Abstract
Potato virus Y (PVY) is a globally and economically important pathogen of potato, tobacco, tomato and other staple crops and caused significant yield losses and reductions in quality.To explore the molecular PVY-host interactions, we analysed changes in the miRNA and mRNA profiles of tobacco in response to PVY infection. A total of 81 differentially expressed miRNAs belonging to 29 families and 8133 mRNAs were identified. The Gene Ontology (GO) enrichment analyses showed that genes encoding the DNA/RNA binding, catalytic activity and signalling molecules were all significantly enriched. Moreover, 88 miRNA-mRNA interaction pairs were identified through a combined analysis of the two datasets. We also found evidence showing that the virus-derived siRNAs (vsiRNAs) from the PVY genome target tobacco translationally controlled tumor protein (NtTCTP) mRNA and mediate plant resistance to PVY. Together, our findings revealed that both miRNA and mRNA expression patterns can be changed in response to PVY infection and novel vsiRNA-plant interactions that may regulate plant resistance to PVY. Both provide fresh insights into the virus-plant interactions.
Similar content being viewed by others
Introduction
Potato virus Y (PVY) is an economically important pathogen of many crops, with many of its strains differing from one another both in their genomic sequences and in the symptoms they produce in their hosts1,2,3. The PVY genome is a positive-sense, single-stranded RNA molecule consisting of approximately 9700 nucleotides. The PVY genome contains two open reading frames (ORFs). The first ORF is translated into a single polyprotein and then processed into individual mature proteins by the viral proteases. The second—and shorter—ORF is translated as the P3N–PIPO fusion protein which is embedded within the P3 cistron of the polyprotein4.
Small RNAs are a group of regulatory molecules that play important roles in diverse biological processes, namely in development, genome maintenance and integrity, and in the adaptive responses to biotic and abiotic stress in most of the eukaryotes. Small RNAs are of two major types: microRNAs (miRNAs) and short interfering RNAs (siRNAs), both function by suppressing the expression of target genes at the transcriptional and/or post-transcriptional level via specific base-pairing with their targets5. As a key compontent of the eukaryotic gene regulatory networks, miRNA has attracted increasing attention with respect to its biogenesis and mechanisms of miRNA-mediated gene regulation6,7,8. It was reported that some animal cellular miRNAs play important roles in the proess of development and the response to pathogens and stresses6, 7. Many animal viruses can down-regulate the expression level of host miRNAs9,10,11,12,13. Deep sequencing has also identified a few new miRNAs induced only in virus infected-cells14, 15. In Arabidopsis, the activation of antiviral RNAi is accompanied by the production of an abundant class of endogenous siRNAs mapped to the exon regions of more than 1,000 host genes and rRNA. These virus-activated siRNAs are predominantly 21 nucleotides in length, with an approximately equal ratio of sense and antisense strands, and they may confer broad-spectrum antiviral activity16.
As a part of small RNA, virus-derived small interfering RNA (vsiRNA), is abundant during the viral infection in plants. Although double-strand replication intermediates (RIs) could form in the process of virus multiplication, the dsRNA-like secondary structures of single stranded viral RNAs were those that most likely contributed to vsiRNA biogenesis17,18,19,20. Analogous to that of endogenous small RNA, the biogenesis of vsiRNA requires not only Dicer-like (DCL) —especially DCL4 which processes the viral dsRNA transcript into primary vsiRNA21, 22—but also (RNA- dependent RNA polymerase (RDR) and Argonaute (AGO) which produced secondary vsiRNA through amplification23,24,25. Biogenesis of vsiRNA during virus infection indicates that vsiRNA may function in many regulation pathways. For instance, vsiRNA could be recruited by diverse AGOs to form RNA-induced silencing complex (RISC) and target viral genome molecules (including viral RNA and viral DNA) through post-transcriptional gene silencing (PTGS)21, 26, and vsiRNA might also target and down-regulate host transcripts that largely determine the virus symptoms in the host27,28,29,30.
To date, the responses of plants to PVY infection have until now been studied at different levels, ranging from morphological to biochemical, and from proteomic to transcriptomic and metabolic31,32,33,34,35. Nevertheless, our knowledge of how plants respond to PVY infection remains rather limited. Hence, further investigation is warranted to fully explore the plant-virus interaction dynamics behind the appearance of disease symptoms and the plant resistance processes.
To acquire a better understanding of how the transcriptome changes in response to viral infection in tobacco, we used high-throughput sequencing technology to simultaneously analyse the miRNA and mRNA expression profiles in virus-infected tobacco plants. We combined these two datasets and identified miRNA-mRNA interactions under PVY infection. We also found that the vsiRNAs from PVY target tobacco NtTCTP mRNA and mideate plant resistance to PVY infection.These integrated high-throughput expression data provide a new and valuable resource for examining global genome expression changes in response to PVY infection. This may contribute to viral symptom development and thereby provide new insights into plant-virus interactions.
Results
Construction and deep sequencing of small RNA and mRNA libraries
To profile the global small RNA and mRNA changes via deep sequencing, the total RNAs were isolated from the PVY-inoculated (three biological replicates: A1, A2, A3) and mock-inoculated (three biological replicates: B1, B2, B3) tobacco plants, and used to construct small RNA and transcriptome libraries. All the data generated by this deep sequencing exercise were uploaded to the SRA database (Accession number: SRP090053).
Small RNA sequencing generated approximately 1 million raw reads from each library. After the 5′ and 3′ adaptors were identified and removed from the raw reads, those reads with a sequence length of 18 to 30 nt were selected for further analysis (Table 1). The distribution of small RNAs among the different categories is summarized in Fig.1. In the PVY-infected plants, 21 nt and 22 nt are the most frequent sizes. By contrast, in the mock-inoculated plants, the most frequent small RNAs were 21 nt and 24 nt in length. More than 75% of the small RNA sequences that had a perfect match to the tobacco genome sequence (https://solgenomics.net) were obtained from each library. In the small RNA libraries of the PVY-infected tobacco plants, a similar percentage of high-quality read and unigene were found well matched to the PVY genome.
Approximately 7G of clean reads were obtained from each mRNA library. The mapping rate of all three PVY-infected libraries (A1, A2, A3) was 77–78%, while it was slightly higher for the mock-inoculated libraries (B1, B2, B3) at 84–89%. Meanwhile, the multi-map rate of the samples was less than 0.1% (Table 2). The gene expression density of all the samples showed a high similarity: there were few differential genes, and these usually would not change the overall gene distribution (Fig. 2A). Both the PVY-infected samples and mock-inoculated samples showed good correlations based on the Hierarchical cluster analysis (Fig. 2C).
Differentially expressed mRNAs in PVY-infected and mock-inoculated tobacco plants. (A) Gene density of mRNAs in the PVY-infected (three biological replicates: A1, A2, A3) and mock-inoculated (three biological replicates: B1, B2, B3) tobacco RNA libraries; (B) Total numbers of up-regulated and down-regulated DEGs; (C) Differentially expressed mRNAs in PVY-infected (A1, A2, A3) and mock-inoculated (B1, B2, B3) tobacco. Every row shows a different gene. Red, black and green indicates expression levels of mRNA slow, medium and high, respectively.
All the data showed good agreement among the replicates. Therefore, the data derived to form high quality small RNA and mRNA libraries were deemed robust for further analysis.
PVY infection selectively altered the expression of tobacco miRNAs
To speculate on how PVY affects the expression of miRNAs, we compared the total reads of the tobacco miRNAs among the six libraries using pooled data from the three independent biological replicates. The analysis of the miRNA data showed that 322 miRNAs were detected in the PVY-infected and mock-inoculated tobacco plants (Supplementary Table S1), However, only 81 miRNAs changed their expression level significantly in the PVY-infected tobacco, of which 24 miRNAs were down-regulated and 57 miRNAs were up-regulated (Supplementary Fig. S1A, Supplementary Table S2). This suggested that some miRNA, or the locus encoding the miRNA precursor, could respond to the PVY infection. To verify the accuracy of the miRNA alterations by data calculation, several characteristic miRNAs, such as mi156g and mi168a, were selected for confirmation by the northern blot method. These results showed that the differences in the heat map were consistent with those of the northern blot (Supplementary Fig. S1B,C).
To obtain those genes that possibly regulated by the differentially expressed miRNAs, targets of the miRNAs were predicted by using psRobot software (Supplementary Table S3). To confirm the predicted results, we sequenced a degradome library constructed using the total RNA isolated from the leaf of PVY-infeted tobacco plants. Over 9.9 million of raw reads were thus generated (Accession number: SAMN06844094). The degradome sequencing datasupported the predicted results for the miRNA targets (Supplementary Table S4). GO enrichment analysis of the target genes of significantly differentially-expressed miRNAs (P adj < 0.05 and |log2(fold_change)| ≥ 1) among the paired group samples was performed. The ensuing GO categorization of the predicted targets of the differentially expressed miRNAs showed that these genes were involved in a broad range of biological processes related to cellular responses to various stimuli; namely, the positive regulation of cellular processes, the negative regulation of growth, the nucleic acid binding and the transcription factors requied for signal transduction (Fig. 3A,B,C).
Global mRNA expression profiles of tobacco in response to PVY infection
To further study the tobacco gene expression profiles responsive to PVY infection, the total RNA samples used for small RNA sequencing were subjected to transcriptome sequencing. Analysing this deep sequencing revealed that a total of 8133 genes were significantly altered by PVY infection, of which 3790 were up-regulated and 4343 were down-regulated (Fig. 2B, Supplementary Table S5). To confirm the RNA sequencing data, we selected 10 up-regulated genes, 2 down-regulated genes and 1 gene that was not changed via qRT-PCR to check their expression patterns compared with RNA-seq data, the qRT-PCR results were consistent with the deep sequencing data (Supplementary Fig. S2). To understand the putative roles of the significantly altered genes, GO analyses were conducted to discover their relevance to diverse biological processes, molecular functions and cellular components. Genes in PVY-infected plant were found to be enriched in metabolic and stress response processes (Fig. 4A,B,C).
Combined analysis of miRNA and mRNA expression network under PVY infection
To explore the miRNA and mRNA expression networks in the PVY-infected tobacco plants, data on the miRNA and mRNA expression profiles were combined for a further correlation analysis. It is well known that miRNAs play pivotal roles in regulating mRNA expression. Correlation analysis of the 81 differentially-expressed miRNAs and the 8133 differentially-expressed mRNAs identified a total of 88 interaction pairs of miRNA and its corresponding targets mRNA (Supplementary Table S6). For each such pair, a one-to-one correspondence between up-regulated miRNA (or down-regulated miRNA) and down-regulated target mRNA (or up-regulated target mRNA) was not necessarily expected. So, to directly demonstrate the relationship between differentially-expressed miRNAs and mRNAs, we then classified these relationships into two categories depending on their regulation mode: either as positive or negative, respectively, for up-regulated (down-regulated) miRNAs versus up-regulated (down-regulated) target genes (Fig. 5A, Supplementary Table S7) or up-regulated (down-regulated) miRNAs versus down-regulated (up-regulated) target genes (Fig. 5B, Supplementary Table S8). The results showed that in addition to the repression caused by miRNA, other mechanisms such as ceRNA regulation might be involved. These data collectively suggested that the interactions between mRNA and miRNA are very complicated in the tobacco plant.
Combined analysis of miRNA and mRNA expression network under PVY infection.(A) Positive regulation of miRNAs versus their target genes; (B) Negative regulation of miRNAs versus their target genes.The triangle represents mRNA, circle represents miRNA, green and red represent down regulate and up regulate, respectively.
PVY-derived siRNA targets the NtTCTP mRNA and mideate plant resistance to PVY infection
To further investigate the plant response to PVY infection, we analysed the PVY-derived siRNAs and their relationships with the tobacco mRNAs. Computer-assisted analyses identified several vsiRNAs derived from the PVY genome that were complementary to the mRNA sequence of the NtTCTP gene (Nicotiana tabacum Translationally Controlled Tumor Protein, mRNA_68091) ORF (Fig. 6A). The most abundant vsRNA that targets NtTCTP was quantified by Northern blotting (Supplementary Fig. S3). Subsequent analysis of the degradome sequencing data also confirmed that the NtTCTP was excised in the PVY-infected plants, thus suggesting that vsiRNAs from PVY may down-regulate NtTCTP expression such that NtTCTP may also have an important role in plant-PVY interactions.
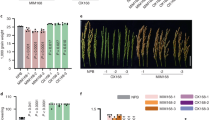
Virus-derived siRNA (vsiRNA) from PVY targets NtTCTP mRNA and mideates plant resistance to PVY infection.(A) VsiRNA from PVY targets NtTCTP mRNA, x-axis represents the read counts of degradome sequencing for the genes that predicted as the targets of vsiRNA from PVY genome, y-axis represents vsiRNA reads from PVY genome;(B) The incidence rate of NtTCTP overexpressing lines O2, O7,wild type and NtTCTP silencing lines Ri16, Ri20; (C) Typical symptoms of NtTCTP overexpressing lines O2, O7,wild type and NtTCTP silencing lines Ri16, Ri20.(D) Virus concentration of healthy plant (H), NtTCTP overexpressing lines O2, O7,wild type and NtTCTP silencing lines Ri16, Ri20 detected by ELISA.
To test this hypothesis, we conducted a PVY infection assay with two NtTCTP-overexpressing lines (NtTCTP-OE: O2 and O7) and two NtTCTP-silencing lines (NtTCTP-RNAi: Ri16 and Ri20) as part of a further analysis36. Seedlings of O2, O7, Ri16, Ri20 and the wild type (variety Xanthi) were challenged with PVY at the four-leaf stage of development. After 2 weeks, PVY-induced symptoms in the systemic leaves of the wild type and in the two NtTCTP-overexpressing lines showed greater sensitivity when compared with the wild type seedlings. In contrast, the two NtTCTP-silencing lines showed high resistance to PVY infection and they did not display any obvious symptoms (Fig.6B,C). ELISA consistently detected high PVY tires in the wild type and NtTCTP-overexpressing lines, whereas PVY was hardly detected in the two NtTCTP-silencing lines (Fig. 6D). These results suggest that PVY-plant interaction involved NtTCTP which acted as a susceptibility factor to promote the PVY infection.
A prior study reported that NtTCTP interacts and stabilizes the ethylene receptor NtHK1 to reduce the plant response to ethylene and to promote plant growth through accelerated cell proliferation36. To see whether NtHK1 also participated in the tobacco plant reponse to the PVY infection, we tested several NtHK1-overexpressing (NtHK1-OE:16-4) and NtHK1-silencing (NtHK1-RNAi:1–8) lines. However, no apparent differences were found among the NtHK1-overexpressing, the NtHK1-silencing and the wild type plants after inoculation with PVY. All of those plants showed typical symptoms and their incidence rate and virus content were almost the same across the three lines (data not shown), which together suggested that NtHK1 had no function in the PVY infection of tobacco. Nonetheless, these results did demonstrate that the involvement of NtTCTP in the PVY-plant interaction did not occur via the ethylene pathway.
Discussion
High throughput sequencing approaches have become powerful tools for analysing global gene expression profiles and for identifying low-abundance novel miRNAs unidentifiable by traditional cloning and sequencing techniques37,38,39. Global expression profiling analysis of miRNAs and mRNAs in the same samples can provide a unique opportunity to enhance our understanding of potential miRNA regulatory mechanisms in host-infection by virus.
RNA-seq analyses have been done for PVY-infected potato plants40, with these studies finding that several genes were expressed differently between the susceptible and resistant varieties. By way of comparison, our sequencing results provide a detailed view of miRNAs and mRNAs expression in tobacco leaf in response to PVY infection, thus adding new information to better understand the virus-host interactions as well as offering novel insights into the impact of viral infection on host small RNA and mRNA exprsssion.
In general, miRNA accumulation will lead to the down-regulation of corresponding mRNA targets, and vice-versa. After an integrated analysis of the differentially-expressed miRNAs and mRNAs, we found several important regulatory miRNAs likely involved in virus infection. For example, miRNA6019a was able to target a “disease resistance protein” (mRNA_90605), which is considered as one of TIR-NBS-LRR family resistance genes. In the PVY infection process, miRNA6019a was down-regulated (Supplementary Fig. S1B,C) while the amount of its target mRNA 90605 increased correspondingly (Supplementary Table S5). This coordinated activity suggests that miRNA-regulated resistance might be promoted during PVY infection.
When studying vsiRNA, how it performs key roles in antiviral resistance and in host transcripts regulation are the core questions. For example, it was reported that a siRNA derived from CMV satellite RNA could target and silence CHLI, a chlorophyll biosynthetic gene, to induce the symptoms of yellowing in virus-infected plants27, 28. Moreover, a vsiRNA derived from RSV (Rice stripe virus) targets eIF4A mRNA in tobacco and down-regulates its expression, resulting in a phenotype of leaf-twisting, deficient flowers and stunting30. In the present study, we identified several vsiRNAs derived from the PVY genome which could target the host gene NtTCTP. The PVY infection assay on the NtTCTP-silencing and over-expressing transgenic lines showed that silencing NtTCTP suppressed the PVY infection, whereas the over-expression of NtTCTP increased plant susceptibility to the PVY infection. Therefore, it is plausible NtTCTP encodes a host factor that is essential to the PVY infection process, not unlike for other host factors recently reviewed41.
TCTP is a highly conserved protein present in all eukaryotes. Its mammalian homologs are perhaps the best studied due to their role in cancer development. TCTP is an important component of the TOR (target of rapamycin) signalling pathway, the major regulator of cell growth in both animals and fungi. Though many studies have revealed that TCTP is involved in cell cycle progression, cell growth, stress protection, maintenance of genomic integrity and apoptosis42, its molecular function remains elusive. Recently, TCTP was suggested as an important host factor in the Pepper yellow mosaic virus (PepYMV) infection of tomato and Nicotiana benthamiana plants. This particular virus interferes with the subcellular localization of this protein, probably due to the involvement of TCTP at some crucial stage of the infection process43. Tobacco TCTP (NtTCTP) encodes a small ER-located protein containing 168 amino acids, and the transcripts of NtTCTP were more abundant in roots than in the other plant organs36. Finally, our research found that tobacco NtTCTP was a target of vsiRNA from the PVY genome and that the silencing of NtTCTP mediated resistance against PVY.
As our primal expectation, the content of PVYN should accumulate increasingly in NtTCTP-silencing lines because vsiRNA derived from PVY targeted to NtTCTP, but the opposite results showed that NtTCTP was required for PVY infection. Why does the virus generate the vsiRNA which has adverse effect on itself? We speculated that targeting of NtTCTP by vsiRNA is a host strategy resistant against PVY infection, but the function of these vsiRNA could be suppressed to some extend by some else mechanism. It might be an episode of host-virus competition series. Further research is planned towards determining the functional role of TCTP in viral infection, and to test whether this gene can be manipulated against PVY, PepYMV and related viruses.
Conclusion
We described the miRNA and mRNA expression profiles in virus-infected tobacco plants by using high-throughput sequencing technology. Combining these two datasets we identified an network consisting of 88 miRNA-mRNA interactions. In so doing, we further found that vsiRNA from PVY target tobacco NtTCTP mRNA to mediate plant resistance to PVY infection. The integrated high-throughput expression datasets we obtained provides a valuable resource to examine global genome expression changes in plant responses to PVY infection, which should also contribute to viral symptom development. This study thus offers new insights into the pathogenicity mechanisms of PVY and associated plant resistance mechanisms.
Materials and Methods
Plant growth conditions and virus infection
Seeds of tobacco (Nicotiana tabacum) were surface sterilized with a 3% sodium hypochlorite solution, rinsed five times with distilled water, immersed in distilled water for two days, and then allowed to germinate for another 2 days at 37 °C. Seedlings were grown in a medium of half-strength growth nutrients under a 16-h light (28 °C)/8-h dark (25 °C) photoperiod. The PVYN (tobacco veinal-necrotic strain) was maintained in potato in green houses at 25 ± 3 °C, 60 ± 5% relative humidity under natural sunlight. Seedlings were challenged with PVY at the four-leaf stage. Two weeks later, when the virus induced symptoms appeared in the systemic leaves of the PVY-infected plants, leaves from the PVY-infected plants (three biological replicates: A1, A2, A3) as well as the mock-inoculated plants (three biological replicates: B1, B2, B3) were collected and immediately frozen in liquid nitrogen until subsequent use.
RNA isolation and sequencing
Total RNA was extracted using the phenol-chloroform method.All samples were assessed for integrity and population size using the Agilent 2100 Bioanalyzer. The concentration and purity of each RNA sample was measured using the Nanodrop spectrophoto meter (Thermo Scientific). Small RNA and mRNA library preparation and sequencing were performed using Small RNA Sample Preparation Kit (Illumina, RS-200-0048) and NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (#E7530L, NEB, USA) following the manufacturer’s recommendations.
Analysis of miRNA and mRNA sequencing data
The filtered sequence of all the samples were mapped to tobacco genome (http://solgenomics.net/) by bowtie144, 45 with –v –a -best, that is, align reads to tobacco genome without any mismatch (because of the short length of miRNA) and the all best alignments were retained for follow-up analysis. The mapped sRNA sequence would be used for the known miRNA, Rfam, repeat and some other annotation. Tobacco miRNA from miRBase 21 was used for known miRNA reference sequence. Rfam 12.0 was used for ncRNA annotation. The unannotated sRNA was used for novel miRNA prediction by miRDeep-P software.To get the differential miRNA for both known miRNA and novel miRNA, DESeq software was used. MiRNAs which satisfied FDR ≤ 0.05 and log2|fold change| > = 1 were considered as significantly differential expressed miRNAs (DEM) between the two groups.Target genes of the DEMs were predicted by software psRobot.
Libraries of RNA were sequenced on the Illumina Hiseq 2500 platform and sequencing reads that contained polyA/T and adapters were discarded, and reads with low quality and high Ns were pre-filtered before mapping, too. Filtered reads were mapped to the reference Tobacco genome sequence (http://solgenomics.net/) by using Tophat with the default parameters.HTseq is applied to generate gene counts. Then gene expression difference was analyzed by DEseq, genes with |log2FoldChange| ≥ 1 and p-value < 0.05 were determined to be statistically significant.
Function and pathway enrichment were analyzed depending on the Gene Ontology database (http://geneontology.org/). The GO term with a q-value lower than 0.05 is determined to be enriched significantly.
Quantitative reverse transcription real-time PCR analysis
Quantitative reverse transcription real-time PCR (qRT-PCR) was performed using a SYBR Real-time PCR Detection System (MJ Research, Waltham, MA, USA) following the manufacturer’s instructions. Each reaction was prepared in a total volume of 20 µl containing 10 µl SYBR Green Mix (Takara), 1.5 µl of diluted cDNA (corresponding to 1.5 ng of reverse-transcribed total RNA) and 0.2 µl of each primer (200 nM working concentration). The reactions were subjected to an initial denaturation step of 95 °C for 10 s, followed by 35 cycles of 95 °C for 5 s, 60 °C for 30 s and 72 °C for 10 s. Each sample was prepared in triplicate. Each sample was prepared in triplicate.The qRT-PCR primers used in this study were list in Supplementary Table S9.
Northern blot hybridization
MiRNA northern blot hybridization was performed as described46 by Guo et al. with 30 micrograms of sRNA loaded for each sample. DNA oligonucleotides complementary to miRNA sequences were end-labeled with r-32P-ATP (5000 Ci mmol−1) using T4 polynucleotide kinase (NEB, Beijing, China) as the probe. Membranes were hybridized for 16 h at 42 °C and were briefly air-dried and were exposed to X- ray film at −80 °C.
Target prediction of the differentially expressed miRNAs (DEMs) and degradome sequencing
To predict the target genes of the DEMs, a PsRobot was used. This tool identifies a particular set of sRNAs with stem-loop shaped precursors (such as microRNAs and short hairpin RNAs), as well as their target genes or transcripts, especially in plants. It predicts their targets using a modified Smith–Waterman algorithm. This program was performed with –ts 2 –gl 10 setting. That means when aligned with the reference genes, those DEMs at position after 10 was allowed at most one gap or bulge, target penalty score should be lower than 2. Mismatches, gaps or bulges are evaluated with a penalty of plus 1, while the G:U pairs are evaluated with a penalty of plus 0.547.
The degradome library was constructed as previously described48 by using the RNA of the A1 treatment as the core material. Firstly, the RNA fragments with poly (A) tail were isolated from the total RNA by using the Oligo texm RNA mini kit (Qiagen), Secondly, a 5′ RNA adapter with a Mme I restriction site at its 3′ end was added to the 5′ ends of the isolated poly(A) RNAs. Thirdly, reverse transcription PCR using oligod (T) as the primer was performed and the PCR products were purified and digested with Mme I. After ligating a double-stranded DNA adapter to the 3′ end of the digested products, the ligated products were further purified and amplified, and then sequenced using the Illumina GAII platform.
References
Gray, S. et al. Potato virus Y: An evolving concern for potato crops in the United States andCanada. Plant Disease 94, 1384–1397 (2010).
Karasev, A. V. & Gray, S. M. Continuous and emerging challenges of potato virus Y in potato. Annual Review of Phytopathology 51, 571–586 (2013).
Nie, X., Singh, R. P. & Singh, M. Molecular and pathological characterization of N:O isolates of the Potato virus Y from Manitoba, Canada. Canadian Journal of Plant Pathology 26, 573–583 (2004).
Ivanov, K. I. et al. Molecular insights into the function of the viral RNA silencing suppressor HCPro. Plant Journal for Cell & Molecular Biology 85, n/a–n/a (2015).
Zamore, P. D. & Haley, B. Ribo-gnome: The Big World of Small RNAs. Science 309, 1519–1524 (2005).
Bartel, D. P. M. R. N. A. genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Mallory, A. C. & Vaucheret, H. Erratum: Functions of microRNAs and related small RNAs in plants. Nature Genetics 38(Suppl), e471–e471 (2006).
Willmann, M. R. & Poethig, R. S. Conservation and evolution of miRNA regulatory programs in plant development. Current Opinion in Plant Biology 10, 503–511 (2007).
Sinclair, S. J., Murphy, K. J. & Birch, C. D. Molecular characterization of quinolinate phosphoribosyltransferase (QPRTase) in Nicotiana. Plant Molecular Biology 44, 603–617 (2000).
Sinclair, S. J., Johnson, R. & Hamill, J. D. Analysis of wound-induced gene expression in Nicotiana species with contrasting alkaloid profiles. Functional Plant Biology 31, 721–729 (2004).
Deboer, K. D., Lye, J. C. & Aitken, C. D. The A622 gene in Nicotiana glauca (tree tobacco): evidence for a functional role in pyridine alkaloid synthesis. Plant Molecular Biology 69, 299–312 (2009).
Frazier, T. P., Xie, F., Freistaedter, A., Burklew, C. E. & Zhang, B. Identification and characterization of microRNAs and their target genes in tobacco (Nicotiana tabacum). Planta 232, 1289–1308 (2010).
Kim, H. J., Baek, K. H., Lee, B. W., Choi, D. & Hur, C. G. In silico identification and characterization of microRNAs and their putative target genes in Solanaceae plants. Genome 54, 91–98 (2011).
Chen, H. M. & Weigel, D. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proceedings of the National Academy of Sciences 107, 15269–15274 (2010).
Howell, M. D. et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19, 926–942 (2007).
Cao, M. et al. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proceedings of the National Academy of Sciences 111, 14613–14618 (2014).
Donaire, L. et al. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392, 203 (2009).
Szittya, G. et al. Structural and Functional Analysis of Viral siRNAs. Plos Pathogens 6, e1000838 (2010).
Wang, X. B. et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proceedings of the National Academy of Sciences 107, 484–489 (2010).
Zhang, C., Wu, Z., Li, Y. & Wu, J. Biogenesis, Function, and Applications of Virus-Derived Small RNAs in Plants. Frontiers in Microbiology 6, 50–69 (2015).
Fusaro, A. F. et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. Embo Reports 7, 1168–1175 (2006).
Garcia-Ruiz, H. et al. Arabidopsis RNA-Dependent RNA Polymerases and Dicer-Like Proteins in Antiviral Defense and Small Interfering RNA Biogenesis during Turnip Mosaic Virus Infection. Plant Cell 22, 481 (2010).
Voinnet, O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends in Plant Science 13, 317–328 (2008).
Mallory, A. C. & Vaucheret, H. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. Embo Reports 10, 521 (2009).
Mallory, A. & Vaucheret, H. Form, function, and regulation of ARGONAUTE proteins. The Plant Cell 22, 3879–3889 (2010).
Moissiard, G. & Voinnet, O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proceedings of the National Academy of Sciences of the United States of America 103, 19593–19598 (2006).
Shimura, H. et al. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. Plos Pathogens 7, 202–221 (2011).
Smith, N. A., Eamens, A. L. & Wang, M. B. Viral Small interfering RNAs target host genes to mediate disease symptoms in plants. Plos Pathogens 7, e1002022 (2011).
Navarro, B. et al. Small RNAs containing the pathogenic determinant of a chloroplast‐replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant Journal for Cell & Molecular Biology 70, 991–1003 (2012).
Shi, B. et al. Identification and regulation of host genes related to rice stripe virus symptom production. New Phytologist 209, 95–102 (2016).
Kogovšek, P. & Ravnikar, M. Physiology of the Potato–Potato Virus Y Interaction. ProgressinBotany, 101–133 (2013).
Goyer, A., Hamlin, L., Crosslin, J. M., Buchanan, A. & Chang, J. H. RNA-Seq analysis of resistant and susceptible potato varieties during the early stages of potato virus Y infection. BMC Genomics 16, 1–13 (2015).
Baebler, Š. et al. Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Yinfection in potato. Journal of Experimental Botany 65, 1095–1109 (2014).
Stare, T., Ramšak, Ž., Blejec, A., Stare, K. & Turnšek, N. Bimodal dynamics of primarymetabolism-related responses in tolerant potato-Potato virus Y interaction. BMC Genomics 1763, 290–295 (2015).
Kogovšek, P. et al. Primary Metabolism, Phenylpropanoids and Antioxidant Pathways Are Regulated in Potato as a Response to Potato virus Y Infection. Plos One 11, e0146135 (2015).
Tao, J. J. et al. Tobacco translationally controlled tumor protein interacts with ethylene receptor tobacco histidine kinase1 and enhances plant growth through promotion of cell proliferation. Plant Physiology 169, 96–114 (2015).
Hsieh, L. C. et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiology 151, 2120–2132 (2009).
Lelandaisbrière, C. et al. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21, 2780–2796 (2009).
Zhang, J., Xu, Y., Huan, Q. & Kang, C. Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10, 449 (2009).
Goyer, A., Hamlin, L., Crosslin, J. M., Buchanan, A. & Chang, J. H. RNA-Seq analysis of resistant and susceptible potato varieties during the early stages of potato virus Y infection. BMC Genomics 16, 472 (2015).
Wang, A. M. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annual Review of Phytopathology 53, 45–66 (2015).
Bommer, U. A. Cellular function and regulation of the translationally controlled tumour protein TCTP. Open Allergy Journal 5, 19–32 (2012).
Bruckner, F. P. et al. Translationally controlled tumor protein (TCTP) from tomato and Nicotiana benthamiana is necessary for successful infection by A Potyvirus. Molecular Plant Pathology (2016).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, 1–10 (2009).
Yang, X., Zhang, H. & Li, L. Global analysis of gene-level microRNA expression in Arabidopsis using deep sequencing data. Genomics 98, 40–46 (2011).
Guo, Y. et al. Identification and characterization of miRNAome in tobacco (Nicotiana tabacum) by deep sequencing combined with microarray. Gene 501, 24 (2012).
Schwab, R. et al. Specific effects of microRNAs on the plant transcriptome. Developmental Cell 8, 277–284 (2005).
German, M. A., Luo, S., Schroth, G., Meyers, B. C. & Green, P. J. Construction of parallel analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nature Protocol 4, 356–362 (2009).
Acknowledgements
We thank Prof. Xiangdong Li (Shandong Agricultural University, China) and members of our lab for helpful discussions. We also would like to acknowledge Prof. Jinsong Zhang (State Key Lab of Plant Genomics, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing all the transgenic lines of tobacco TCTP and HK1 genes. This work was financially supported by the Natural Science Foundation of China (31360431), Guizhou tobacco company project (Grant No. 2015-04), the Natural Science Foundation of Guizhou province of China (qiankehe J[2014] 2117, 2118), the Doctorate staff Foundation at the Guizhou Normal University (000-030111037024).
Author information
Authors and Affiliations
Contributions
M.A.J. and I.H.S. conceived and designed the experiments, Y.S.G., M.A.J., Y.M.Y., L.C.L., L.L.Z., X.F.C., J.Y.C. and J.Z. conducted the experiments and analysed the data, Y.S.G., M.A.J, J.Y., T.L., Q.F. I.H.S and J.H.Z. wrote the paper. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, Y., Jia, Ma., Yang, Y. et al. Integrated analysis of tobacco miRNA and mRNA expression profiles under PVY infection provids insight into tobacco-PVY interactions. Sci Rep 7, 4895 (2017). https://doi.org/10.1038/s41598-017-05155-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-05155-w
This article is cited by
-
Identification of miRNA Targets by AtFT Overexpression in Tobacco
Plant Molecular Biology Reporter (2020)
-
Integrated mRNA and microRNA transcriptome analyses provide insights into paclobutrazol inhibition of lateral branching in herbaceous peony
3 Biotech (2020)
-
High-degree and broad-spectrum resistance mediated by a combination of NIb siRNA and miRNA suppresses replication of necrotic and common strains of potato virus Y
Archives of Virology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.