-TK inhibition. Also the interaction data supported this expectation as 7l and 7m showed maximum number of hydrogen bonds within the receptor binding site and along with hydrophobic regions as compared to other ligands (Table 5).
The binding mode between EGFR tyrosine kinase domain target and the molecules 7a to 7o including ERL, the structures in complex derived from the best dock results are given in Fig. 6. The hydrogen bond donor, acceptor along with hydrophobic regions of the compounds 7a to 7o was found to be oriented within the active center of target protein binding site. The top ligands 7l, 7m, 7e, and 7f, evaluated by least binding affinity more than −8.82 Kcal/mol which indicated the effective binding. The moderate binding was observed in the case of remaining ligands with binding energy found to be in the range of −7 to −8.80 kcal/mol. From the docking, it was suggested that there are key H-bonds created via interacting with MET769, ASP831, LYS721 and CYS773 amino acids in the EGFR-TK - 7l/7m complexes (Table S1(l,m)), while 7e and 7f (Table S1(e,f)) has H-bonds with THR766, GLN767, THR830 apart from the other key amino acid residues. The reference molecule Erlotinib (ERL) (Table S1(ERL)) have one key H-bond with MET769 AA. Among the binding residues, those involved in hydrogen bonds with the inhibitors are detailed with type of hydrogen bond formed (in context of hydrogen bond donor and acceptor) and distance between the atoms forming them. 3D representation of 7f and 7m compounds are given in Fig. 7. A 2D depiction of the interactions of binding residues of receptor protein with the ligands and H-bond distances are represented in Table S1 of Supporting Information. Results indicated that 7f, 7l and 7m intensively bind EGFR-TK through H-bonds and hydrophobic interactions. Molecules including 7l, 7m, 7f, 7g had longer and broader structures when compared with the co-crystal ligand ERL and the protein binding pocket size. This was considerable reason for slight discrepancies in the binding energies while comparing with experimental data. Therefore compounds like 7l and 7g showed significantly decreased binding energies and favourable inhibition. Docking simulation was performed to provide a molecular level foundation to illustrate and position compounds into the EGFR active site to determine the probable binding conformation at the active site of EGFR tyrosine kinase.
3D representation of the receptor-ligand interactions of 7f (A) and 7m (B) compounds. Interaction details of all compounds are given in Table S1 of SI.
In vivo pharmacology
Encouraged by the excellent EGFR-TK inhibitory activity and anticancer effect of compound 7l against breast cancer cells which are further substantiated with docking analysis, prompted us to further analyze the effect of compound 7l in the in-vivo system. Therefore, we have utilized 7,12-Dimethylbenz(a)anthracene (DMBA), a polycyclic aromatic hydrocarbon, to induce the mammary carcinogenesis in the experimental animals to quantify the potential protective effect of compound 7l. The mechanism behind its action to induce the cancer is based on its metabolized product, i.e. the epoxide that readily forms DNA adducts which is responsible for malignant alteration. At the first instance, the effect of compound 7l was analyzed on the effect of body weight of the experimental animals. As shown in Fig. 8, the body weight of the DMBA treated group showed considerable decline in the weight as compared to the control due to reduction in the normal metabolic process needed for maintaining the energy balance. Moreover, the body weight of the experimental animals has been significantly improved in the case of 7l treated group in a dose-dependent manner.
The effect of compound 7l was also investigated on the percentage of tumour incidence and on the tumour volume. The results of the study was clearly elucidated in the Fig. 9, which indicated that, compound 7l was efficient in reducing the percentage of incidence of tumour in cancer bearing animals. Moreover, 7l was also showed significant reduction in the tumour volume of the cancer induced experimental groups. The results suggests clearly suggest the broad protective effect of compound 7l against the mammary cancer.
The next part of the study was aimed at elucidating the effect of the compound 7l on the antioxidant status of the experimental animals. Previous studies have confirmed the role of antioxidant system as a primary line of defence against cancer inducing agent. Therefore, this study has vital implication to understand the role of 7l against the breast cancer because; the effectiveness of endogenous anti-oxidant system has been greatly compromised and often aberrantly deregulated. In the present study, we observe a significant decline in the level of endogenous anti-oxidative enzymes, such as, SOD, CAT, GSH and GPx. As shown in fig 10 and 11 in plasma and mammary tissues, respectively, the level of these enzymes in DMBA alone treated group was significantly reduced as compared to the control. Whereas, on admistration of compound 7l showed significant improvement in the antioxidant status of the DMBA treated animals. However, when these results are correlated with tumour incidence and volume, it has been suggested, the compound 7l showed considerable protection against the tumour and exert its protective action against breast cancer probable via scavenging the free radical. The lipid peroxidation has been greatly influenced by the generation of reactive oxygen species (ROS) and have profound role in the progression of the tumour. Therefore, in the next instance, the effect of compound 7l was determined on the level of TBARS and LOOH, which considered as critical biomarker for the assessment of peroxidation of membrane lipids. As evident from the Figs 10(A and B) and 11(A & B), it was clear that, the level of TBARS and LOOH has been found to be significantly elevated as compared to control. Moreover, it was marked to note that, the administration of 7l causes significant reduction in the level of these investigated markers, which further found to be suitably correlated with improvement in antioxidant system.
The metabolic enzymes are well known for their ability to clear the metabolic by-products generated by the cancer causing agents. In a classical metabolic pathway of the DMBA, it has been found that, CYP450 dependent oxygenases, including other Phase I enzymes causes metabolic degradation of the DMBA into reactive species which are indicated to be responsible for cancer inducing effect. Whereas, the enzymes involved in Phase II metabolic process, such as, GST and GR causes conversion of DMBA to water soluble conjugates which will be easily cleared-off and acts as a protective mechanism to prevent its cancer inducing effect. Consequently, it is imperative to determine the effect of compound 7l on these enzymes in different treated group to determine its beneficial effect against mammary tumour. The Figs 12 and 13 clearly showed the expression of various biotransformation enzymes in the liver and mammary tissues of the treated and control animals, for instance, phase I (CYP450, Cyt-b5) and phase II (GST, GR). It has been found that, in DMBA treated group, the level of CYP450 and Cyt-b5 were found to be significantly elevated as compared to the control. Whereas, the level of Phase II enzymes, e.g. GST and GR were found to be significantly reduced. In the case of 7l treated animals, the marked decline in the activity was reported in the concentration of Phase I enzymes (CYP450 and Cyt-b5). Moreover, a significant upsurge was reported in the concentration of both GST and GR. From the study, it has been corroborated that, compound 7l was significantly effective in modulating the concentration of both Phase I and Phase II enzymes in dose-dependent manner. These results were also found in accordance with earlier studies, which may be indicated as possible mechanism of compound 7l to exert potent anti-breast cancer activity.
Effect of compound 7l on the EGFR activation and downstream signalling
The extensive analysis of pharmacological activity of compound 7l in both in-vitro and in-vivo studies together with docking analysis as presented above showed the excellent inhibitory activity against breast cancer via inhibition of EGFR-TK. However, despite of excellent inhibitory activity of 7l against the EGFR-TK, its mechanism behind this phenomena has not been elucidate till now, thus, in this part of the study, we are interested in exploring the effect of compound 7l on the activation and downstream signalling pathway of the EGFR. Towards this, the effect of 7l has been determined on the expression of p-EGFR and EGFR, p-Akt and Akt levels of MCF-7 by immunoblotting experiments. Results as presented in Fig. 14 confirmed the effect of 7l on the expression of p-EGFR and p-Akt. It has been found that, the level of both p-EGFR and p-Akt were significantly reduced in concentration-dependent manner under influence of 7l. Results clearly indicate the ability of 7l to inhibit the phosphorylation of EGFR which in turn decreases Akt (the downstream target protein of p-EGFR).
Conclusion
A novel series of hybrid analogues of monastrol-1,3,5-triazine were designed and developed via three-component, one-pot synthesis and proved as effective anticancer agent via inhibition of EGFR-TK along with excellent in-vivo activity. These molecules will certainly hold a promise for the future drug development initiative.
Experimental
Chemistry
Melting points of the synthesized compounds was determined in an open capillary tube Hicon Melting point apparatus and are uncorrected. Thin layer chromatography (TLC) was performed on silica gel-G coated plates to detect the completion of reaction. The diverse mobile phase was selected in different proportion according to the assumed polarity of the products. The spots was visualised by exposure to the Iodine vapour. Infra-Red (IR) spectra were recorded in KBr on Biored FTs spectrophotometer and the reported wave numbers are given in cm−1. 1H NMR spectra were recorded in DMSO on Bruker Model D9RX-400MHz spectrometer. Chemical shifts were reported as δ (ppm) relative to TMS as internal standard. Mass spectra were obtained on VG-AUTOSPEC spectrometer equipped with electrospray ionization (ESI) sources.
General Procedure
Step 1: The synthesis of the various intermediates was performed in accordance with the earlier reported procedure28.
Step 2: Multicomponent one-pot synthesis of target derivatives, 7 (a–o)
A solution of an appropriate ethyl acetoacetate (1.2 mmol), corresponding aldehyde (1.0 mmol), urea or thiourea (1.2 mmol) in ethanol (5 ml) was heated under reflux in the presence of a catalytic amount of Bi(NO3)3 (10 mol%) till the completion of reaction as indicated by TLC. The reaction mixture was washed thoroughly with water, filtered and further purified by column chromatography to afford pure products. Elemental analysis of C, H and N was performed on a Vario EL III CHNOS elemental analyzer. The spectral data of the compounds are given below.
Ethyl 1-(4,6-bis(phenylamino)-1,3,5-triazin-2-yl)-6-methyl-4-phenyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7a
Yield: 68%; M.p: 225–227 °C; MW: 537.64; R f : 0.63; FTIR (νmax; cm−1 KBr): 3292 (N–H secondary), 3058 (C–H broad), 1716 (C=O), 1688–1652 (C=N aromatic), 1653 (C=C), 1368–1052 (C-N aromatic), 1272 (C=S), 1085 (CO stretching), 693, 548; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.52–7.51 (m, 4 H, 4 × CH, Ar-H), 7.31–7.30 (m, 2 H, 2 × CH, Ar-H), 7.26–7.20 (m, 7H, 7×CH, Ar-H), 6.81–6.79 (m, 2H, 2×CH, Ar-H), 4.38 (q, 2 H J = 7.08 Hz, CH2 oxoacetate), 4.28 (s, 1H, pyrimidin), 3.87 (br,s, 2H, 2 × NH), 2.56 (s, 3H, CH3 pyrimidin), 1.89 (s, 1H, NH pyrimidin), 1.48 (t, 3H, J = 6.93 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.14, 168.73, 167.39, 167.28, 158.42, 143.32, 138.96, 129.52, 128.53, 126.92, 126.52, 122.42, 117.82, 104.28, 61.82, 58.68, 18.16, 14.23; Mass: 538.68 (M + H)+; Elemental analysis for C29H27N7O2S: Calculated: C, 64.79; H, 5.06; N, 18.24. Found: C, 64.81; H, 5.04; N, 18.27.
Ethyl 1-(4,6-bis((4-nitrophenyl)amino)-1,3,5-triazin-2-yl)-6-methyl-2-thioxo-4-(p-tolyl)-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7b
Yield: 63%; M.p: 183–185 °C; MW:641.66; R f : 0.48; FTIR (νmax; cm−1 KBr): 3286 (N–H secondary), 3046 (C–H broad), 1712 (C=O), 1672–1637 (C=N aromatic), 1605 (C=C), 1527 (NO2), 1347–1023 (C-N aromatic), 1276 (C=S), 1093 (CO stretching), 610, 558; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 8.12–8.08 (m, 4 H, 4 × CH, Ar-H), 7.34–7.31 (m, 4 H, 4 × CH, Ar-H), 6.89 (m, 4H, 4 × CH, Ar-H), 4.32 (s, 3H, CH3 pyrimidin), 4.12 (s, 1H, CH pyrimidin), 4.38 (q, 2H J = 7.08 Hz, CH2 oxoacetate), 3.85 (br,s, 2H, 2 × NH), 2.43 (s, 3H, CH3), 1.89 (s, 1H, NH pyrimidin), 1.42 (t, 3H, J = 6.94 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.23, 168.92, 167.32, 167.21, 158.43, 145.23, 140.31, 137.9, 136.40, 128.82, 126.90, 124.72, 119.21, 104.20, 61.80, 58.40, 21.36, 18.46, 14.21; Mass: 642.65 (M + H)+; Elemental analysis for C30H27N9O6S: Calculated: C, 56.15; H, 4.24; N, 19.65. Found: C, 56.18; H, 4.26; N, 19.68.
Ethyl 1-(4,6-bis((2-nitrophenyl)amino)-1,3,5-triazin-2-yl)-6-methyl-2-thioxo-4-(p-tolyl)-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7c
Yield: 68%; M.p: 232–234 °C; MW: 641.66; R f : 0.54; FTIR (νmax; cm−1 KBr): 3288 (N–H secondary), 3048 (C–H broad), 1715 (C=O), 1674–1639 (C=N aromatic), 1611 (C=C), 1528 (NO2), 1347–1026 (C-N aromatic), 1278 (C=S), 1091 (CO stretching), 628, 562; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 8.08 (m, 2H, 2 × CH, No2- Ar-H), 7.59–7.51 (m, 4H, 4 × CH, No2-Ar-H), 7.34 (s, 1H Ar-H), 7.13–7.11 (m, 3H, 3 × CH, Ar-H), 6.89 (m, 2H, 2 × CH, Ar-H), 4.52 (s, 1H, CH pyrimidin), 4.46 (q, 2H J = 7.06 Hz, CH2 oxoacetate), 3.87 (br,s, 1H, NH), 2.36 (s, 3H, CH3 Ar), 2.13 (s, 3H, CH3 pyrimidin), 1.88 (s, 1H, NH), 1.36 (t, 3H J = 6.89 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.21, 168.82, 167.36, 167.25, 158.42, 144.60, 140.32, 137.14, 136.43, 129.10, 128.82, 126.76, 125.84, 119.64, 110.58, 104.24, 61.71, 58.60, 21.28, 18.52, 14.21; Mass: 642.68 (M + H)+; Elemental analysis for C30H27N9O6S: Calculated: C, 56.15; H, 4.24; N, 19.65. Found: C, 56.18; H, 4.25; N, 19.64.
Ethyl 1-(4,6-bis((3-chlorophenyl)amino)-1,3,5-triazin-2-yl)-4-(4-hydroxyphenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7d
Yield: 71%; M.p: 161–163 °C; MW: 622.52; R f : 0.42; FTIR (νmax; cm−1 KBr): 3478 (OH), 3282 (N–H secondary), 3053 (C–H broad), 1721 (C=O), 1674–1642 (C=N aromatic), 1618 (C=C), 1349–1023 (C-N aromatic), 1268 (C=S), 1082 (CO stretching) 1014 (C-Cl stretching), 678, 549; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.68–7.61 (m, 2H, 2 × CH, Ar-H), 7.14–7.08 (m, 4H, 4 × CH, Ar-H), 6.87–6.82 (m, 4H, 4 × CH, Ar-H), 6.63–6.58 (m, 2H, 2 × CH, Ar-H), 5.46 (s, 1H, Ar-OH), 4.48 (q, 2H J = 7.06 Hz, CH2 oxoacetate), 4.08 (s, 1H, pyrimidin), 3.89 (br,s, 2H, 2 × NH), 2.34 (s, 3H, CH3 pyrimidin), 1.94 (s, 1H, NH), 1.36 (t, 3H J = 6.89 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.24, 168.80, 167.32, 167.24, 158.42, 156.54, 143.84, 135.92, 135.10, 130.90, 126.74, 122.46, 116.68, 115.90, 115.74, 104.26, 61.78, 58.48, 18.24, 14.26; Mass: 623.56 (M + H)+; Elemental analysis for C29H25Cl2N7O3S: Calculated: C, 55.95; H, 4.05; N, 15.75. Found: C, 55.98; H, 4.09; N, 15.77.
Ethyl 1-(4,6-bis((3-bromophenyl)amino)-1,3,5-triazin-2-yl)-4-(2-hydroxyphenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7e
Yield: 69%; M.p: 245–247 °C; MW: 711.43; R f : 0.48; FTIR (νmax; cm−1 KBr): 3472 (OH), 3286 (N–H secondary), 3048 (C–H broad), 1716 (C=O), 1676–1638 (C=N aromatic), 1621 (C=C), 1342–1018 (C-N aromatic), 1271 (C=S), 1086 (CO stretching) 916 (C-Br stretching), 669, 538; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.57–7.54 (m, 2 H, 2 × CH, Ar-H), 7.09–7.06 (m, 2H, 2 × CH, Ar-H), 6.96–6.92 (m, 4H, 4 × CH, Ar-H), 6.76–6.64 (m, 4H, 4 × CH, Ar-H), 5.44 (s, 1H, Ar-OH), 4.46 (q, 2H J = 7.06 Hz, CH2 oxoacetate), 4.12 (s, 1H, pyrimidin), 3.94 (br,s, 2H, 2 × NH), 2.32 (s, 3H, CH3 pyrimidin), 1.96 (s, 1H, NH pyrimidin), 1.38 (t, 3 H J = 6.91 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.10, 168.84, 167.30, 167.21, 158.40, 154.03, 144.68, 130.69, 128.42, 128.10, 123.86, 122.74, 121.54, 121.10, 116.86, 115.84, 115.52, 104.23, 61.70, 52.41, 18.23, 14.16; Mass: 712.46 (M + H)+; Elemental analysis for C29H25Br2N7O3S: Calculated: C, 48.96; H, 3.54; N, 13.78. Found: C, 48.98; H, 3.52; N, 13.81.
Ethyl 1-(4,6-bis((2-chlorophenyl)amino)-1,3,5-triazin-2-yl)-4-(4-methoxyphenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7f
Yield: 72%; M.p: 91–93 °C; MW: 636.55; R f : 0.48; FTIR (νmax; cm−1 KBr): 3286 (N–H secondary), 3052 (C–H broad), 1721 (C = O), 1672–1646 (C = N aromatic), 1614 (C = C), 1348–1026 (C-N aromatic),1264 (C = S), 1084 (COO stretching) 1024 (C-Cl stretching), 676, 538; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 8.08–8.04 (m, 2 H, 2 × CH, Ar-H), 7.45–7.38 (m, 4 H, 4 × CH, Ar-H), 7.12–7.08 (m, 2 H, 2 × CH, Ar-H), 6.85–6.78 (m, 4 H, 4 × CH, Ar-H), 4.38 (q, 2 H J = 7.04 Hz, CH2 oxoacetate), 4.18 (s, 1 H, pyrimidin), 3.89 (br,s, 2 H, 2 × NH), 3.74 (s, 3 H, Ar-OCH3), 2.42 (s, 3 H, CH3 pyrimidin), 1.92 (s, 1 H, NH pyrimidin), 1.32 (t, 3 H J = 6.93 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.12, 168.94, 167.34, 167.23, 158.62, 158.41, 136.42, 135.61, 130.72, 127.60, 125.72, 125.41, 122.83, 122.24, 114.12, 104.24, 61.71, 58.65, 55.82, 18.30, 14.24; Mass: 637.58 (M + H)+; Elemental analysis for C30H27Cl2N7O3S: Calculated: C, 56.61; H, 4.28; N, 15.40. Found: C, 56.66; H, 4.32; N, 15.42.
Ethyl 1-(4,6-bis((3-fluorophenyl)amino)-1,3,5-triazin-2-yl)-4-(2-methoxyphenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7g
Yield: 63%; M.p: 132–135 °C; MW: 603.64; R f : 0.62; FTIR (νmax; cm−1 KBr): 3283 (N–H secondary), 3056 (C–H broad), 1723 (C = O), 1678–1651 (C = N aromatic), 1618 (C = C), 1349–1028 (C-N aromatic),1262 (C = S), 1084 (CO stretching), 978 (C-F stretching), 685, 548; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.76–7.74 (m, 2 H, 2 × CH, Ar-H), 7.38–7.26 (m, 4 H, 4 × CH, Ar-H), 7.02–6.89 (m, 3 H, 3 × CH, Ar-H), 6.82 (d, 1 H J = 5.45 Hz, Ar-H), 6.58–6.56 (m, 2 H, 2 × CH, Ar-H), 4.32 (q, 2 H J = 7.06 Hz, CH2 oxoacetate), 4.24 (s, 1 H, pyrimidin), 3.87 (br,s, 2 H, 2 × NH), 3.81 (s, 3 H, Ar-OCH3), 2.46 (s, 3 H, CH3 pyrimidin), 1.98 (s, 1 H, NH pyrimidin), 1.29 (t, 3 H J = 6.96 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.12, 168.86, 167.38, 167.21, 163.72, 158.44, 156.58, 144.18, 131.12, 127.89, 127.73, 121.24, 120.87, 113.46, 112.38, 110.54, 104.86, 104.21, 61.76, 56.14, 52.64, 18.20, 14.26; Mass: 604.65 (M + H)+; Elemental analysis for C30H27F2N7O3S: Calculated: C, 59.69; H, 4.51; N, 16.24. Found: C, 59.68; H, 4.52; N, 16.26.
Ethyl 1-(4,6-bis((4-nitrophenyl)amino)-1,3,5-triazin-2-yl)-4-(4-chlorophenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7h
Yield: 64%; M.p: 146–147 °C; MW: 662.08; R f : 0.65; FTIR (νmax; cm−1 KBr): 3289 (N–H secondary), 3052 (C–H broad), 1721 (C = O), 1678–1656 (C = N aromatic), 1621 (C = C), 1528 (NO2), 1356–1036 (C-N aromatic), 1268 (C = S), 1089 (CO stretching) 1019 (C-Cl stretching), 689, 552; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 8.01–7.98 (m, 4 H, 4 × CH, Ar-H), 7.38–7.36 (m, 4 H, 4 × CH, Ar-H), 6.89–6.87 (m, 4 H, 4 × CH, Ar-H), 4.36 (q, 2 H J = 7.06 Hz, CH2 oxoacetate), 4.23 (s, 1 H, pyrimidin), 3.89 (br,s, 2 H, 2 × NH), 2.49 (s, 3 H, CH3 pyrimidin), 1.94 (s, 1 H, NH pyrimidin), 1.36 (t, 3 H J = 6.84 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.18, 168.82, 167.39, 167.22, 158.39, 145.21, 141.46, 137.92, 132.38, 128.64, 126.17, 124.74, 119.26, 104.24, 61.72, 58.68, 18.18, 14.26; Mass: 663.10 (M + H)+; Elemental analysis for C29H24ClN9O6S: Calculated: C, 52.61; H, 3.65; N, 19.04. Found: C, 52.64; H, 3.68; N, 19.02.
Ethyl 1-(4,6-bis((2-nitrophenyl)amino)-1,3,5-triazin-2-yl)-4-(3-bromophenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7i
Yield: 78%; M.p.: 185–186 °C; MW: 706.53; R f : 0.59; FTIR (νmax; cm−1 KBr): 3292 (N–H secondary), 3058 (C–H broad), 1716 (C = O), 1682–1658 (C = N aromatic), 1628 (C = C), 1518 (NO2), 1355–1032 (C-N aromatic),1258 (C = S), 1085 (CO stretching) 985 (C-Br stretching), 695, 558; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 8.08–8.06 (m, 2 H, 2 × CH, Ar-H), 7.64–7.62 (m, 2 H, 2 × CH, Ar-H), 7.42–7.38 (m, 4 H, 4 × CH, Ar-H), 7.22–7.19 (m, 2 H, 2 × CH, Ar-H), 6.89–6.87 (m, 2 H, Ar-H), 4.38 (q, 2 H J = 7.04 Hz, CH2 oxoacetate), 4.22 (s, 1 H, pyrimidin), 3.84 (br,s, 2 H, 2 × NH), 2.51 (s, 3 H, CH3 pyrimidin), 1.96 (s, 1 H, NH pyrimidin), 1.42 (t, 3 H J = 6.94 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.21, 168.86, 167.36, 167.23, 158.47, 145.56, 144.68, 137.12, 131.78, 129.94, 129.21, 129.00, 125.94, 125.38, 122.92, 119.67, 110.58, 104.21, 61.72, 57.93, 18.21, 14.16; Mass: 707.56 (M + H)+; Elemental analysis for C29H24BrN9O6S: Calculated: C, 49.30; H, 3.42; N, 17.84. Found: C, 49.32; H, 3.41; N, 17.86.
Ethyl 1-(4,6-bis((4-chlorophenyl)amino)-1,3,5-triazin-2-yl)-4-(2-chlorophenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7j
Yield: 59%; M.p: 112–114 °C; MW: 640.97; R f : 0.49; FTIR (νmax; cm−1 KBr): 3296 (N–H secondary), 3052 (C–H broad), 1726 (C=O), 1684–1652 (C=N aromatic), 1634 (C=C), 1358–1026 (C-N aromatic),1252 (C=S), 1078 (CO stretching), 1019 (C-Cl stretching), 698, 565; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.73–7.68 (m, 5H, 5 × CH, Ar-H), 7.24–7.18 (m, 7H, 7 × CH, Ar-H), 4.36 (q, 2H J = 7.02 Hz, CH2 oxoacetate), 4.24 (s, 1H, pyrimidin), 3.86 (br,s, 2H, 2 × NH), 2.56 (s, 3H, CH3 pyrimidin), 1.94 (s, 1H, NH pyrimidin), 1.47 (t, 3H J = 6.92 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.11, 168.89, 167.32, 167.24, 158.42, 142.86, 137.18, 132.28, 129.64, 128.60, 128.34, 128.12, 127.78, 126.62, 122.17, 104.24, 61.72, 53.58, 18.21, 14.18; Mass: 641.98 (M + H)+; Elemental analysis for C29H24Cl3N7O2S: Calculated: C, 54.34; H, 3.77; N, 15.30. Found: C, 54.38; H, 3.78; N, 15.32.
Ethyl 1-(4,6-bis((3-bromophenyl)amino)-1,3,5-triazin-2-yl)-6-methyl-4-(4-nitrophenyl)-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7k
Yield: 64%; M.p: 167–165 °C; MW: 740.43; R f : 0.58; FTIR (νmax; cm−1 KBr): 3298 (N–H secondary), 3048 (C–H broad), 1723 (C=O), 1684–1656 (C=N aromatic), 1638 (C=C), 1526 (NO2), 1361–1028 (C-N aromatic),1257 (C=S), 1075 (CO stretching), 987 (C-Br stretching), 678, 569; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 8.06–8.05 (m, 2 H, 2 × CH, Ar-H), 7.54–7.49 (m, 4 H, 4 × CH, Ar-H), 6.96–6.94 (m, 4 H, 4 × CH, Ar-H), 6.64–6.62 (m, 2H, 2 × CH, Ar-H), 4.34 (q, 2H J = 7.08 Hz, CH2 oxoacetate), 4.34 (s, 1H, pyrimidin), 3.87 (br,s, 2H, 2 × NH), 2.58 (t, 3H J = 6.88 Hz, CH3 pyrimidin), 1.92 (s, 1H, NH pyrimidin), 1.38 (s, 3H, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.16, 168.86, 167.32, 167.12, 158.45, 149.46, 145.92, 144.67, 130.62, 128.32, 123.92, 121.62, 116.81, 115.54, 104.28, 61.79, 58.62, 18.16, 14.22; Mass: 741.48 (M + H)+; Elemental analysis for C29H24Br2N8O4S: Calculated: C, 47.04; H, 3.27; N, 15.13. Found: C, 47.01; H, 3.29; N, 15.11.
Ethyl 1-(4,6-bis((3-fluorophenyl)amino)-1,3,5-triazin-2-yl)-4-(4-fluorophenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7l
Yield: 68%; M.p: 171–172 °C; MW: 591.61; R f : 0.54; FTIR (νmax; cm−1 KBr): 3294 (N–H secondary), 3048 (C–H broad), 1721 (C=O), 1686–1654 (C=N aromatic), 1642 (C=C), 1367–1038 (C-N aromatic),1259 (C=S), 1078 (CO stretching), 978 (C-F stretching), 689, 572; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.72–7.70 (m, 2H, 2 × CH, Ar-H), 7.40–7.38 (m, 2H, 2 × CH, Ar-H), 7.21–7.14 (m, 6H, 6 × CH, Ar-H), 6.63–6.62 (m, 2H, 2 × CH, Ar-H), 4.38 (q, 2H J = 7.04 Hz, CH2 oxoacetate), 4.32 (s, 1H, pyrimidin), 3.87 (br,s, 2H, 2 × NH), 2.58 (s, 3H, CH3 pyrimidin), 1.87 (s, 1H, NH pyrimidin), 1.42 (t, 3H J = 7.04 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.24, 168.81, 167.38, 167.23, 163.79, 160.92, 158.42, 144.18, 138.92, 131.12, 128.50, 115.32, 113.42, 110.52, 104.86, 104.22, 61.72, 58.68, 18.18, 14.24; Mass: 592.64 (M + H)+; Elemental analysis for C29H24F3N7O2S: Calculated: C, 58.88; H, 4.09; N, 16.57. Found: C, 58.90; H, 4.11; N, 16.56.
Ethyl 1-(4,6-bis((3-chlorophenyl)amino)-1,3,5-triazin-2-yl)-4-(4-fluorophenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7m
Yield: 72%; M.p: 153–155 °C; MW: 624.52; R f : 0.63; FTIR (νmax; cm−1 KBr): 3296 (N–H secondary), 3042 (C–H broad), 1718 (C=O), 1684–1652 (C=N aromatic), 1648 (C=C), 1367–1042 (C-N aromatic),1265 (C=S), 1082 (CO stretching), 992 (C-Cl stretching), 972 (C-F stretching), 678, 578; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.78–7.76 (m, 2H, 2 × CH, Ar-H), 7.45–7.44 (m, 2H, 2 × CH, Ar-H), 7.28–7.21 (m, 6H, 6 × CH, Ar-H), 7.07–7.06 (m, 2H, 2 × CH, Ar-H), 4.35 (q, 2H J = 7.05 Hz, CH2 oxoacetate), 4.28 (s, 1H, pyrimidin), 3.84 (br,s, 2H, 2 × NH), 2.58 (s, 3 H, CH3 pyrimidin), 1.89 (s, 1H, NH pyrimidin), 1.48 (t, 3H J = 6.94 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.14, 168.86, 167.32, 167.22, 160.92, 158.84, 143.82, 138.92, 135.18, 130.92, 128.52, 122.34, 116.72, 115.92, 115.36, 104.24, 61.74, 58.68, 18.21, 14.25; Mass: 625.53 (M + H)+; Elemental analysis for C29H24Cl2FN7O2S: Calculated: C, 55.77; H, 3.87; N, 15.70. Found: C, 55.79; H, 3.91; N, 15.72.
Ethyl 1-(4,6-bis(p-tolylamino)-1,3,5-triazin-2-yl)-6-methyl-2-thioxo-4-(o-tolyl)-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7n
Yield: 61%; M.p: 205–207 °C; MW: 579.72; R f : 0.52; FTIR (νmax; cm−1 KBr): 3292 (N–H secondary), 3048 (C–H broad), 1719 (C=O), 1686–1658 (C=N aromatic), 1652 (C=C), 1418 (-CH3), 1365–1048 (C-N aromatic), 1263 (C=S), 1087 (CO stretching), 696, 512; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.33–7.32 (m, 4H, 4 × CH, Ar-H), 7.18 (s, 1H, 1 × CH, Ar-H), 7.08–7.05 (m, 7H, 7 × CH, Ar-H), 4.38 (q, 2H J = 7.08 Hz, CH2 oxoacetate), 4.32 (s, 1H, pyrimidin), 3.87 (br,s, 2H, 2 × NH), 2.57 (s, 3H, CH3 pyrimidin), 2.23–2.21 (s, 9H, 3 × CH3 Ar-CH3), 1.89 (s, 1 H, NH pyrimidin), 1.48 (t, 3H J = 6.93 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.34, 168.81, 167.38, 167.21, 158.87, 142.72, 135.98, 134.78, 131.12, 130.28, 129.82, 126.62, 125.58, 124.92, 120.37, 104.26, 61.72, 56.13, 21.31, 19.28, 18.16, 14.24; Mass: 580.74 (M + H)+; Elemental analysis for C32H33N7O2S: Calculated: C, 66.30; H, 5.74; N, 16.91. Found: C, 66.32; H, 5.78; N, 16.92.
Ethyl 1-(4,6-bis(p-tolylamino)-1,3,5-triazin-2-yl)-4-(4-hydroxyphenyl)-6-methyl-2-thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate 7o
Yield: 48%; M.p: 131–133 °C; MW: 581.69; R f : 0.58; FTIR (νmax; cm−1 KBr): 3296 (N–H secondary), 3051 (C–H broad), 1723 (C=O), 1683–1654 (C=N aromatic), 1658 (C=C), 1365–1057 (C-N aromatic),1268 (C=S), 1093 (CO stretching), 699, 538; 1H NMR (400 MHz, DMSO, TMS) δ ppm: 7.33–7.31 (m, 4H, 4 × CH, Ar-H), 7.06–7.04 (m, 6H, 6 × CH, Ar-H), 6.67–6.65 (m, 2H, 2 × CH, Ar-H), 5.28 (s, 1H, Ar-OH), 4.37 (q, 2H J = 7.03 Hz, CH2 oxoacetate), 4.36 (s, 1H, pyrimidin), 3.89 (br,s, 2H, 2 × NH), 2.59 (s, 3H, CH3 pyrimidin), 2.23–2.21 (s, 6H, 2 × CH3 Ar-CH3), 1.89 (s, 1H, NH pyrimidin), 1.48 (t, 3H J = 6.91 Hz, CH3 oxoacetate); 13C NMR (100 MHz, DMSO) δ ppm: 176.24, 168.87, 167.32, 167.21, 158.46, 156.42, 136.52, 135.92, 131.23, 129.82, 126.12, 120.32, 115.78, 104.22, 61.76, 58.63, 21.38, 18.12, 14.23; Mass: 582.69 (M + H)+; Elemental analysis for C31H31N7O3S: Calculated: C, 64.01; H, 5.37; N, 16.86. Found: C, 64.04; H, 5.36; N, 16.89.
Biological Evaluation
In vitro Anticancer Evaluation
The target compounds were subjected to in vitro cytotoxicity bioassay. In vitro cytotoxicity was determined using a standard MTT assay with protocol appropriate for the individual test system29,30,31. The four human cancer cell lines HeLa, MCF-7, HL-60, HepG2 and MCF-12A were cultured in the MEM medium supplemented with 10% FBS, 1% glutamine and 50 mM/ml gentamicin sulfate in a CO2 incubator in a humidified atmosphere of 5% CO2 and 95% air. The test compounds were prepared prior to the experiment by dissolving in 0.1% DMSO and diluted with medium. The cells were then exposed to different concentrations of drugs (1–100 mM) in the volume of 100 mM/well. Cells in the control wells received the same volume of medium containing 0.1% DMSO. After 24 h, the medium was removed and cell cultures were incubated with 100 ml MTT reagent (1 mg/ml) for 5 h at 37 °C. The known number of cells (1.0 ± 105) was incubated in a 5% CO2 incubator at 37 °C in the presence of different concentrations of test compounds. After 48 h of drug incubation, the MTT solution was added in each well and absorbance was recorded at 540 nm by an ELISA reader. The experiment was performed in triplicate. Cell survival was calculated as the percentage of MTT inhibition as % growth inhibition = 100 − (mean OD of individual test Group/Mean OD of each Control Group) × 100.
EGFR tyrosine kinase inhibitory activity
Kinase activity was determined using Kinase-Glo Plus luminescence kinase assay kit, by quantitating the amount of ATP remaining in the solution of kinase reaction32. The luminescent signal is correlated with the residual amount present and it was inversely related with kinase activity. The tested compounds were diluted to 100 mM in 10% DMSO, then 5 mL of the dilution was added to a 50 mL reaction. All of the enzymatic reactions were performed at 30 °C for 40 min, 50 mL of reaction mixture contains 10 mM MgCl2, 40 mM Tris, pH 7.4, 0.1 mg/mL BSA, 0.2 mg/mL Poly (Glu, Tyr) substrate, 10 mM ATP and EGFR. Incubate the plate for 5 min at r.t. then add 50 mL of Kinase-GloPlus Luminescence kinase assay to each reaction. ADP-Glo assay kit is the protein kinase assays used to determine IC50 values in which ADP generation was measured as it leads to an increase in luminescence signal. The reaction mixture was incubated in a 96-well plate at 30 °C for 30 min, after the incubation period add 25 mL of ADP-Glo reagent to terminate the assay. Shake the 96-well plate for 30 min at ambient temperature then incubate it, then add 50 mL of kinase detection reagent. Read the 96-well plate using the ADP-Glo Luminescence reader. All the assay components were added to the blank control except the substrate. By removing the blank control value you can obtain the corrected activity for each protein kinase target.
In Vivo Anticancer evaluation
Chemicals. The chemicals used in the study were purchased from Sigma-Aldrich Chemicals (USA) and were of analytical grade.
Animal model. The female Sprague-Dawley rats (100–120 g) were obtained from the Institutional animal house after approval from the Institutional Animal Ethics Committee. The animals were kept in suitable cages with free access to food and water under a controlled temperature (24 ± 2 °C), humidity (60 ± 10%) with alternate light/dark cycle of 12 h. The entire experimental procedures involving animals were approved by the Institutional Animal Ethics Committees (IAEC) of SHUATS, in accordance and duly approved with CPCSEA (Committee for the Purpose of Control and Supervision of Experiments on Animals, Government of India. The authors confirm that all experiments were performed in accordance with relevant guidelines and regulations of SHUATS.
Initiation of breast carcinogenesis. The mammary carcinogenesis was induced in experimental rat using one-time subcutaneous administration of DMBA (25 mg) in emulsified form with the help of sunflower oil and saline to each rat.
Selection of test compound dose. The dose of compound 7l was estimated using an acute toxicity study in healthy Sprague Dawley rats (6–8 weeks old). The animals were observed for 48 hours after the administration of drug for the onset of clinical or toxicological symptoms. Mortality, if any, was observed over a period of 2 weeks. It has been found that, upto 40 mg/kg, no mortality has been reported. Thus, dose of 5 mg/kg, 10 mg/kg and 15 mg/kg was selected in the present study.
Design of experiment. Experimental design. Total numbers of 30 animals were randomly divided into five groups and each group contains 6 animals. The various groups received different treatment as follows:
Group I: Untreated control
Groups II: DMBA
Group III: DMBA + 7l (5 mg/kg)
Group IV: DMBA + 7l (10 mg/kg)
Group V: DMBA + 7l (15 mg/kg)
The DMBA (s.c.; 25 mg/rat) was administered to each rats near mammary gland at the end of the first week. The test drug was administered once in a day and was continued till the end of the experimental period. At the end of 16th week, the rats were fasted overnight and sacrificed by the cervical decapitation under influence of anesthesia. The blood samples were collected in and after centrifugation, the plasma was used for the further biochemical analysis. The liver and mammary tissues were excised immediately from the experimental rats and homogenized with suitable buffer, centrifuged and the resulting supernatant was used for biochemical estimations.
Biochemical Estimation. The remaining excised liver and mammary tissues were rinsed in ice-cold saline and known amount of the tissue were homogenized in 0.1 M Tris-HCl buffer (pH 7.4) at 4°C. The supernatant was collected as tissue homogenate and was used for the estimation of various biochemical parameters. The microsomal protein concentration was estimated by the method of Lowry et al.33; while, the concentration of plasma and mammary TBARS was estimated by the earlier reported method34, 35 and the concentration of plasma and mammary tissue LOOH were estimated by the previous method36. Moreover, the activity of the antioxidant enzyme system, such as SOD, CAT, GPX, GSH, CYP450, Cyt-b5, GST and GR activity in plasma and mammary tissue were determined by the earlier method given elsewhere37,38,39,40,41,42,43.
Lipid profile. The estimation of lipid parameters was performed using the lipid extracted from plasma and mammary tissues by previous reported method of ref. 44. Moreover, the level of TC, TG, FFA, PL, LDL, VLDL-C, HDL-C and cholesterol in the plasma and mammary tissue were approximated using the earlier reported method45,46,47,48,49,50.
Western blot analysis
After incubation with the test compound, the cells were harvested and washed with PBS, and then lysed in lysis buffer. After centrifugation at 12,000 × g for 5 min, the supernatant was collected as total protein. The concentration of the protein was determined by a protein assay kit. The protein samples were separated by 10% SDS–PAGE and subsequently electro-transferred onto a PVDF (Millipore, Bedford, MA, USA). The membrane was blocked with 5% skimmed milk for 2 h at room temperature. The blocked membrane was probed with the indicated primary antibodies overnight at 4°C, and then incubated with a horse radish peroxidase (HRP)-coupled secondary antibody. The antibodies were purchased from Cell Signaling Technology (MA, USA) Thus, the expression of p-EGFR, EGFR, p-Akt (pS473) and Akt were visualized by enhanced chemiluminescence (ECL) (Thermofischer Scientific Inc. USA) kit and band intensity was analyzed by image j software.
Molecular docking analysis
EGFR-TK inhibitor
It was worthwhile to perform molecular docking of compounds to define precise key interaction with EGFR. Molecular docking analysis were undertaken to evaluate the most preferred geometry of protein–ligand complexes. In this study, we used Autodock 4.2 for molecular docking studies51, 52. EGFR tyrosine kinase domain with 4-anilinoquinazoline inhibitor erlotinib (ERL) (PDB ID:1M17)53 was considered as the target protein. The protein was prepared as a receptor for docking process by protonating54, 55 and the prepared protein was then loaded to MGL – AutoDock Tools to identify the binding sites of EGFR-TK domain using the co-crystal ligand and assign polar hydrogen atoms including Kollman charges to the minimized protein56. The program AutoGrid was used to generate the grid maps for the binding site of the protein. Grid was generated based on the amino acid residues in the active center and co-crystal ligand bound interaction points, further in order to match different interaction types like Br, Cl, F, SA, OA for all ligands, the grid generation parameter was manually updated to generate the custom grid maps for the protein binding site. Binding site includes Met 769, Gly 772, Leu 694, Leu 768, Lys 721, Thr 766, Thr 830 and Asp 831 amino acid residues in protein kinases ATP-binding region and Tyrosine protein kinases specific active-site regions.The ligands 7a-7o, and erlotinib (ERL) was drawn using Marvin Sketch software, converted to 3D using MM2 and conformational search was performed using MMFF94 forcefield57. The lowest energy conformer was further optimized using MOPAC with AM1 parameterization58, 59. The optimized ligands were loaded to MGL Tools one by one to re-assign Gasteiger partial charges and non-polar hydrogen atoms. All torsions (rotatable bonds) were auto detected for ligands and assigned to generate different poses during docking procedure.
Docking of EGFR-TK inhibitors was parameterized with flexible residues to locate the optimum binding conformations of 7a-7o in EGFR-TK protein. Each docking calculation was iterated 100000 times with 250 GA runs, resulting 250 docked poses. For all ligands, random initial positions, random conformers and torsions were used. The translation, quaternion and torsion steps were considered from default values indicated in AutoDock user guide. Lamarckian genetic algorithm was used for minimization using optimum parameters to generate all possible energies to rank the conformers. The parameters for the grid generation and genetic algorithm based docking calculations are shown in Table S2 that is given in supplementary information. The docking parameterization was validated and optimized based on the docking analysis on the active conformation of co-crystal ligand AQ4 - [6,7-Bis(2-Methoxy-Ethoxy)Quinazoline-4-Yl]-(3-Ethynylphenyl)Amine named as Erlotinib (ERL). Re-docking with Autodock Vina was performed for more accuracy based on global optimization and better search algorithm. Best hits were identified and the determination of binding energy to assess the binding affinity of ligands was calculated by employing highest stable ligand-receptor complex through MGL Tools to extract the energy data.
Statistical analysis
All the data were expressed as the mean ± SEM and analysis of variance (ANOVA) was used for the statistical analysis using Graph Pad Prism version 5.0. The values were considered to be significant when the P value was p < 0.05.
References
World Health Organisation (WHO) Antimicrobial Resistance, 194, http://www.who.int/mediacentre/factsheets/fs194/en/(2014).
Thun, M. J., DeLancey, J. O., Center, M. M., Jemal, A. & Ward, E. M. The global burden of cancer: priorities for prevention. Carcinogenesis 31, 100–110 (2010).
Reeler, A.V. & Mellstedt, H. Cancer in developing countries: challenges and solutions. Ann Oncol viii7–viii8 (2006).
Wells, A. EGF receptor. Int. J. Biochem.Cell Biol 31, 637–643 (1999).
Partanen, A. Epidermal growth factor and transforming growth factor-alpha in the development of epithelial-mesenchymal organs of the mouse. Curr. Top. Dev. Biol 24, 31–55 (1990).
Adamson, E. D. Developmental activities of the epidermal growth factor receptor. Curr. Top. Dev. Biol 24, 1–29 (1990).
Dutta, P. R. & Maity, A. Cellular responses to EGFR inhibitors and their relevance to cancer therapy. Cancer Letters 254, 165–177 (2007).
Grandis, J. R. & Sok, J. C. Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacol. Ther 102, 37–46 (2004).
Ciardiello, F. & Tortora, G. A novel approach in the treatment of cancer: targeting the epidermal growth factor receptor. Clin Cancer Res 7, 2958–70 (2001).
Mendelsohn, J. & Baselga, J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol 21, 2787–2799 (2003).
Normanno, N. et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366, 2–16 (2006).
Bai, F. et al. Discovery of novel selective inhibitors for EGFR-T790M/L858R. Bioorg. Med. Chem. Lett 22, 1365–1370 (2012).
Zuccotto, F., Ardini, E., Casale, E. & Angiolini, M. Through the “gatekeeper door”: exploiting the active kinase conformation. J. Med.Chem 53, 2681 (2010).
Liu, Y. & Gray, N. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol 2, 358 (2006).
Singh, B., Bhat, H. R., Kumawat, M. K. & Singh, U. P. Structure-guided discovery of 1,3,5-triazine-pyrazole conjugates as antibacterial and antibiofilm agent against pathogens causing human diseases with favorable metabolic fate. Bioorg Med Chem Lett 24, 3321–3325 (2014).
Bhat, H. R. et al. 4-Aminoquinoline-1,3,5-triazine: Design, synthesis, in vitro antimalarial activity and docking studies. New J.Chem 37, 2654–2662 (2013).
Kumar, S., Bhat, H. R., Kumawat, M. K. & Singh, U. P. Design and one-pot synthesis of hybrid thiazolidin-4-one-1,3,5-triazines as potent antibacterial agents against human disease-causing pathogens. New J. Chem 37, 581–584 (2013).
Bhat, H. R. et al. Antimalarial activity and docking studies of novel bi-functional hybrids derived from 4-aminoquinoline and 1,3,5-triazine against wild and mutant malaria parasites as pf-DHFR inhibitor. RSC Adv 3, 2942–2952 (2013).
Singh, U. P., Pathak, M., Dubey, V., Bhat, H. R. & Singh, R. K. Design, synthesis, antibacterial activity, and molecular docking studies of novel hybrid 1,3-thiazine-1,3,5-triazine derivatives as potential bacterial translation inhibitor. Chem Biol Drug Des 80, 572–583 (2012).
Dubey, V., Pathak, M., Bhat, H. R. & Singh, U. P. Design, facile synthesis, and antibacterial activity of hybrid 1,3,4-thiadiazole-1,3,5-triazine derivatives tethered via -S- bridge. Chem Biol Drug Des. 80, 598–604 (2012).
Gahtori, P. et al. Antimalarial evaluation and docking studies of hybrid phenylthiazolyl-1,3,5-triazine derivatives: a novel and potential antifolate lead for Pf-DHFR-TS inhibition. Exp Parasitol 130, 292–299 (2012).
Singh, U. P., Bhat, H. R. & Gahtori, P. Antifungal activity, SAR and physicochemical correlation of some thiazole-1,3,5-triazine derivatives. J MYCOL MED 22, 134–144 (2012).
Domling, A. & Ugi, I. Multicomponent Reactions with Isocyanides. Angew. Chem. Int. Ed. Engl 39, 3168 (2000).
Bienayme, H., Hulme, C., Oddon, G. & Schmitt, P. Maximizing synthetic efficiency: multi-component transformations lead the way. Chem. Eur. J. 6, 3321 (2000).
Orru, R. V. A. & Greef, M.de. Recent advances in solution-phase multicomponent methodology for the synthesis of heterocyclic compounds. Synthesis 1471 (2003).
Tempest, P. A. Recent advances in heterocycle generation using the efficient Ugi multiple-component condensation reaction. Curr. Opin. Drug Discovery Dev. 8, 776 (2005).
Stamos, J., Sliwkowski, M. X. & Eigenbrot, C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J.Biol.Chem 277, 46265–46272 (2002).
Singh, U. P. et al. Synthesis and antibacterial evaluation of series of novel tri-substituted-s-triazine derivatives. Med. Chem. Res 20, 1603–1610 (2011).
Denizot, F. & Lang, R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. lmmunol, Methods 89, 271–277 (1986).
Zuccotto, F., Ardini, E., Casale, E. & Angiolini, M. J. Through the “gatekeeper door”: exploiting the active kinase conformation. Med.Chem 53, 2681 (2010).
Liu, Y. & Gray, N. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol 2, 358 (2006).
Balzano, D., Santaguida, S., Musacchio, A. & Villa, F. A general framework for inhibitor resistance in protein kinases. Chem. Biol 18, 966–975 (2011).
Lowry, O. H., Rosebrough, M. J., Farr, A. L. & Randall, R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem 193, 265–275 (1951).
Yagi, K. Lipid peroxides and human diseases. Chem. Phys. Lipids 45, 337–351 (1987).
Ohkawa, H., Ohishi, N. & Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem 95, 351–358 (1979).
Jiang, Z. Y. & Hunt, J. V. & wolff, S.P. Ferrous ion oxidation in the presence of xylenol orange for detection of lipid hydroperoxide in low density lipoprotein. Anal. Biochem 202, 384–389 (1992).
Kakkar, P., Das, B. & Viswanathan, P. N. A modified spectrophotometric assay of superoxide dismutase. Indian J. Biochem. Biophys 21, 130–132 (1984).
Sinha, A. K. Colorimetric assay of catalase. Anal. Biochem 47, 389–394 (1972).
Rotruck, J. T. et al. Selenium: biochemical role as a component of glutathione peroxidase. Science 179, 588–590 (1973).
Beutler, E. & Kelley, B. M. The effect of sodium nitrite on red cell GSH. Experientia 29, 96–97 (1963).
Omura, T. & Sato, R. the carbon monoxide-binding pigment of liver microsomes. i. evidence for its hemoprotein nature. J. Biol. Chem 239, 2370–2378 (1964).
Habig, W. H., Pabst, M. J. & Jakoby, W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem 249, 7130–7139 (1974).
Carlberg, I. & Mannervik, B. Glutathione reductase. Methods Enzymol 113, 484–490 (1985).
Folch, J., Lees, M. & Sloane, S. G. H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem 226, 497–509 (1957).
Zlatkis, A., Zak, B. & Boyle, G. J. A new method for the direct determination of serum cholesterol. J. Clin. Med 41, 486–492 (1953).
Foster, L. B. & Dunn, R. T. Stable reagents for determination of serum triglycerides by a colorimetric Hantzsch condensation method. Clin. Chem 19, 338–340 (1973).
Falholt, K., Lund, B. & Falholt, W. An easy colorimetric micromethod for routine determination of free fatty acids in plasma. Clin. Chim. Acta 46, 105–111 (1973).
Zilversmit, D. B. & Davis, A. K. Microdetermination of plasma phospholipids by trichloroacetic acid precipitation. J. Lab. Clin. Med 35, 155–160 (1950).
Burnstein, M., Scholnic, M. R. & Mortin, R. Rapid method for the isolation of lipoproteins from human serum by precipitation with polyanions. J. Lipid Res 11, 583–595 (1970).
Friedwalt, W. T., Levy, R. I. & Fredricken, D. S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem 18, 499–502 (1972).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem 30, 2785–91 (2009).
Morris, G. M. & Lim-Wilby, M. Molecular docking. Methods Mol. Biol 443, 365–82 (2008).
Stamos, J. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem 277, 46265–46272 (2002).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–12 (2004).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem 25, 1157–74 (2004).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model 25, 247–60 (2006).
Halgren, T. A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem 17, 490–519 (1996).
Stewart, J. J. P. MOPAC: a semiempirical molecular orbital program. J. Comput. Aided. Mol. Des 4, 1–103 (1990).
Dewar, M. J. S., Zoebisch, E. G., Healy, E. F. & Stewart, J. J. P. AM1: A New General Purpose Quantum Mechanical Molecular Model. J. Am. Chem. Soc 107, 3902–3909 (1985).
Acknowledgements
Authors are thankful to SAIF, Punjab University, India for providing spectroscopic data of compounds synthesized herein and SHUATS, India for necessary infrastructural facilities.
Author information
Authors and Affiliations
Contributions
Conduct of the study, data collection, analysis and interpretation (Srivastava J.K., Bhat H.R., Verma A., Pillai G.G., Singh U.P.); Manuscript design, preparation and review (Singh U.P.). All Authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Srivastava, J.K., Pillai, G.G., Bhat, H.R. et al. Design and discovery of novel monastrol-1,3,5-triazines as potent anti-breast cancer agent via attenuating Epidermal Growth Factor Receptor tyrosine kinase. Sci Rep 7, 5851 (2017). https://doi.org/10.1038/s41598-017-05934-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-05934-5
This article is cited by
-
Lariciresinol Induces Apoptosis and Inhibits Migration-Invasion of Patient-Derived Hypertrophic Scar Fibroblasts via Preventing Accumulation of Collagen
Revista Brasileira de Farmacognosia (2024)
-
Eco-friendly green synthesis of N‑pyrazole amino chitosan using PEG-400 as an anticancer agent against gastric cancer cells via inhibiting EGFR
In Vitro Cellular & Developmental Biology - Animal (2024)
-
Protective effect of avicularin against lung cancer via inhibiting inflammation, oxidative stress, and induction of apoptosis: an in vitro and in vivo study
In Vitro Cellular & Developmental Biology - Animal (2024)
-
A concise review on anti-breast cancer effectiveness of s-triazines through EGFR kinase inhibition
Medicinal Chemistry Research (2024)
-
Quinazoline clubbed 1,3,5-triazine derivatives as VEGFR2 kinase inhibitors: design, synthesis, docking, in vitro cytotoxicity and in ovo antiangiogenic activity
Inflammopharmacology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.