Abstract
Bacillus thuringiensis (Bt) Cry toxins play an important role in the management of insect pests. Resistance to Bt toxins has been reported in many pest insects but the mechanism responsible for this resistance in rice crop pests remains largely unknown. Cadherin is one of several Bt toxin receptors. At present, only one cadherin gene, CsCAD1, has been documented in the striped rice stem borer, Chilo suppressalis. We amplified a nearly full-length transcript of another C. suppressalis cadherin gene, CsCAD2, and found that it has a different expression pattern to CsCAD1. CsCAD1 was highly expressed in fifth and sixth instar larvae, especially in the midgut, while the expression levels of CsCA2 were equably in each developmental stage. Newly hatched larvae were fed on rice smeared with synthesized siRNA to knockdown either CsCAD1 or CsCAD2, and then were fed transgenic rice expressing either the Cry2A or Cry1C toxins. The siRNA-treatment groups had lower mortality and higher survival rates than the control group, suggesting that reduced expression of CsCAD1 or CsCAD2 increased resistance to Cry2A and Cry1C. We conclude that CsCAD1 and CsCAD2 interact with Bt toxins in C. suppressalis and that this interaction could be the mechanism underlying Bt resistance in this insect.
Similar content being viewed by others
Introduction
Insecticidal Cry proteins from Bacillus thuringiensis (Bt) have been widely used to develop transgenic crops that have become an important part of agricultural pest management1,2,3. Cry toxins are ingested by digestive proteases in the midgut of insects where the activated toxins interact with midgut brush border membrane proteins, including cadherin4, ABC type C transporters (ABCCs)5, 6, alkaline phosphatase (ALP)7, 8, and aminopeptidase N (APN)8, 9. Cry toxins are integrated into the membrane, leading to pore formation, cell lysis and insect death10. However, the development of resistance to Bt toxins in many pest insects threatens to make transgenic Bt crops redundant11.
The development of resistance to Bt toxins in insects has been associated with mutation, down-regulation, or deletion, of Bt receptors2, 12, 13. Cadherin, a calcium-dependent cell adhesion protein14, is thought to be one of several such receptor proteins that bind to Cry toxins15, 16. The first such cadherin protein to be identified was the Cry1A toxin-binding protein in Manduca sexta 17, which was then found to be involved in binding Cry toxins in other Lepidopteran, coleopteran and dipteran insects4, 18,19,20,21,22,23. However, the affinity of cadherins for different Cry toxins varies in different insects. Fox example, some Cry toxins are not lethal to the Coleoptera or Lepidoptera24, 25.
The striped rice stem borer, Chilo suppressalis Walker, is one of the most destructive rice pests in China and other Asian countries. Transgenic rice strains expressing the Cry toxins Cry2A and Cry1C have been developed to protect rice crops from this notorious pest. However, it is likely that C. suppressalis will develop resistance to these toxins once transgenic rice becomes more widely grown. It is, therefore, important to understand the mechanisms that confer resistance to Cry toxins in this species.
A cadherin-like C. suppressalis gene (CsCAD1, AY118272) has been deposited in the NCBI GenBank26,27,28. We used Rapid-amplification of cDNA ends (RACE) to clone another C. suppressalis cadherin gene, which we named CsCAD2, and investigated the expression of both genes in different C. suppressalis developmental stages and tissues. We found that knockdown of these genes reduced sensitivity to both Cry2A and Cry1C in C. suppressalis.
Results
Amplifying the CsCAD2 gene
We obtained a fragment of a cadherin gene from the transcriptome of C. suppressalis and, after mapping this fragment onto the C. suppressalis genome29, found that this gene had not previously been reported. We named this newly discovered gene CsCAD2. By amplifying the transcript with RACE and incorporating information from the C. suppressalis genome, we obtained a nearly full-length transcript of CsCAD2, including a 5′ untranslated coding region (UTR), open reading frame (ORF). The CsCAD2 ORF was 4,912 bp, encoding 1,493 amino acids. The CsCAD2 protein sequence had high identity with other insect cadherins, for example 88% with that of Bombyx mori and 86% with that of Plutella xylostella.
Gene structure and phylogenetic analysis of CsCAD1 and CsCAD2
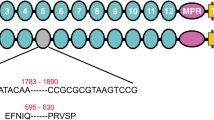
The nucleic acid sequences of CsCAD1 and CsCAD2 were aligned with the genome of C. suppressalis to obtain the structures of both genes (Fig. 1A). The two genes are located in different scaffolds of the genome. CsCAD1 had 41 exons and 40 introns and a length of 44,762 bp, whereas CsCAD2 had only 24 exons and 23 introns and a length of 316,095 bp. Surprisingly, the first CsCAD2 intron was very long; 253,600 bp. Conserved domain analysis indicates that both CsCAD1 and CsCAD2 have characteristics that are conserved in other cadherin proteins. CsCAD1 had eight cadherin repeat domains, two Ca2+ binding sites and one trans-membrane region, whereas CsCAD2 had seven cadherin repeat domains, five Ca2+ binding sites and one trans-membrane region (Fig. 1B). Comparison of CsCAD1 and CsCAD2 with nine other cadherin genes of three well-studied insects; B. mori, P. xylostella and D. melanogaster, indicated that CsCAD1 was conserved in Lepidopteran. However, CsCAD2 was not clustered with other Lepidopteran CADs but was adjacent with Drosophila CADs, which is worthy of further investigations (Fig. 1C).
Structure (A), predicted domain structure (B) and phylogeny (C), of the Chilo suppressalis cadherin genes CsCAD1 and CsCAD2. CR = characteristic cadherin repeat domains, S = signal peptide, TM = transmembrane domain. The phylogenetic tree of the relationships between CsCAD1 and CsCAD2 and eleven other Lepidopteran and Dipteran cadherin genes (C) was constructed using the neighbor-joining method in MEGA 6.0 with 1000 bootstrap replications. The GenBank accession numbers of the genes compared are AAM78590.1 and AGG36450.1 for C. suppressalis (CsCAD1 and CsCAD2), NP_001036368.2, NP_650554.3, NP_731649.2, NP_788635.3 and NP_731930.2 for Drosophila melanogaster (DmeCAD-1, DmeCAD-2, DmeCAD-3, DmeCAD-4 and DmeCAD-5), ADD92322, ABU41413.1 and NP_001036368.2 for Plutella xylostella (PxyCAD-1, PxyCAD-2, and PxyCAD-3), and NP_001037682.1 for Bombyx mori (BmoCAD).
Temporal and spatial expression of CsCAD1 and CsCAD2
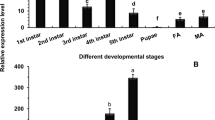
The expression patterns of both CsCAD1 and CsCAD2 were compared using quantitative PCR (qPCR). The house-keeping gene E2F and the G3PDH gene were used as internal controls. qPCR analyses of the expression of CsCAD1 and CsCAD2 in first to sixth instar larvae, and in adults, indicated that CsCAD1 expression peaked in 6th instar larvae and decreased markedly in adults. In contrast, CsCAD2 was expressed in all developmental stages without significant difference (Fig. 2). CsCAD1 expression was almost exclusively confined to the midgut and was detected only at very low levels in other tissues, including the head, epidermis and fat body (Fig. 3A). In contrast, although CsCAD2 expression was also highest in the midgut, it was also highly expressed in the head and epidermis. Furthermore, although CsCAD2 expression was lowest in the fat body, its expression in that organ was nearly half that in the midgut (Fig. 3B). This indicates that although CsCAD1 expression is largely confined to the midgut, CsCAD2 was expressed in all tested tissues. This was consistent with phylogenetic analysis, indicating that CsCAD1 was conserved in Lepidopteran insects, while CsCAD2 was a more ancient protein and conserved in almost all insects.
Knockdown of CsCAD1 and CsCAD2 by RNA interference
In order to investigate the functions of CsCAD1 and CsCAD2 we used siRNAs to silence both genes and a random sequence siRNA without any targets in C. suppressalis as a negative control (siNC). Newly hatched larvae were put into petri dishes containing rice stems smeared with siRNAs and the expression levels of CsCAD1 and CsCAD2 were measured with qPCR after 48 hrs (Fig. 4). The relative abundance of target gene transcripts was 40% of that in the control group, indicating that both target genes were successfully downregulated. The expression of non-target homolog genes was not affected, indicating that RNAi knockdown of CsCAD1 and CsCAD2 was achieved without influencing non-target genes.
Relative expression of the Chilo suppressalis cadherin genes CsCAD1 and CsCAD2 24 hrs after siRNA treatment. Results are means ± SE. Bars with the same lowercase letter are not significantly different; NC = negative control, siCAD1 = CsCAD1 knockdown treatment group, siCAD2 = CsCAD2 knockdown treatment group.
Silencing CsCAD1 and CsCAD2 reduced sensitivity to Bt toxins
The RNAi-treated larvae were transferred to feed on one of three rice varieties; transgenic Cry2A rice, transgenic Cry1C rice, and non-transgenic Minghui 63 (negative control).
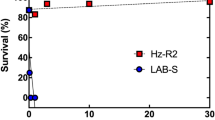
The death rate of the siNC treatment group was about 50% after feeding on transgenic Cry2A rice two days. In contrast, the mortality of the siCAD1 treatment group was just 10%, and that of siCAD2 treatment group only 5%, after two days (p < 0.05, Tukey’s HSD). Over the following three days, significant differences in mortality were observed between the two siRNA treatment groups and the siNC group. Over 70% of the siNC group were dead after 5 days whereas <30% of both siRNA treatment groups died over the same time period (Figs 5A and 6).
COX proportional hazard models for survival analysis; siNC-non-transgenic rice = negative control group feeding on non-transgenic rice, siCAD1-non-transgenic rice = the CsCAD1 knockdown treatment group feeding on non-transgenic rice, siCAD2-non-transgenic rice = the CsCAD2 knockdown treatment group feeding on non-transgenic rice, siNC-Transgenic Cry2A rice = the negative control group feeding on transgenic rice expressing Cry2A; siCAD1-Transgenic Cry2A rice = the CsCAD1 knockdown group feeding on rice expressing Cry2A; siCAD2-Transgenic Cry2A rice = the CsCAD2 knockdown treatment group feeding on transgenic rice expressing Cry2A, siNC-Transgenic Cry1C rice = negative control group feeding on transgenic rice expressing Cry1C, siCAD1-Transgenic Cry1C rice = the CsCAD1 knockdown treatment group feeding on transgenic rice expressing Cry1C, siCAD2-Transgenic Cry1C rice = CsCAD2 knockdown treatment group feeding on transgenic rice expressing Cry1C.
Similar results were obtained in experiments in which larvae were fed transgenic rice expressing the Cry1C toxin. In this case mortality in the siCAD1 and siCAD2 treatment groups were 10% and 20%, respectively, significantly lower than in the NC group (p < 0.05, Tukey’s HSD) (Figs 5B and 6). Interestingly, knockdown of CsCAD2 conferred higher resistance to the Cry2A toxin than knockdown of CsCAD1 (Fig. 5A). Conversely, silencing CsCAD1 conferred higher resistance to the Cry1C toxin than to CsCAD2 (Fig. 5B). This suggests that these two cadherin genes have different binding affinities for different Bt toxins.
Discussion
Phylogenetic analysis of a newly cloned C. suppressalis cadherin gene, CsCAD2, suggests that this is most closely related to D. melanogaster cadherin genes. In contrast to CsCAD1, which was almost exclusively expressed in the midgut, CsCAD2 were highly expressed in a number of different organs. This suggests that these two genes have different functions.
Recent studies have confirmed that Cry1A toxins interact with at least one type of receptor in the midgut30, 31. After the Cry1A toxin has been activated by enzymes in the midgut, it binds to the first receptor (midgut cadherin) with high affinity, which facilitates oligomerization of the toxins via a proteolytic process resulting in cell lysis. Previous studies have already indicated that cadherin is a receptor of Cry toxins in M. sexta, B. mori, Diatraea saccharalis and Heliothis virescens 4, 32. However, most studies have focused on cadherins that are specifically expressed in the midgut. Our results demonstrate that CsCAD2 is both expressed in organs other than the midgut, and that knockdown of this gene increased resistance to both Cry1C and Cry2A. This suggests that this widely expressed gene is also probably a Bt toxin receptor.
We found that silencing either CsCAD1 or CsCAD2 reduced the sensitivity of C. suppressalis to Bt toxins, suggesting that both genes interact with Bt toxins in C. suppressalis. However, their differential expression in different tissues suggests that they could have different functions. This hypothesis is supported by the results of the RNAi experiment. The mortality of the siCAD1 treatment group on transgenic Cry2A rice was slightly higher than that of the siCAD2 treatment group, suggesting that CsCAD2 has higher affinity for Cry2A than CsCAD1. However, when larvae were fed on transgenic Cry1C rice the mortality of the siCAD2 treatment group was higher than that of the siCAD1 treatment group, suggesting that the affinity of CsCAD1 for Cry1C was higher than that of CsCAD2.
Materials and Methods
Insects and rice
C. suppressalis larvae of a strain susceptible to Bt toxins were collected in Wenzhou, Zhejiang province, China. Larvae were raised on rice seedlings in a laboratory at 25 ± 1 °C, 16/8 h light/dark and >80% humidity. The midguts of fourth instar larvae were dissected, immediately frozen in liquid nitrogen and stored at −70 °C until required for RACE.
Two transgenic rice strains, one expressing Cry2A and the other Cry1C, were used in experiments. These strains were derived from the same parental strain, Minghui 63, which was used as the negative control. Rice seeds were soaked for three days and then germinated in petri dishes on wet filter paper. All rice seeds were kindly provided by Prof. Yong-Jun Lin of Huazhong Agricultural University.
Total mRNA isolation and cDNA synthesis
Whole bodies of different developmental stages of C. suppressalis (2nd to 6th instar larvae and adults), and specific body parts (head, midgut, epidermis, fat body), were first homogenized in a tissue grinder. TRIzol reagent (GIBCO, USA) was then used to isolate total mRNA from these samples according to the manufacturer’s protocol. Genomic DNA was removed from total RNA with a DNA-free kit (Ambion, USA). The integrity of the RNA obtained was checked by electrophoresis on a 1.5% agarose gel. The 260/280 nm absorbance ratios of all RNA samples were between 1.8 and 2.2. First strand cDNA was synthesized using M-MLV reverse transcriptase (Takara, Japan) with Oligo (dT18) as the anchor primer. The reaction mixtures were incubated at 70 °C for 10 min followed by 42 °C for one hour and 70 °C for 15 min. The cDNA was stored at −20 °C for further use.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was carried out with a SYBR Premix Ex Taq kit (Takara) using an ABI Prism 7300 (Applied Biosystems, USA) to detect the expression of CsCAD1 and CsCAD2 in the midgut of different development stages, and in different body parts. Primers were designed with Beacon Designer 7 (Table S1) and dissolution curves and gel electrophoresis were used to determine primer specificity. The amplification efficiency of all primers was checked with a cDNA dilution gradient, after which 2 μL of cDNA template was used in the PCR reaction according to the PCR kit’s protocol. The qPCR began at 95 °C for 30 secs, followed by 40 cycles of 95 °C for 5 secs, annealing at 60 °C for 31 sec, ending with cycles of 95 °C for 15 sec, 60 °C for 60 sec, and 95 °C for 15 sec. The specificity of the qPCR reactions was monitored with melting curve analysis using SDS software (version 1.4) and gel electrophoresis. Amplification efficiencies were determined by a series of template dilutions. All experiments were repeated in triplicate. The raw Ct values were obtained using ABI 7300 SDS software (Version 1.4). The standard Delta-Delta-Ct method was used to analysis the qPCR data. The housekeeping genes E2F and GAPDH (GenBank No.: DQ311161.1 and AB262581.1) were used as the internal controls. Significant differences among multiple means were determined using Tukey’s HSD (P < 0.05).
RACE amplification of CsCAD2
Total RNA was extracted from the midgut of the 3rd instar larvae. RACE amplification was carried out with a SMARTer RACE cDNA Amplification Kit (Takara) according to the manufacturer’s protocol. Fragments of CsCAD2 cDNA were obtained from the transcriptome data used in previous studies33. The primers CTCATTACCTCCCTCCCACTCGGCAG (5′ RACE) and TGACAATCCACCACATTTCACGCAGG (3′ RACE) were designed, based on the sequences obtained, to amplify the full-length of the CsCAD2 gene. The end-to-end primers (5′ AAACTTAATAGGCTTACTCGTTCTACC and 3′ GCTGTTCCCTGTCAAATGTCAC) were designed to amplify the full length of the CsCAD2 gene. PCR products were inserted into vector (Takara, Dalian, China) and sequenced by the Nanjing Genscript Company, China. The transcriptome and genome data29 were used to obtain the full-length transcript of CsCAD2. The resultant sequence was submitted to GenBank (Accession No. JQ747493).
RNA interference of cadherins
Two types of siRNA, siCAD1 and siCAD2, were used to silence the CsCAD1 and CsCAD2 genes, respectively, and a random sequence siRNA was included as a negative control (siNC) (Table S2). All siRNAs were synthesized by the GenePharma Company. Larvae were treated with siRNAs in petri dishes with wet filter paper on the bottom. The siRNA was smeared onto 4 cm-long sprouts of non-transgenic rice and about 1000 newly hatched larvae were then put in the petri dishes to feed on the treated rice sprouts. Rice sprouts were replaced every 4 hrs for three days. All experiments were conducted at 27 °C and were repeated in triplicate so that there about 3000 insects were used per treatment and nearly 10 thousand in total.
Larvae from each treatment were used to assess both susceptibility to Bt transgenic rice and investigate gene expression. Susceptibility to transgenic rice was assessed in a randomly selected group of 90 larvae. These were subdivided into three groups of 30 which were randomly assigned to feed on either transgenic rice expressing Cry2A toxin, transgenic rice expressing Cry1C toxin, or non-transgenic rice. For another part, to examine the gene expression change, 50 insects were randomly selected at the 2nd day after the insects were fed on transgenic rice. All experiments were repeated in triplicate.
Gene evolution and domain analysis
Phylogenetic analysis was conducted in MEGA (v6.0) using the Neighbor-joining method with 1000 bootstrap replications. Domain structures of candidate cadherin genes were analyzed using CD Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Signal peptides were predicted with SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) and transmembrane domains with TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). Cox’ proportional hazard model implemented in Program R (R Core Team R version 3.2.3) was used to analyze changes in survival from the 2nd to 5th day after larvae had commenced feeding on transgenic rice plants.
References
Prado, J. R. et al. Genetically engineered crops: from idea to product. Annual review of plant biology 65, 769–790, doi:10.1146/annurev-arplant-050213-040039 (2014).
Pardo-Lopez, L., Soberon, M. & Bravo, A. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS microbiology reviews 37, 3–22, doi:10.1111/j.1574-6976.2012.00341.x (2013).
Gao, Y., Oppert, B., Lord, J. C., Liu, C. & Lei, Z. Bacillus thuringiensis Cry3Aa toxin increases the susceptibility of Crioceris quatuordecimpunctata to Beauveria bassiana infection. J Invertebr Pathol 109, 260–263, doi:10.1016/j.jip.2011.12.003 (2012).
Gahan, L. J., Gould, F. & Heckel, D. G. Identification of a Gene Associated with Bt Resistance in Heliothis virescens. Science 293, 857–860 (2001).
Gahan, L. J., Pauchet, Y., Vogel, H. & Heckel, D. G. An ABC Transporter Mutation Is Correlated with Insect Resistance to Bacillus thuringiensis Cry1Ac Toxin. Plos Genetics 6, e1001248 (2010).
Park, Y. et al. ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biology 12, 1–15 (2014).
Juratfuentes, J. L. & Adang, M. J. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. European Journal of Biochemistry 271, 3127–3135 (2004).
Guo, Z. et al. MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. 11, e1005124 (2015).
Wang, X. Y. et al. RNAi in the striped stem borer, Chilo suppressalis, establishes a functional role for aminopeptidase N in Cry1Ab intoxication. J Invertebr Pathol 143, 1–10, doi:10.1016/j.jip.2016.11.004 (2017).
Adang, M. J., Crickmore, N. & Jurat-Fuentes, J. L. Chapter Two - Diversity of Bacillus thuringiensis Crystal Toxins and Mechanism of Action. (Elsevier Science & Technology, 2014).
An, J., Gao, Y., Lei, C., Gould, F. & Wu, K. Monitoring cotton bollworm resistance to Cry1Ac in two counties of northern China during 2009–2013. Pest Management Science 71, 377–382 (2014).
Jin, L. et al. Large-scale test of the natural refuge strategy for delaying insect resistance to transgenic Bt crops. Nat Biotechnol 33, 169–174, doi:10.1038/nbt.3100 (2015).
Zhang, H. et al. Diverse genetic basis of field-evolved resistance to Bt cotton in cotton bollworm from China. Proc Natl Acad Sci USA 109, 10275–10280, doi:10.1073/pnas.1200156109 (2012).
Hirano, S., Nose, A., Hatta, K., Kawakami, A. & Takeichi, M. Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. 105, 2501–2510 (1988).
Bravo, A. et al. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochimica Et Biophysica Acta 1667, 38 (2004).
Zhang, H. et al. Cadherin mutation linked to resistance to Cry1Ac affects male paternity and sperm competition in Helicoverpa armigera. Journal of Insect Physiology 70, 67–72 (2014).
Vadlamudi, R. K., Weber, E., Ji, I., Ji, T. H. & B. L., Jr. Cloning and Expression of a Receptor for an Insecticidal Toxin of Bacillus thuringiensis. Journal of Biological Chemistry 270, 5490 (1995).
Nagamatsu, Y. et al. Cloning, sequencing, and expression of the Bombyx mori receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin. Bioscience Biotechnology & Biochemistry 62, 727–734 (1998).
Xu, X., Yu, L. & Wu, Y. Disruption of a cadherin gene associated with resistance to Cry1Ac {delta}-endotoxin of Bacillus thuringiensis. Helicoverpa armigera. Applied & Environmental Microbiology 71, 948–954 (2005).
S, M. et al. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proceedings of the National Academy of Sciences 100, 5004 (2003).
Flannagan, R. D. et al. Identification, cloning and expression of a Cry1Ab cadherin receptor from European corn borer, Ostrinia nubilalis (Hübner) (Lepidoptera: Crambidae). Insect Biochemistry & Molecular Biology 35, 33–40 (2005).
Hua, G., Zhang, R., Abdullah, M. A. F. & Adang, M. J. Anopheles gambiae Cadherin AgCad1 Binds the Cry4Ba Toxin of Bacillus thuringiensis israelensis and a Fragment of AgCad1 Synergizes Toxicity. Biochemistry 47, 5101 (2008).
Hua, G., Park, Y. & Adang, M. J. Cadherin AdCad1 in Alphitobius diaperinus larvae is a receptor of Cry3Bb toxin from Bacillus thuringiensis. Insect Biochemistry & Molecular Biology 45, 11–17 (2013).
Tan, S. Y. et al. RNAi induced knockdown of a cadherin-like protein (EF531715) does not affect toxicity of Cry34/35Ab1 or Cry3Aa to Diabrotica virgifera virgifera larvae (coleoptera: chrysomelidae). Insect Biochemistry & Molecular Biology 75, 117 (2016).
Guo, Z. et al. The midgut cadherin-like gene is not associated with resistance to Bacillus thuringiensis toxin Cry1Ac in Plutella xylostella (L.). Journal of Invertebrate Pathology 126, 21–30 (2015).
Hong-Kun et al. Gene cloning and expression of aminopeptidase N and cadherin from midgut of the rice stem borer, Chilo suppressalis. Insect Science 17, 393–399 (2010).
Alcantara, E. P., Aguda, R. M., Curtiss, A., Dean, D. H. & Cohen, M. B. Bacillus thuringiensis -endotoxin binding to brush border membrane vesicles of rice stem borers (p 169–177). Archives of Insect Biochemistry & Physiology 55, 169–177 (2010).
Avisar, D. et al. The Bacillus thuringiensis delta-endotoxin Cry1C as a potential bioinsecticide in plants. Plant Science 176, 315–324 (2009).
Yin, C. et al. ChiloDB: a genomic and transcriptome database for an important rice insect pest Chilo suppressalis. Database the Journal of Biological Databases & Curation 2014, 92–108 (2014).
Bravo, A. et al. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-formingtoxin to aminopeptidase N receptor leading to insertion into membrane microdomains, Biochim. Biophys. Acta 1667, 38–46. Biochimica Et Biophysica Acta 1667, 38–46 (2004).
Pigott, C. R. & Ellar, D. J. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiology & Molecular Biology Reviews Mmbr 71, 255–281 (2007).
Yang, Y. et al. Down regulation of a gene for cadherin, but not alkaline phosphatase, associated with Cry1Ab resistance in the sugarcane borer Diatraea saccharalis. Plos One 6, e25783 (2011).
Ma, W. et al. Correction: Exploring the Midgut Transcriptome and Brush Border Membrane Vesicle Proteome of the Rice Stem Borer, Chilo suppressalis (Walker). Plos One 7, e38151 (2012).
Acknowledgements
This work was partially supported by grants from the National Key Research and Development Program [2016YFC1200600], the Chinese Ministry of Agriculture [2016ZX08001001], the Chinese National Basic Research Program [2013CB127600] and the Ministry of Education Science and Technology Research Project [V201308]. The use of trade names and commercial products in this article does not imply their recommendation or endorsement by the authors.
Author information
Authors and Affiliations
Contributions
Z.Z. analyzed the data, made the figures and tables and wrote the manuscript. X.L.T. carried out experiments and analyzed the data. W.H.M. contributed to the discussion. F.L. designed the project, analyzed the data and also contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Z., Teng, X., Ma, W. et al. Knockdown of two Cadherin genes confers resistance to Cry2A and Cry1C in Chilo suppressalis . Sci Rep 7, 5992 (2017). https://doi.org/10.1038/s41598-017-05110-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-05110-9
This article is cited by
-
Analysis of the Genetic Stability of Insect and Herbicide Resistance Genes in Transgenic Rice Lines: A Laboratory and Field Experiment
Rice (2023)
-
Differences in midgut transcriptomes between resistant and susceptible strains of Chilo suppressalis to Cry1C toxin
BMC Genomics (2020)
-
Cadherin CsCad plays differential functional roles in Cry1Ab and Cry1C intoxication in Chilo suppressalis
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.