Abstract
Kalanchoe daigremontiana reproduces asexually by producing plantlets along the leaf margin. The aim of this study was to identify the function of the SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 gene in Kalanchoe daigremontiana (KdSOC1) during plantlet morphogenesis. In this study, KdSOC1 gene expression was detected at stem cell niche during in vitro somatic embryogenesis and plantlet morphogenesis. Disrupting endogenous auxin transportation suppressed the KdSOC1 gene response. Knockdown of the KdSOC1 gene caused a defect in cotyledon formation during the early heart stage of somatic embryogenesis. Over-expression (OE) of the KdSOC1 gene resulted in asymmetric plantlet distribution, a reduced number of plantlets, thicker leaves, and thicker vascular fibers. Higher KdPIN1 gene expression and auxin content were found in OE plant compared to those of wild-type plant leaves, which indicated possible KdSOC1 gene role in affecting auxin distribution and accumulation. KdSOC1 gene OE in DR5-GUS Arabidopsis reporting lines resulted in an abnormal auxin response pattern during different stages of somatic embryogenesis. In summary, the KdSOC1 gene OE might alter auxin distribution and accumulation along leaf margin to initiate plantlet formation and distribution, which is crucial for plasticity during plantlet formation under various environmental conditions.
Similar content being viewed by others
Introduction
Since auxin was discovered almost 70 years ago1, auxin- and cytokinin-mediated plant growth, including embryogenesis, organ initiation, phyllotaxy, and formation of leaf shape have been studied intensively2. An excellent review regarding auxin and cytokinin as a Yin-Yang model (traditional Chinese model in which Yin and Yang balance everything) treats auxin and cytokinin as key regulators of different plant developmental events3.
The stem cell niche and formation of leaf shape are excellent examples of elucidating how these two hormone signaling pathways and their downstream gene networks regulate changes during multiple phases of plant growth4, 5. Plants harbor two stem cell niches, such as the shoot apical meristem (SAM) and root apical meristem (RAM), which are well-organized structures that regulate above- and below-ground development6. The way in which stem cell fate is regulated in the SAM and RAM by different environmental cues, including photoperiod, temperature fluctuations, and environmental stress is the key mechanism that initiates formation of different organs7, 8. Many master genes have been identified in past decades, including those regulating stem cell identity maintenance in the SAM (WUSCHEL [WUS], SHOOTMERISTEMLESS [STM], and ZWILLE/PINHEAD/AGO10) and RAM (WUSCHEL HOMEOBOX, SHORTROOT, and SCARECROW) for initiating lateral organ formation in both stem cell niches (PINFORMED [PIN1], CUP-SHAPED COTYLEDON [CUC], and PLETHORA). The auxin and cytokinin signaling pathways and downstream networks crosstalk through positive and negative loops that regulate stem cell fate, the SAM and RAM structural patterns, and initiate organogenesis sequences9. “Go with the response” describes how auxin metabolism and transport control a developmental event, particularly during morphogenesis of leaf shape10. The leaf shape formation process is initiated in the SAM and shows how polar transportation of auxin is mediated by the PIN1 gene in association with the CUC2 and ASYMMETRIC LEAVES 1 (AS1) genes, resulting in the oval shape of mature leaves in Arabidopsis 11, 12. Most studies have focused on discovering the molecular clues to further explain the details of the interaction between auxin and cytokinin.
Kalanchoe daigremontiana reproduces asexually to produce plantlets and is regarded as a model plant13 in crassulacean acid metabolism research. Kalanchoe daigremontiana plantlet morphogenesis is the result of somatic embryogenesis and organogenesis. Knockdown of the SHOOTMERISTEMLESS gene in Kalanchoe daigremontiana (KdSTM) abolishes the formation of plantlets by erasing the pedestal site (the central stem cell niche zone) between leaf blades14. An evolutionary mutation in the LEAFY COTYLEDON 1 (LEC1) gene results in aborted seed maturation during zygotic embryogenesis, which is associated with plantlet propagation15. Closely following plantlet morphogenesis revealed similar traits, such as hormone or related gene expression, during the well-elucidated zygote embryogenesis of Arabidopsis, suggesting that auxin- and cytokinin-mediated growth could also be key during plantlet morphogenesis16.
We reported previously that the KdSOC1 gene, which is a member of the MINICHROMOSOME MAINTENANCE 1, AGAMOUS, DEFICIENS, SERUM RESPONSE FACTOR (MADS) box transcription factor family, is highly expressed during plantlet formation under a long day photoperiod17. After years of research on AtSOC1 gene function in Arabidopsis, the AtSOC1 network of genes controlling flowering was elucidated18, 19. However, other studies have reported that SOC1 affects the duration of dormancy rather than stimulates flowering in kiwifruit20. Kalanchoe daigremontiana depends on the formation of plantlets for propagation, suggesting that the KdSOC1 gene might play a novel function during plantlet morphogenesis. In addition, determining whether auxin signaling controls KdSOC1 expression during plantlet morphogenesis would deepen our understanding of this issue.
In this study, the expression patterns of the KdSOC1 gene during in vitro and in vivo somatic embryogenesis and organogenesis were analyzed. The phenotypes and physiological properties after over-expression (OE) and knockdown (RNAi) of the KdSOC1 gene in Kalanchoe daigremontiana transgenic lines were analyzed, including hormone content, relative gene expression profiles, and physiological parameters. In addition, the auxin response pattern and the potential role of the KdSOC1 gene in auxin transport during somatic embryogenesis were also monitored by the DR5 reporter. The goal was to identify KdSOC1 gene function during plantlet morphogenesis.
Results
KdSOC1 gene expression patterns during tobacco callus and shoot induction and plantlet formation in Kalanchoe daigremontiana
The activation of the KdSOC1 gene during plantlet morphogenesis in Kalanchoe daigremontiana raises an interesting question as to why a gene that stimulates flowering would be involved in asexual reproduction. We speculated that the KdSOC1 gene might regulate plantlet morphogenesis through leaf margin somatic embryogenesis. Therefore, we checked the spatial expression of the KdSOC1 gene by deploying GUS gene expression under its own promoter during induction of the tobacco callus (in vitro somatic embryogenesis during induction of the callus is similar to plantlet morphogenesis in Kalanchoe daigremontiana) and plantlet morphogenesis in Kalanchoe daigremontiana.
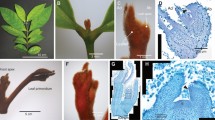
Interestingly, substantial KdSOC1 gene expression was detected in the entire early domed-like somatic embryo (similar to globular stage during zygotic embryogenesis) of the tobacco leaf callus (Fig. 1c). However, during the callus shooting stage before the cotyledon extends (similar to the heart stage of zygotic embryogenesis), the GUS protein stained areas were focused in the margin and center of the SAM of the tobacco somatic embryo (Fig. 1f). When the cotyledon and hypocotyl extended, KdSOC1 gene expression was detected within the SAM area of the newly formed tobacco plantlet (Fig. 1g–i). Taken together, the KdSOC1 gene was expressed in the globular to heart stages of tobacco somatic embryogenesis and was mainly localized to the SAM.
KdSOC1 gene expression profiles during somatic embryogenesis in tobacco callus induction by GUS staining. (a–c) Callus of tobacco somatic embryogenesis. (d–f) Shooting stage of tobacco somatic embryogenesis. (g–i) Mature plantlet regenerated from tobacco callus. Red arrow indicates the GUS staining site of KdSOC1 gene. SAM: shoot apical meristem, CL: cotyledon. HL: hypocotyls.
The KdSOC1 gene expression profiles during Kalanchoe daigremontiana plantlet morphogenesis were also analyzed with the same GUS protein staining method. KdSOC1 gene expression was detected at the center between two leaf serrations when leaf serrations began to form along the leaf margin of Kalanchoe daigremontiana (Fig. 2b). After the pedestal site (which forms from the two leaf serrations and harbors the “stem cell niche” to initiate plantlet formation) appeared with the torpedo-like stage somatic cell, KdSOC1 gene expression vanished from the entire pedestal site region (Fig. 2d). Because of the unequal growth rate of plantlets, which formed sequentially from the tip side to the bottom side of the leaf, KdSOC1 gene expression was only detected between leaf serration where no pedestal site formed (Fig. 2f, red arrow).
Taken together, the KdSOC1 gene was expressed within the SAM area before the late heart stage of tobacco callus or before the formation of the Kalanchoe daigremontiana pedestal site, but expression decreased sharply after the cotyledon expanded in tobacco shoot formation and Kalanchoe daigremontiana plantlet formation. Thus, this gene may have an role during the early development stage of somatic embryogenesis.
Disrupting auxin distribution arrests KdSOC1 gene expression during tobacco callus and shoot induction
According to the well-studied auxin signal pathway model during embryogenesis, we wondered whether disrupting auxin distribution could affect the spatial expression of the KdSOC1 gene during somatic embryogenesis. Thus, N-1-naphthylphthalamic acid (NPA) and 2,3,5-triidobenzoid acid (TIBA), which are auxin transport inhibitors, were used in tobacco callus and shoot induction media to test whether the KdSOC1 gene promoter driving the GUS reporter gene would remain activated (Fig. 3).
TIBA and NPA disrupted the spatial expression of KdSOC1 gene promoter driving GUS reporter gene during tobacco somatic embryogenesis. (a) Normal callus induction medium (CK) without TIBA or NPA. (b) Callus induction medium with 10 μM NPA. (c) Callus induction medium with 40 μM TIBA. (d) Normal shoot induction medium (CK) without TIBA or NPA. (e) Shoot induction medium with 10 μM NPA. (f) Callus induction medium with 40 μM TIBA.
During the tobacco somatic embryo domed-like stage, 10 μM NPA (Fig. 3b) and 40 μM TIBA (Fig. 3c) totally suppressed GUS staining around the somatic cell compared to that under the normal (CK) condition (Fig. 3a), whereas 10 μM NPA (Fig. 3e) and 40 μM TIBA (Fig. 3f) repressed the GUS staining area around the SAM-like area during the tobacco callus shooting stage (near the somatic embryo torpedo stage), compared to those under the CK condition (Fig. 3d).
In general, disrupting the auxin distribution during tobacco somatic embryogenesis interrupted spatial expression of the KdSOC1 gene, suggesting that the proper auxin concentration between cells is a precondition for synthesizing the KdSOC1 transcript.
KdSOC1 gene knockdown results in failed regeneration of transgenic plants
After the KdSOC1 gene knockdown (RNAi) line transformation was accomplished, most calluses regenerated from infected leaves arrested in the globular stage, even on shoot-inducing medium (Figure S1b,d) compared to that during the control (used as empty vector) regeneration procedure (Figure S1a,c). Few calluses initiated cotyledons, but the cotyledons dried out (Figure S1d) and failed to maintain further organ development, which resulted in the absence of the KdSOC1 gene RNAi lines of Kalanchoe daigremontiana.
Thus, the KdSOC1 gene was essential for somatic embryogenesis, and knockdown prevented cotyledon maintenance and further organ development.
Asymmetric plantlet formation and leaf morphological parameters in OE plants
We speculated that KdSOC1 might be a key regulator of plantlet formation in Kalanchoe daigremontiana based on its expression pattern. We over-expressed the KdSOC1 gene with the 35 S promoter to identify the mechanism for modulating plantlet morphogenesis. After screening the positive transformation lines (compared to wild-type [WT] plants) using the KdSOC1 expression level and quantitative real-time polymerase chain reaction (RT-qPCR), differences in plantlet formation were found along the leaf margins in three OE individual lines (OE lines 2, 9, and 10; OE 2, OE 9, and OE 10) (Fig. 4f).
Asymmetric distribution of plantlets along leaf margin in KdSOC1 OE lines. (a) Normal symmetrical plantlets distribute along WT plant leaf margin. (b) Normal symmetrical plantlets distribute along NC plant leaf margin. (c–e) Asymmetric distribution of plantlets along OE line 2, 9, 10 plant leaf margin. (f) KdSOC1 gene expression in WT, NC and OE plant leaves. (g–i) Leaf thickness, plantlet number and leaf length in WT, NC and OE plants. White pentagram indicates the loss of pedestal site for plantlet formation and green pentagram indicates the symmetric plantlet. Dark pentagram indicated significantly different from the WT (P < 0.05). Errors bars represent ± SD (standard deviations) of three independent replications.
The WT (Fig. 4a) and negative control (NC: transformed with empty vector; Fig. 4b) plants developed normally shaped leaves and a symmetrical distribution of plantlets formed along the leaf margin with almost equal expression of the KdSOC1 gene (Fig. 4f). However, a fleshy leaf shape and asymmetrical distribution of plantlets were found along the leaf margin in the lines with KdSOC1 OE genes 2, 9, and 10 (Fig. 4c–e) (white pentagrams in Fig. 4c–e). KdSOC1 expression levels in the OE lines were higher than those in WT or NC plants (Fig. 4, right panel); however, the higher expression level of the gene did not positively correlate with the severity of the alternate leaf and symmetrical plantlet distributions (Fig. 4c,e). Mean leaf thickness in OE lines 2, 9, and 10 was much higher than that in WT and NC plants (Fig. 4g). Mean leaf length in OE lines 2, 9, and 10 was much shorter than that in WT and NC plants (Fig. 4i). To our surprise, the mean numbers of plantlets in OE lines 2, 9, and 10 plants were four times lower (p < 0.05) than those of WT and NC plants (Fig. 4h).
The defect in symmetrical plantlet distribution in the KdSOC1 gene OE plants suggested that the pedestal site with the “stem cell niche”, which gives birth to plantlets, might be missing. The symmetrical plantlet formation pedestal sites were detected by scanning electron microscopy along the leaf margins of WT plants (Fig. 5a,b, red arrow). The leaf margins of OE line 2 (Fig. 5c) contained only one pedestal site (dark pink arrow), whereas the other zygomorphous pedestal site (Fig. 5d) was abolished (green pentagram). The same side of the leaf margin in OE line 9 had two non-isometric plantlet formation pedestal sites (Fig. 5e, dark pink arrow) and one missing pedestal site (Fig. 5f, green) between them. The early developing stage of OE line 10 leaf also manifested a zigzag margin (Fig. 5g, dark pink arrow) and a missing zigzag margin (Fig. 5h, green pentagram).
Asymmetric plantlet distribution was formed along the leaf margin of Kalanchoe daigremontiana by SEM scanning. (a,b) Normal zygomorphous distribution of plantlet was along WT leaf margin. (c,e,g) Asymmetric plantlet distribution was formed along KdSOC1 gene OE plant leave. (d,f,h) Plantlet formation pedestal site was disappeared beside leaf margin. Dark pink arrow indicates normal zygomorphous or non-isometric plantlet formation site. Green pentagram indicated absence of pedestal site for plantlet formation.
Applying exogenous auxin to the leaf vein stimulates the formation of vascular tissue and influences vascular patterning21. We were interested in determining whether OE of the KdSOC1 gene in the leaves would change the vascular structure.
The thickness of vascular fiber around the leaf serrations in cross-sections of the KdSOC1 OE line (Figure S2b) and WT leaves (Figure S2a) showed significant difference. The average thickness of vascular fiber near leaf serration in KdSOC1 OE leaf was higher than that of in WT leaf (Figure S2c).
In summary, KdSOC1 gene OE resulted in an asymmetric plantlet distribution phenotype because the essential plantlet formation pedestal site was aborted. Besides, the KdSOC1 gene OE resulted in increased thickness of vascular fiber near leaf serration.
High auxin content and PIN1 gene expression in OE plants
CUC2, the KANADI family genes, and the PIN1 gene control Arabidopsis leaf number, leaf shape, the vascular pattern, and responder organ formation by regulating auxin response factor and auxin response transport activities22, 23. In addition, key master genes for maintaining the SAM, such as WUSHEL (WUS), and SHOOTMERISTEMLESS (STM), are also essential for controlling leaf shape patterns24, 25. Therefore, we checked the indole-3-acetic acid (IAA) and zeatin contents as well as KdPIN1, KdCUC1, KdSTM, and KdWUS gene expression profiles in WT, NC, and OE plant leaves.
IAA concentrations produced by OE lines 2, 9, and 10 were slightly higher than those of the WT and NC plants (Fig. 6a). However, zeatin concentrations in OE lines 2, 9, and 10 were slightly lower than those of WT and NC plants (Fig. 6b). Higher KdPIN1 gene expression levels were detected in the OE lines than in WT and NC plants, which was consistent with the higher IAA concentrations in the OE lines, which may have re-arranged the auxin response along the leaf margin. Surprisingly, expression levels of the KdCUC1, KdSTM, and KdWUS genes, which are well-known key regulators for maintaining SAM identity and the SAM lateral organ boundary, were not different in WT, NC, and OE plant leaves (Fig. 6d–f).
In addition, WUS, STM, PIN1, AS1, and KNOTTED 1 (Kn1) were also analyzed during KdSOC1 gene OE and WT tobacco callus embryogenesis. The WUS and STM genes, which maintain the meristem identity of somatic cells during the globular stage of embryogenesis in the callus, were expressed at significantly lower levels in the KdSOC1 gene OE callus at the globular stage compared to that in the WT callus (Figure S3a,b). The AS1 gene, which is a lateral organ initiation regulator, exhibited no significant difference (Figure S3c) in expression level at this stage, but expression of the auxin response efflux controller PIN1 decreased tremendously (Figure S3d). The WUS, STM, AS1, and PIN1 genes all showed high level expression levels in KdSOC1 gene OE callus (Figure S3c) during the callus late heart stage, compared to those in the WT callus, whereas Kn1 was not expressed (Figure S3d).
Taken together, higher IAA and lower zeatin concentrations, including higher KdPIN1 gene expression levels in the OE lines might explain the missing pedestal site to initiate plantlet formation and the asymmetric distribution of plantlets by breaking the hormone balance. KdSOC1 gene does have the potential ability to affect the auxin response through different ways.
The KdSOC1 gene changes the auxin response pattern during somatic embryogenesis of Arabidopsis
Considering the asymmetric distribution of plantlets and the relatively high auxin concentration in the KdSOC1 gene OE lines, we hypothesized that the normal auxin response pattern required for somatic embryogenesis might be dysfunctional. Therefore, OE of the KdSOC1 gene was generated in Arabidopsis DR5-GUS reporting lines to observe the auxin response pattern between 35S-KdSOC1-DR5-GUS and DR5-GUS callus genotypes (WT was used as control).
The auxin response during the early stage of DR5-GUS callus formation (Fig. 7b, red arrow) was concentrated in the SAM-like tissue, whereas the auxin response in the 35S-KdSOC1-DR5-GUS callus was dispersed in a much wider area (Fig. 7c, red arrow) around the SAM-like structure. The auxin response at a later stage was concentrated more between two unstretched cotyledons in the DR5-GUS callus (Fig. 7e, red arrow), whereas the auxin response in the 35S-KdSOC1-DR5-GUS callus (Fig. 7f) was biased within a single area. The auxin response in the DR5-GUS callus (Fig. 7h, red arrow) was typically distributed along the cotyledon margin at the early shooting stage of somatic embryogenesis, but the auxin response of the 35S-KdSOC1-DR5-GUS callus was similar to the last stage. The growth phase changed more slowly in the 35S-KdSOC1-DR5-GUS callus than in the DR5-GUS callus (data not shown), which may have been the result of the KdSOC1 gene OE disturbance.
Over-expression of KdSOC1 gene disturbed the auxin flow during Arabidopsis embryogenesis. (a–c) Early globular stage of Arabidopsis somatic embryogenesis. (d–f) Early heart stage of Arabidopsis somatic embryogenesis. (g–i) Torpedo stage of Arabidopsis somatic embryogenesis. Red arrow indicated the high auxin concentration site and white arrow indicated the missing auxin concentration site. SAM: shoot apical meristem, CL: cotyledon.
In summary, KdSOC1 gene OE changed the auxin response pattern during Arabidopsis somatic embryogenesis, which was essential for initiating organ formation and the change in growth phase during somatic embryogenesis.
Discussion
Our results suggest that plantlets formation accompanied with the leaf serration and pedestal site appearance might be possible with proper auxin response. The leaf shape in Arabidopsis is regulated by PIN1 (auxin efflux transporter) and CUC2 (stimulated by PIN1-dependent auxin flow and inhibited by auxin)26. According to this classical model, plantlet morphogenesis process in Kalanchoe daigremontiana also reflects similar characteristics (the SAM structure harbored in the pedestal site between two leaf serrations appeared and plantlet initiated). Thus, we deduced that a KdSOC1-auxin module orchestrated plantlet formation in Kalanchoe daigremontiana.
The plantlet formation process was possibly completed by the KdSOC1 gene-mediated auxin response pattern through the PIN1 gene (Fig. 8a). The KdSOC1 gene up-regulated the KdPIN1 gene expression through a unknown way (Fig. 8a). Then, KdPIN1 gene might function as a transporter to control auxin flow direction which was essential for shaping leaf serration that harbored the pedestal site and initiated plantlet. The certain auxin flow distribution at leaf pedestal site caused certain auxin response which was crucial for the development of stem niche within pedestal site and further leaf development. The KdSOC1 gene OE in the leaf might disturb the auxin efflux through the excessive KdPIN1 gene expression (Fig. 8b). The change in auxin efflux possibly resulted in certain auxin response which was unfavorable for initiating the stem niche. Therefore, OE plant leaves had no stem niche pedestal site, resulting in the plantlet formation defective phenotype. Thus, KdSOC1 gene function during plantlet formation process is possibly to tune the auxin response. Future work on KdSOC1 gene network is another step further to study plantlet formation.
According to previous study in observing endogenous hormones concentrations during plantlet formation in Kalanchoe daigremontiana, increasing accumulation of both auxin and cytokinin at the asexual reproduction site (pedestal site) indicated their positive roles in plantlet development16. In addition, this study also found KdCYCB1 gene (plays essential role in regulating cell G2/M and G1/S phase transition27) up-regulation during plantlet initiation, which suggested that the auxin and cytokinin accumulation in pedestal site might reactivate the cell cycle16. While, other extensive studies also showed that auxin signaling was positive for cell division28. Thus, increasing KdSOC1 gene expression might coincide with the auxin signaling during early plantlet formation in Kalanchoe daigremontiana.
Another gene function study in Arabidopsis axr2-1 [INDOLE-3-ACETIC ACID 7 (IAA7)/ AUXIN RESISTANT 2 (AXR2), one auxin signaling component that regulated flowering time in Arabidopsis] mutant also suggested reduction of free auxin might act to inhibit the timing of floral transition under short days by negatively regulating the expressions of the SOC1 and other related genes29. Therefore, SOC1 gene expression might respond to proper auxin signaling, which was essential for the organ development in right timing. In this study, the KdSOC1 gene was mainly expressed within the SAM until the cotyledon extends or the pedestal site appears, suggesting that KdSOC1 gene assistance was only needed during the globular to heart stage of somatic embryogenesis in the SAM of the callus or before the pedestal site was initiated. The AtSOC1 gene expression pattern detected previously by in situ hybridization shows that it first appears in the SAM and leaf primordia and then is only detected in the floral meristem but not the inflorescence meristem30. This AtSOC1 gene expression pattern was elucidated by its multiple cis-acting element in the promoter31, 32. Therefore, the KdSOC1 and AtSOC1 gene expression patterns suggest that initiating vegetative or sexual organ formation might both be affected by auxin signal transduction. The downstream auxin responsive genes are key, as they control different developmental events during plant growth.
The KdSOC1 gene might affect SAM differentiation like the AGAMOUS (AG) gene (MADS domain protein) function in Arabidopsis floral stem cell fate. A mutation in the AtAG gene results in a defect in floral stem cell fate termination and the floral organ identity specification, which identifies the AG protein together with Polycomb group proteins repress WUS expression by methylating histone H3 Lys-27 of WUS 33. We also detected repressed WUS and STM gene expression during the globular stage of somatic embryogenesis in the KdSOC1 gene OE tobacco callus (Figure S3), suggesting that the KdSOC1 gene terminates the stem cell fate and promotes leaf initiation for a subsequent growth stage during somatic embryogenesis. Therefore, a potential KdSOC1 gene function of stimulating a change in the growth phase during somatic embryogenesis may occur by manipulating the auxin response, which could possibly change cytokinin/auxin ratio in the SAM.
A mutation in the AtSOC1 gene caused bolting arrest and late flowering in Arabidopsis compared to those in WT plants30. However, knockdown of the KdSOC1 gene severely disturbed organ initiation after somatic embryogenesis. Such different phenotypes resulting from a homologous gene mutation suggest that Kalanchoe daigremontiana possesses a distinct regulation module for plantlet morphogenesis. The lack of function of the KdLEC1 gene demonstrates that not all essential growth-related genes are required in all species15. Although genetic information might be inherited from the same ancestor, the continuous adaptation of Kalanchoe daigremontiana in tropical areas may have conferred the KdSOC1 gene with the novel function to affect plantlet formation34. Therefore, further research is needed to discover the KdSOC1 gene network and related hormone signaling to understand the detailed molecular mechanism of plantlet morphogenesis.
Conclusion
This study focused on KdSOC1 gene function during asexual leaf plantlet morphogenesis in Kalanchoe daigremontiana. KdSOC1 gene expression was detected for the first time during in vitro somatic embryogenesis and plantlet morphogenesis. Knockdown of the KdSOC1 gene resulted in a defect in cotyledon formation during the early heart stage of somatic embryogenesis. OE of the KdSOC1 gene resulted in asymmetric plantlet distribution. KdSOC1 gene OE in Arabidopsis DR5-GUS reporting lines resulted in different auxin response patterns during different stages of somatic embryogenesis. In summary, the KdSOC1 gene was determined to regulate the auxin responsive pattern and affected Kalanchoe daigremontiana plantlet formation. Such gene function might be crucial for the plasticity of plantlet formation under various environmental conditions.
Experimental procedures
Plant material and growth conditions
Tobacco (Nicotiana tabacum) and Kalanchoe daigremontiana were grown in substrate (peat:perlite = 3:1) under a 16/8-h light (250 μmol m−2 s−1) cycle at 25 °C at 50–70% relative humidity in a growth room. Arabidopsis thaliana plants were grown under 16/8-h light (250 μmol m−2 s−1) cycle at 20 °C at 75–80% relative humidity in a growth room.
Vector construction and transformation
KdSOC1 gene complete opening reading frame (ORF) cDNA sequence was introduced into pBIN43835 plasmid driven by enhanced CaMV35s promoter as over-expression (OE) construct; RNAi construct of KdSOC1 gene was designed by introducing antisense and sense ORF of KdSOC1 gene into pZH0136 plasmid with partial GUSA gene as intron between them; tissue expression construct of KdSOC1 gene (PKdSOC1-GUS) was designed by introducing its 1.2 kb promoter sequence to replace CaMV35s promoter in pBI12137 plasmid and drives the expression of GUS gene (empty pBI121 plasmid [35S-GUS] was used as positive control). Then all the plasmids mentioned above were all transformed in Agrobacterium tumefeciens strain LBA4404.
Tobacco transformation (PKdSOC1-GUS and 35S-GUS constructs were used) procedure was undergoing with previously described method38. The selection of tobacco positive transformant after LBA4404 infection was divided into shoot induction (with 50 mg ml−1 Kanamycin for selection pressure) and root induction (with 100 mg ml−1 Kanamycin for selection pressure) processes38. Kalanchoe daigremontiana transformation (OE, RNAi and PKdSOC1-GUS constructs were used) method was followed by previously described by Garces39. The selection of Kalanchoe daigremontiana positive transformant after LBA4404 infection was divided into shoot induction (with 50 mg ml−1 Kanamycin for selection pressure) and root induction (with 100 mg ml−1 Kanamycin for selection pressure) processes39. Arabidopsis plant (harboring DR5-GUS reporting genes) transformation (OE construct was used) using the Agrobacterium-mediated floral dip method40.
Confirmation of positive transgenic plant
The confirmation of positive transformation in tobacco T1 generation of each construct was performed by PCR amplification of the KdSOC1 gene promoter. The confirmation of positive transformation in Kalanchoe daigremontiana was performed by RT-qPCR amplification of the KdSOC1 gene ORF. The confirmation of positive OE transformation in DR5-GUS Arabidopsis T1 generation was performed by PCR amplification of the KdSOC1 gene ORF.
Tissue culture of transgenic tobacco and Arabidopsis
The top leaves from T1 generation OE and PKdSOC1-GUS positive transformants were surfaced sterilized39 and cut into 0.5 cm diameter leaf discs. Each leaf disc was first subjected on callus induction Murashige and Skoog (MS) medium with 1 mg ml−1 6-BA, 0.5 mg ml−1 NAA and 50 mg ml−1 Kanamycin for 2 weeks (for auxin distribution repression assay, 40 μM TIBA and 10 μM NPA were added respectively). The newly formed callus was then transferred to MS medium with 1 mg ml−1 6-BA, 0.1 mg ml−1 NAA and 50 mg ml−1 Kanamycin for 3 weeks until the shoot appeared (for auxin distribution repression assay, 40 μM TIBA and 10 μM NPA were added respectively).
The immature seed of Arabidopsis transgenic plants (DR5-GUS; 35S-KdSOC1-DR5-GUS; WT) were used as explants in somatic tissue culture follow the previously described method41.
Microscopy and photography
Scanning electron microscopy for Kalanchoe daigremontiana was performed as previously described42. Phenotype of transgenic plant pictures were captured by Canon digital camera (Japan) The photos of different inducing stage of tobacco callus were taken by Leica MDG41 microsystem (Singapore). All photographs were adjusted assembled into figures using Adobe Photoshop 4.0 (AdobeSystems, San Jose, CA, USA). The vascular fiber thickness was measured based on ImageJ software.
GUS staining assay
Histochemical localization of GUS activity was performed as previously described43. Images are representative of > 10 observed samples stained in three independent experiments.
RT-qPCR analysis of transgenic plants
RNA extractions were performed on three biological replicates of T1 generation transgenic tobaccos expanding leaves using the TRIzol® (Invitrogen, USA), while RNA extractions for Kalanchoe daigremontiana were performed44. All samples were treated using RQ DNAse (Promega, USA) and were ethanol precipitated. cDNA synthesis was performed using GoScriptTM Reverse Transcription Systemfor RT-PCR (Promega, USA) with 2 µg RNA and using the oligo (dT) primer. RT-qPCR was performed using the TransStart® Tip Green qPCR SuperMix (TransGenBiotech, China), following the manufacturer instructions and an ABI Step One PCR instrument (Applied Biosystems, USA) was used. 200 ng cDNA was used for each reaction. The results were calculated using the 2−∆∆CT method45. The KdActin were used as housekeeping gene and the primers used in RT-qPCR were listed in Table S1.
Hormone detection in transgenic plants
Raw hormone mixtures were extracted from 0.1 g of tissue from the uppermost fully developed leaf with 1 mL of pH 7.0 PBS. Indole-3-acetic acid (IAA) and cytokinin concentrations were measured using a colorimetric ELISA kit containing plates pre-coated with antibody specific to IAA or cytokinin. (Winter Song Boye Biotechnology Co. Ltd., Beijing). Samples were diluted six-fold prior to the tests. The standard curve was generated based on a series of known IAA or zeatin standard sample reactions to 180, 120, 60, 30, 15, and 0 ng mL−1. Three individual lines from each transgenic plant and WT were used.
Statistical analyses
ANOVA and mean comparisons were performed using SPSS version 20.0 software. The value of 2−ΔΔCT was also proceeded with SPSS version 20.0 software. Error bars represent standard deviation. Black stars letters indicate statistically significant differences between OE and WT at P < 0.05 based on Duncan’s multiple range test.
Data availability
The authors declare to provide all the data when requested.
References
Abel, S. & Theologis, A. Odyssey of auxin. Cold Spring Harb Perspect Biol 2, a004572 (2010).
Ljung, K. Auxin metabolism and homeostasis during plant development. Development 140, 943–50 (2013).
Schaller, G. E., Bishopp, A. & Kieber, J. J. The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63 (2015).
Aichinger, E., Kornet, N., Friedrich, T. & Laux, T. Plant stem cell niches. Annual Review Of Plant Biology 63, 615–36 (2012).
Sablowski, R. Plant stem cell niches: from signalling to execution. Current Opinion In Plant Biology 14, 4–9 (2011).
Ten, H. C., Lu, K. J. & Weijers, D. Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142, 420–30 (2015).
Bennett, T., van den Toorn, A., Willemsen, V. & Scheres, B. Precise control of plant stem cell activity through parallel regulatory inputs. Development 141, 4055–64 (2014).
Gaillochet, C. & Lohmann, J. U. The never-ending story: from pluripotency to plant developmental plasticity. Development 142, 2237–49 (2015).
Vanstraelen, M. & Benkova, E. Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol 28, 463–87 (2012).
Peer, W. A., Blakeslee, J. J., Yang, H. & Murphy, A. S. Seven things we think we know about auxin transport. Mol Plant 4, 487–504 (2011).
Bar, M. & Ori, N. Leaf development and morphogenesis. Development 141, 4219–30 (2014).
Bilsborough, G. D. et al. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA 108, 3424–9 (2011).
Garces, H. and Sinha, N. The ‘mother of thousands’ (Kalanchoe daigremontiana): a plant model for asexual reproduction and CAM studies. Cold Spring Harb Protoc 2009, pdb.emo133 (2009).
Garces, H. M. et al. Evolution of asexual reproduction in leaves of the genus Kalanchoe. Proc Natl Acad Sci USA 104, 15578–83 (2007).
Garces, H. M., Koenig, D., Townsley, B. T., Kim, M. & Sinha, N. R. Truncation of LEAFY COTYLEDON1 protein is required for asexual reproduction in Kalanchoe daigremontiana. Plant Physiology 165, 196–206 (2014).
Guo, J. et al. Origination of asexual plantlets in three species of Crassulaceae. Protoplasma 252, 591–603 (2015).
Liu, C. Zhu, C. and Zeng, H. M. Key KdSOC1 gene expression profiles during plantlet morphogenesis under hormone, photoperiod, and drought treatments. Genet Mol Res 15 (2016).
Lee, J. & Lee, I. Regulation and function of SOC1, a flowering pathway integrator. Journal Of Experimental Botany 61, 2247–54 (2010).
Liu, C. et al. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135, 1481–91 (2008).
Voogd, C., Wang, T. & Varkonyi-Gasic, E. Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. Journal Of Experimental Botany 66, 4699–710 (2015).
Cano-Delgado, A., Lee, J. Y. & Demura, T. Regulatory mechanisms for specification and patterning of plant vascular tissues. Annu Rev Cell Dev Biol 26, 605–37 (2010).
Pekker, I., Alvarez, J. P. & Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17, 2899–910 (2005).
Yang, F. et al. A maize glutaredoxin gene, Abphyl2, regulates shoot meristem size and phyllotaxy. Plant Cell 27, 121–31 (2015).
Aguilar-Martinez et al. Transcriptional, posttranscriptional, and posttranslational regulation of SHOOT MERISTEMLESS gene expression in Arabidopsis determines gene function in the shoot apex. Plant Physiology 167, 424–42 (2015).
Dolzblasz, A. et al. Stem cell regulation by Arabidopsis WOX genes. Mol Plant. (2016).
Bilsborough, G. D. & Estelle, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proceedings of the National Academy of Sciences of the United States of America 108(8), 3424–9 (2011).
De, V. L. & Joubes, J. D. Plant cell cycle transitions. Current Opinion in Plant Biology 6(6), 536–543 (2003).
Del Pozo, J. C. & Manzano, C. Auxin and the ubiquitin pathway. two players-one target: the cell cycle in action. Journal of Experimental Botany 65(10), 2617–2632.3 (2013).
Yan-Xia, L. & Wang, H.-Q. Yang A gain-of-function mutation in iaa7/axr2 confers late flowering under short-day light in Arabidopsis. Journal of Integrative Plant Biology 53(6), 480–492 (2011).
Borner, R. et al. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant Journal 24, 591–9 (2000).
Lee, J., Oh, M., Park, H. & Lee, I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant Journal 55, 832–43 (2008).
Liu, C. et al. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135, 1481–91 (2008b).
Liu, X. et al. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23, 3654–70 (2011).
Rast-Somssich et al. Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev 29, 2391–404 (2015).
Attia, K. A. et al. Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Current issues in molecular biology 11(Suppl 1), i29–34 (2009).
Chen, D. X. et al. Genetic transformation of rice with pi-d2, gene enhances resistance to rice blast fungus Magnaporthe oryzae. Rice Science 17(1), 19–27 (2010).
Li, L. L. et al. Functional characterization of the ginkgo biloba chalcone synthase gene promoter in transgenic tobacco. Genet Mol Res 13(2), 3446–3460 (2014).
Horsch, R. B. et al. A simple and general method for transferring genes into plants. Science, 227, 1229–1234 (1985).
Garces, H. and Sinha, N. Transformation of the plant Kalanchoe daigremontiana using Agrobacterium tumefaciens. Cold Spring Harb Protoc 2009, pdb.prot5303 (2009).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal for Cell & Molecular Biology 16(6), 735–43 (1998).
Ikeda-Iwai, M., Satoh, S. & Kamada, H. Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. Journal Of Experimental Botany 53, 1575–80 (2002).
Garces, H. and Sinha, N. Fixing and sectioning tissue from the plant Kalanchoe daigremontiana. Cold Spring Harb Protoc 2009, pdb.prot5301 (2009).
Su, Y. H. et al. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant Journal 59, 448–60 (2009).
Garces, H. and Sinha, N. Extraction of RNA from the plant Kalanchoe daigremontiana. Cold Spring Harb Protoc 2009, pdb.prot5305 (2009).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25, 402–8 (2001).
Acknowledgements
This research was supported by The State Bureau of Forestry 948 project “Techniques of Euonymus alatus ‘Compactus’ triploid breeding” (NO. 2012-4-58), National Natural Science foundation of China (NSFC, grant no. 31370598), National High Technology Research and Development Program of China (863 Program) subject “Research and application of functional genomics of Eucommia ulmoides Oliv. (subject number: 2013AA102605–05) and the Fundamental Research Funds for the Central University (BLYJ201506). We thank Dr. Jie Zhang (associate professor from state key laboratory of plant genomics, institute of microbiology chinese academy of sciences) for critical reading and advisement in experiment design; and Dr. Helena Garcês for technical assistance for Kalanchoe daigremontiana transformation; and Dr. Dayong Li for vector providing.
Author information
Authors and Affiliations
Contributions
Chen Zhu designed the experiments, Chen Zhu and Li Wang performed the experiments, Jinhua Chen and Chenglan Liu analysed the data, Huiming Zeng and Huafang Wang fund the research.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, C., Wang, L., Chen, J. et al. Over-expression of KdSOC1 gene affected plantlet morphogenesis in Kalanchoe daigremontiana . Sci Rep 7, 5629 (2017). https://doi.org/10.1038/s41598-017-04387-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-04387-0
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.