Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the selective and progressive loss of motor neurons. The purpose of this study was to clarify effects of brazilian green propolis and the active ingredient against ALS-associated mutant copper-zinc superoxide dismutase (SOD1)-mediated toxicity. Ethanol extract of brazilian green propolis (EBGP) protected N2a cells against mutant SOD1-induced neurotoxicity and reduced aggregated mutant SOD1 by induction of autophagy. Kaempferide and kaempferol, the active ingredients of EBGP, also inhibited mutant SOD1-induced cell death and reduced the intracellular mutant SOD1 aggregates. Both kaempferide and kaempferol significantly suppressed mutant SOD1-induced superoxide in mitochondria. Western blot analysis showed that kaempferol potentially induced autophagy via the AMP-activated protein kinase (AMPK) - the mammalian target of rapamycin (mTOR) pathway. These results suggest that EBGP containing the active ingredient against mutant SOD1-mediated toxicity is a promising medicine or health food for prevention and treatment of ALS.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is characterized by the degeneration of motor neurons and by the formation of intracellular protein aggregations that form in motor neurons. While 95% of ALS is sporadic, 5% is inherited. Among the inherited ALS (also known familial ALS (FALS)), the mutations in copper-zinc superoxide dismutase (SOD1) are the major autosomal dominant inherited cause for ALS1. Mutant SOD1 proteins form insoluble aggregations with components of the ubiquitin proteasome system (UPS) and autophagy pathway in motor neurons2, 3. The relationship between motor neuron death and mutant SOD1 aggregations remain elusive. The excess of mutant SOD1 aggregations decreased the elimination capacity of the UPS3,4,5. The impairment of the UPS by the excess aggregations induced endoplasmic reticulum (ER) stress, mitochondrial dysfunction, and oxidative stress3, 6. On the other hand, several evidences have suggested that autophagy activation alleviates mutant SOD1-linked toxic insults7, 8. In addition, Mahogunin ring finger-1 (MGRN1) E3 ubiqutin ligase, which catalyzes mono-ubiquitination to the substrate, contributes to the clearance of mutant SOD1 aggregations likely via autophagic pathway8.
Propolis is made from a sticky substance that honeybees produce by mixing their own waxes with resinous sap obtained from the bark and leaf-buds of certain trees. The color of propolis can be green, red, brown, or almost black depending on the plants from which the resinous substance is collected. The properties and constituents of propolis also differ with its geographical origin. Propolis presents numerous biological and pharmacological properties, such as anti-bacterial, anti-inflammatory, and anti-oxidative activity9,10,11,12,13. The previous our study also showed that propolis promoted the advantage of the conditioned medium of dental pulp cells via producing neurotrophic factors13. However, the effect of propolis and the active components against ALS-associated mutant SOD1-mediated toxicity is not well known.
In the present study, to examine whether propolis and the active components have neuroprotective effect against mutant SOD1-induced neurotoxicity in a cellular model, we used the ethanol extract of Brazilian green propolis (EBGP). In Japan, Brazilian green propolis has recently come to be used as a health food, which is of sufficiently good quality to provide experimental reproducibility. The ethanol extract of propolis is the major form used in health food14. We also further investigate whether autophagy is involved in the neuroprotection of kaempferol and kaempferide against mutant SOD1-related neurotoxicity via the AMP-activated protein kinase (AMPK) - the mammalian target of rapamycin (mTOR) pathway.
Results
EBGP reduced these intracellular aggregates of SOD1G85R and prevented SOD1G85R-induced neurotoxicity
Currently, more than 150 types of pathogenic mutations in SOD1 gene have been identified in ALS patients15. Among these mutations, the pathogenic SOD1G85R mutation has been frequently studied16. The transgenic mice having transgene of SOD1G85R mutation show rapidly progressive motor neuron degeneration. The aggregate formation was confirmed in SOD1G85R-transfected N2a cells based on previous study8, 17,18,19. As shown in Fig. 1A, the mCherry-fused SOD1G85R (thereafter SOD1G85R) formed intracellular aggregates in approximately 15% of total transfected N2a cells, whereas mCherry-fused SOD1WT (thereafter SOD1WT) was distributed evenly in the cytoplasm (Fig. 1A,C). Immunostaining was performed to confirm that the signal of mCherry was SOD1. As shown in Fig. 1B, the signal of mCherry colocalized with the signal of SOD1. Similarly to previous studies17, 18, western blot analysis showed that triton X-100-insoluble SOD1G85R was increased (Fig. 1D). In order to examine SOD1G85R-mediated toxicity to differentiated N2a cells, we performed MTT assay. Although SOD1WT did not affect cell survival rate, SOD1G85R induced cell death (Fig. 1E). These results suggest that SOD1G85R caused cell death via the formation of insoluble aggregates.
SOD1G85R caused neurotoxicity via the formation of insoluble aggregates. (A) Representative fluorescent microscopy images of N2a cells expressing mCherry-SOD1WT or mCherry-SOD1G85R. The arrowheads indicate the intracellular SOD1 aggregates. (B) Colocalization of SOD1 and mCherry in the N2a cells. (C) Quantified data of intracellular SOD1 aggregates are expressed as mean ± S.E.M from three independent experiments. In each experiment, at least 200 cells were counted. (D) Immunoblot analysis of SOD1 aggregates. N2a cells expressing SOD1 constructs were lysed with 1% TritonX-100 (Triton soluble fraction; S). Triton-insoluble fraction (I) were resuspended with 2% SDS and then analyzed with immunoblotting by anti-mCherry antibody. (E) N2a cells expressing mCherry-SOD1WT or mCherry-SOD1G85R were incubated for 48 h in the differentiation medium. The cell viability was measured by MTT assay. **p < 0.01, ***p < 0.001. Scale bar: 50 µm.
Next, to investigate the effect of EBGP against SOD1G85R aggregates, we evaluated the number of intracellular SOD1G85R aggregates. EBGP significantly reduced these intracellular aggregates of SOD1G85R to approximately 30% of untreated cells (Fig. 2A,B). Western blot analysis also showed that triton X-100-insoluble SOD1G85R was significantly decreased by treatment of EBGP, although triton X-100-soluble SOD1G85R did not change (Fig. 2C,D). In addition, EBGP prevented SOD1G85R-induced neurotoxicity (Fig. 2E). There results suggest that the active components of EBGP have significant effects for the neuroprotection against SOD1G85R-induced neurotoxicity.
EBGP protected SOD1G85R-induced neurotoxicity via the reduction of the aggregates of SOD1G85R. (A) Representative fluorescent microscopy images of N2a cells expressing mCherry-SOD1G85R incubated for 24 h with 20 ng/mL EBGP. The arrowheads indicate the intracellular SOD1 aggregates. (B) Quantified data of intracellular SOD1 aggregates are expressed as mean ± S.E.M from three independent experiments. In each experiment, at least 200 cells were counted. (C) Reduction of Triton-insoluble mutant SOD1G85R by EBGP. After treatment of 20 ng/mL EBGP, N2a cells expressing SOD1G85R were lysed with 1% TritonX-100. Triton-insoluble fraction was resuspended with 2% SDS and analyzed with immunoblotting. (D) The density of Triton-insoluble mutant SOD1G85R is given as mean ± S.E.M from three independent experiment, based on the density of Triton-insoluble mutant SOD1WT. (E) N2a cells expressing mCherry-SOD1G85R were treated with 20 ng/mL EBGP. The cell viability was measured by MTT assay. ***p < 0.001. Scale bar: 50 µm.
The active component of EBGP, kaempferol and kaempferide, are involved in neuroprotection against mutant SOD1-related toxicity
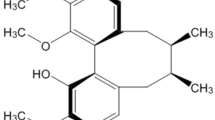
Previous study reported that HPLC analysis shows the major chemical constituents of the EBGP are phenolic acids/flavonoids, such as artepillin C, baccharin, drupanin, p-coumaric, Betuletol, kaempferide and kaempferol20. Among these components, we chose two flavonoids, kaempferol and kaempferide (Fig. 3A). We examined effects of kaempferol and kaempferide against SOD1G85R aggregates. As shown in representative photomicrographs, kaempferol and kaempferide remarkably reduced the intracellular aggregates of SOD1G85R (Fig. 3B). We further examined the effect of kaempferol and kaempferide on SOD1 aggregation by using a series of concentrations based on previous studies (Fig. 3C)21, 22. As results, kaempferol (from 0.3 µM to 10 µM) and kaempferide (from 5 µM to 50 µM) significantly decreased the aggregates. Western blot analysis also showed that triton X-100-insoluble SOD1G85R was significantly decreased by treatment of kaempferol and kaempferide (Fig. 3D,E). Moreover, we showed that kaempferol and kaempferide prevented SOD1G85R-induced neurotoxicity (Fig. 3F and Supplementary Fig. S1). These results suggest that kaempferol and kaempferide are involved in neuroprotection against mutant SOD1-related toxicity.
Kaempferol and kaempferide prevented SOD1G85R-related neurotoxicity. (A) Chemical structures of kaempferol and kaempferide. (B) Representative fluorescent microscopy images of N2a cells expressing mCherry-SOD1G85R incubated for 24 h with 3 µM kaempferol or 15 µM kaempferide. (C) Quantified data of intracellular SOD1 aggregates in N2a cells expressing mCherry-SOD1G85R incubated for 24 h with kaempferol (from 0.3 µM to 10 µM) or kaempferide (from 5 µM to 50 µM) are expressed as mean ± S.E.M from three independent experiments. In each experiment, at least 200 cells were counted. (D) Reduction of Triton-insoluble mutant SOD1G85R by kaempferol and kaempferide. After treatment of 3 µM kaempferol or 15 µM kaempferide, Triton-insoluble fraction of N2a cells expressing SOD1G85R was resuspended with 2% SDS and analyzed with immunoblotting. (E) The band density of Triton-insoluble mutant SOD1G85R group is given as mean ± S.E.M from three independent experiments, based on the band density of Triton-insoluble non-treated mutant SOD1G85R group. (F) N2a cells expressing mCherry-SOD1G85R were treated with kaempferol (from 0.3 µM to 10 µM) or kaempferide (from 5 µM to 50 µM). The cell viability was measured by MTT assay. Data is expressed as mean ± S.E.M from three independent experiments, based on SOD1WT group. *p < 0.05; **p < 0.01; ***p < 0.001. Scale bar: 20 µm.
The anti-oxidant activity is involved in the neuroprotection of kaempferol and kaempferide against mutant SOD1-related toxicity
Previous study showed that flavonoid exerts an anti-oxidant activity23. Because oxidative stress is well recognized as an important pathogenic event in ALS, the effects of kaempferol and kaempferide against SOD1G85R-induced ROS were examined using MitoSOX Red, a mitochondrial-targeted superoxide sensitive fluorogenic probe24. The increase in MitoSOX Red fluorescence in N2a cells transfected GFP-SOD1G85R was significantly attenuated by kaempferol and kaempferide (Fig. 4A,B). To clarify the direct scavenging effect of kaempferol and kaempferide against ROS such as hydroxyl radical (•OH), we performed electron spin resonance (ESR) analysis (Fig. 4C,D). As result, both kaempferol and kaempferide presented potent scavenging activity against •OH. These results demonstrate that the anti-oxidant activity is involved in the neuroprotection of both kaempferol and kaempferide against mutant SOD1-related neurotoxicity.
Both kaempferol and kaempferide suppressed SOD1G85R-induced superoxide in mitochondria. (A) N2a cells expressing mCherry-SOD1G85R were treated with 3 µM kaempferol or 15 µM kaempferide for 24 h. Subsequently, MitoSOX® Red was added to the cell culture to a final concentration of 5 μM to the cells and incubated for 30 min at 37 °C. (B) The relative fluorescence intensity was quantified by computerized image analysis with ImageJ. Data are expressed as mean ± S.E.M from three independent experiment, based on the fluorescence intensity of SOD1WT. (C) Traces show typical spectra of DMPO-OH spin adducts generated from H2O2 plus Fe2+ in the absence (control) or presence of kaempferol and kaempferide. (D) The amount of •OH was semi-quantitatively measured as the formation of DMPO-OH spin adducts by ESR spectrometry. Data is expressed as the mean ± S.E.M of three determinations, based on control. *p < 0.05; **p < 0.01. ***p < 0.001. Scale bar: 50 µm.
Kaempferol, but not kaempferide, reduced intracellular aggregate via the activation of autophagy
Previous study reported that flavonol induced autophagy21. To further investigate whether autophagy is involved in the neuroprotection of kaempferol and kaempferide against mutant SOD1-related neurotoxicity, western blot analysis was performed using LC3 antibody, a selective marker of autophagy. Western blot analysis showed that kaempferol, but not kaempferide, enhanced the formation of LC3-II (Fig. 5A,B). Western blot analysis was also performed using p62 antibody, another selective marker of autophagy. As shown in Fig. 5C–E, the expression of LC-3-II/LC-3-I was increased while that of p62 was decreased after treatment with kaempferol. In addition, the change of LC-3-II/LC-3-I and p62 levels was improved by 3-methyladenine (3-MA), an inhibitor of autophagy, indicating the autophagy is activated by kaempferol. To confirm the relationship between neuroprotective effect of kaempferol against mutant SOD1-mediated toxicity and induction of autophagy, the MTT assay was performed with 3-MA. The protective effect of kaempferol was significantly abolished by 3-MA treatment (Fig. 5F). In addition, immunofluorescence staining showed that the reduction of intracellular aggregates of SOD1G85R caused by kaempferol was significantly abolished by 3-MA treatment (Fig. 5G,H). These results suggest that kaempferol, but not kaempferide, reduced intracellular aggregate via the activation of autophagy and then inhibited mutant SOD1-induced neurotoxicity.
Kaempferol, but not kaempferide, reduced the intracellular aggregates via the activation of autophagy. (A) Induction of LC-3-II by kaempferol, but not kaempferide. N2a cells were transfected with SOD1G85R, and then incubated with 3 µM kaempferol or 15 µM kaempferide for 24 h. The lysates were analyzed by immunoblotting with anti-LC-3 and anti-β-actin antibodies. (B) Relative levels of LC-3-II/LC-3-I normalized by the expression of β-actin were quantified, based on the band density of SOD1WT. (C–E) Immunoblotting analysis of autophagy. N2a cells expressing SOD1G85R were treated with kaempferol for 24 h with and without 3-MA. The lysates were analyzed by immunoblotting with antibodies for LC-3, p62 and β-actin. Relative levels of LC-3-II/LC-3-I and p62 normalized by the expression of β-actin were quantified, based on the band density of SOD1WT. (F) Representative fluorescent microscopy images of N2a cells expressing mCherry-SOD1G85R incubated for 24 h with 3 µM kaempferol with and without 3-MA. (G) Quantified data of intracellular SOD1 aggregates are expressed as mean ± S.E.M from three independent experiments. In each experiment, at least 200 cells were counted. (H) N2a cells expressing SOD1G85R was incubated for 24 h with 3 µM kaempferol with and without 3-MA. Cell viability was determined. Data is expressed as mean ± S.E.M from three independent experiments. *p < 0.05; **p < 0.01. ***p < 0.001.
Kaempferol increases AMPK phosphorylation
Previous study reported that flavonol inhibited the mammalian target of rapamycin (mTOR) complex, which suppresses autophagy22. However, the mechanisms through which kaempferol inhibits mTOR are still unclear. To identify the signal transduction pathway mediated by kaempferol, we investigated AMPK and AKT pathways. AMPK inactivates mTOR, and AKT induced mTOR activity. Kaempferol significantly inhibited mTOR phosphorylation (Fig. 6A,B) and significantly increased AMPK phosphorylation (Fig. 6A,C). On the other hand, AKT phosphorylation was not affected by kaempferol (Fig. 6A,D). These results suggest that kaempferol potentially induces autophagy via the AMPK-mTOR pathway.
Kaempferol increases AMPK phosphorylation. (A) Immunoblotting analysis of autophagy regulators. N2a cells expressing SOD1G85R were treated with 3 µM kaempferol for 24 h. The lysates were analyzed by immunoblotting with antibodies for phosphorylated mTOR (p-mTOR), mTOR, phosphorylated AMPK (p-AMPK), phosphorylated AKT (p-AKT), β-actin. (B) Relative levels of p-mTOR normalized by the expression of mTOR were quantified, based on the density of SOD1WT. (C,D) Relative levels of p-AMPK and p-AKT normalized by the expression of β-actin were quantified, based on the band density of SOD1WT. *p < 0.05. N.S.; not significant.
Discussion
Many studies have been conducted to elucidate the health benefits and the mechanisms of EBGP, including studies on its effects on bacteria9, viruses10, inflammation11, and cancer12. However, little has been reported on effects of EBGP for neurodegenerative disease such as ALS. Dominant mutations in SOD1 are frequent cause of inherited ALS1. Misfolded and aggregated SOD1 is present in both sporadic and familial ALS, although its pathobiology remains unclear25, 26. A reduction of misfolded and aggregated SOD1 might be one of potential therapeutic approaches for ALS17. Therefore, in the present study, we examine whether EBGP have neuroprotective effect against SOD1G85R -induced neurotoxicity in a cellular model for ALS. EBGP prevented SOD1G85R-induced neurotoxicity. Brazilian green propolis has been reported to contain many biological active compounds27. There results suggest that the active components of EBGP play an important for the neuroprotection against SOD1G85R-induced neurotoxicity.
Propolis is collected by honeybees from various plants. There are different types of propolis according to its botanical origin and geographical region. Brazilian green propolis has been reported to contain many biological active compounds, including cinnamic acid derivatives (e.g., artepillin C, baccharin, drupanin, and p-coumaric acid) and flavonoids (e.g., kaempferol, and kaempferide)20, 27. Flavonoids are considered as an important part of the anti-oxidants in food. Kaempferol and kaempferide are known to have an anti-oxidative activity23. In addition, previous study reported that kaempferol induced autophagy, although the mechanism is unclear21, 22. Therefore, because oxidative stress and autophagy are well recognized as an important pathogenic event in ALS, we chose two flavonoids, kaempferol and kaempferide among these components in Brazilian green propolis, and examined effects of these flavonoids against mutant SOD1-related toxicity.
In the present study, separation of protein fraction by using triton X-100 was performed according to published protocols17, 18. We also performed immunocytochemical assay based on previous studies8, 17,18,19. In addition, previous studies have shown that progressive accumulation of mutant SOD1, which is correlated with a decrease of its solubility in triton X-100, is observed in the spinal cord of SOD1 tg mice, and evident aggregates of SOD1 are found at a late stage of the disease in SOD1 tg mice28, 29. Considering the evidence of these previous studies, we selected the triton X-100 as a detergent in the present study.
Numerous studies have identified significant mitochondrial abnormalities in mutant SOD1-associated ALS24, 30, 31. In addition, mutant SOD1 forms insoluble aggregates in mitochondria at the surface of outer membrane, raising the prospect of a direct cause-effect mechanism by which mutant SOD1 impact mitochondrial function32, 33. Taken together, these studies provide reasonable evidence for the involvement of mitochondrial-produced oxidants and mitochondrial oxidative damages in mutant SOD1-related ALS. The cause and effect of increased mitochondrial produced ROS in mutant SOD1-induced mitochondrial dysfunction requires further study. However, the present results herein showing that the active components such as kaempferol and kaempferide of Brazilian green propolis inhibited mutant SOD1-induced superoxide suggest that Brazilian green propolis and the active components might be utilized to promote the therapeutic strategy for ALS.
The propensity of a flavonoid to inhibit free-radical mediated events is governed of the chemical structure. Previous study showed that flavonoids with 3-hydroxy (OH) group play a positive role in anti-oxidative activity23. Kaempferol and kaempferide are contained in 3-OH group. Therefore, the chemical structure of flavonoid having the 3-OH group may contribute to the anti-oxidative activity against mutant SOD1-induced ROS. On the other hand, kaempferol, but not kaempferide, induces autophagy via the AMPK-mTOR pathway in the present study. The difference in the chemical structure of flavonoid is only functional group in the B ring between kaempferol and kaempferide. Kaempferol and kaempferide frame with the B ring 4′-OH group and 4′-methoxy group, respectively (Fig. 3). There are differences in beneficial actions such as anti-oxidative activity according to the structure of the B ring of flavonoids23. Therefore, kaempferol having 4′-OH group in the B ring may be related to the autophagy, although further studies are needed to elucidate the relation between the chemical structure of flavonoid and the autophagy. On the other hand, to the best of our knowledge, there is no report that flavonoids including kaempferol and kaempferide bind to mutant SOD-1 like a chemical chaperone. In the present study, it is unknown whether kaempferol and kaempferide directly bind to mutant SOD1. It remains in future study. Further research is required to provide more detail of interaction experiments using recombinant mutant SOD1.
In the present study, kaempferol increased AMPK phosphorylation to inhibit mTOR phosphorylation, and subsequent induced autophagy. Previous study demonstrated that AMPK was inactivated both in the in vitro and in vivo models of SOD1-associated ALS and that the reduced activation of AMPK by mutant SOD1 was recovered by cystain C treatment17. In addition, resveratrol protective effects were associated with increased expression and activation of AMPK in spinal cords of SOD1G93A mice32. Moreover, riluzole, an approved ALS drug, leads to AMPK phosphorylation33. However, several previous studies demonstrated that the reduced activated of AMPK improved the mutant SOD1-induced motor neuron death in vitro and the motor activity of neurons in vivo 34. These results suggest that an imbalance in energy metabolism should be considered an important factor in both progression and potential treatment of ALS and that AMPK play a key role in the disease. Therefore, kaempferol as well as EBGP containing the active ingredient against mutant SOD1-mediated toxicity will be an efficient source of new treatments and prevention for ALS.
Methods
Plasmid, cell culture, and transfection
Wild-type human SOD1 cDNA was purchased from TransOMIC Technologies, and subcloned into pmCherry-N1 (Clontech Laboratories Inc., CA, USA) between the HindIII/BamHI sites (pmCherry-SOD1WT). The mutant SOD1 gene was generated by Quick Change site-directed mutagenesis (Stratagene, La Jolla, CA, USA) according to the manufacturer’s protocol (pmCherry-SOD1G85R). The primer pairs were as follows: 5′-GTTAAGCTTATGGCGACGAAGGCCGTGTGC-3′ and 5′-GCAGGATCCGGTTGGGCGATCCCAATTACACC-3′ for pmCherry-SOD1WT, 5′-GACTTGCGCAATGTGACTGCTGACAAA-3′ and 5′-CACATTGCGCAAGTCTCCAACATGCCT-3′ for pmCherry-SOD1G85R.
N2a cells were maintained in Dulbecco’s modified Eagle medium (DMEM, Wako Pure Chemical Industries, Ltd., Osaka Japan) containing 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Inc., Waltham, MA, USA) under a humidified atmosphere of 5% CO2 at 37 °C. The cells were passaged by trypsinization every 3–4 days. Transient expression of each plasmid in N2a cells was accomplished with Lipofectamine 2000 according to the manufacturer’s protocol (Thermo Fisher Scientific Inc., Waltham, MA, USA).
Aggregation rate analysis
After 24 h of transfection of each vector to N2a cells, the cells were treated with or without EBGP (a gift from the Api Co., Ltd., Gifu, Japan), kaempferol (Wako Pure Chemical Industries Ltd., Osaka, Japan), or kaempferide (INDOFINE Chemical Company) in the presence or absence of 3-methyladenine (3-MA) for 24 h. Subsequently, the cells were washed with PBS for 5 min twice and fixed with 4% paraformaldehyde for 15 min. Fluorescent microscopy images were acquired using a confocal fluorescence microscope (LSM700, Carl Zeiss, Jena, Germany) and the images were analyzed using ZEN software (Carl Zeiss). Aggregates were defined as bright non-homogeneities with the cytoplasm. To quantify aggregate formation, the intensity of the even and diffuse fluorescence was set as threshold and the number of cells in each field showing fluorescence intensities over the threshold was counted using imaging software. Following the threshold analysis, the presence of aggregates was confirmed by eye in each field, based on previous studies8, 17,18,19.
Western blot analysis
After 24 h of transfection to N2a cells each vector, the cells were treated with or without EBGP, kaempferol, or kaempferide. The cells were lysed with TNE lysis buffer (50 mM Tris-HCl (pH. 7.4), 150 mM NaCl, 1 mM ethylenediamineteraacetic acid, protease inhibitor cocktail) containing 1% Triton-X and then were centrifuged at 15,000 × g for 5 min at 4 °C. The supernatant of the lysate was defined as Triton soluble fraction. After centrifuge, the remaining deposition was resuspended with TNE lysis buffer containing 2% sodium dodecyl sulfate (SDS) (defined as Triton-insoluble fraction). Protein concentrations were measured using a BCA protein assay kit (Thermo Fisher Scientific Inc.) with bovine serum albumin (BSA) as a standard. Lysates were mixed with sample buffer containing 10% 2-mercaptoethanol, and subjected to 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The 20 µg of protein was applied to SDS-PAGE in order to separate proteins as certain molecular weight. SDS-PAGE was performed under constant voltage at 200 V at room temperature for 40 min. The separated proteins in poly acrylamide gel were transferred to PVDF membrane in transfer buffer (0.3% Tris, 1.44% glycine, 20% methanol) under constant voltage at 100 V at 4 °C for 90 min. The membranes were incubated with 5% BSA (Wako) for 60 min, and then with following primary antibodies for overnight: mouse monoclonal antibodies against mCherry (1:1000, Clontech), β-actin (1:2000, Santa Cruz Biotechnology, Dallas, TX USA),; rabbit polyoclonal antibodies: LC-3 (1:1000, Cell Signaling Technology, Danvers, MA USA), p-AKT (1:1000, Cell Signaling Technology), AKT (1:1000, Cell Signaling Technology), p-AMPK (1:1000, Cell Signaling Technology), p-mTOR (1:1000, Cell Signaling Technology), mTOR(1:1000, Cell Signaling Technology), p62 (1:1000, Cell Signaling Technology). After the primary antibody reaction, membrane was incubated in the second antibody (goat anti-rabbit antibody conjugated with HRP (1:2500, Santa Cruz Biotechnology), goat anti-mouse HRP antibody conjugated with HRP (1:2500, Santa Cruz Biotechnology)). The membrane was incubated in ECL prime (GE Healthcare, Buckinghamshire, UK) to generate the chemiluminescence from HRP antibodies. The chemiluminescence was detected by using LAS3000 mini (Fuji film, Tokyo, Japan). The band density was measured by ImageJ software (NIH, New York, NY, USA).
Neurotoxicity assays
N2a cells were seeded at 2.0 × 105 cells/ml in 96-well plates in DMEM containing 10% FBS. After transfection of each plasmid, the cells were differentiation for 48 h in low glucose (1.0 g/l glucose) DMEM supplemented with 2% FBS and 2 mM N,N-dibutyladenosine 3′,5′-phosphoric acid (dbcAMP; Nacalai tesque, Kyoto, Japan) with or without EBGP, kaempherol, or kaempheride in the presence or absence of 3-MA. The number of live cells was estimated by Cell Counting Kit-8, following the manufacturer’s instructions (Wako Pure Chemical Industries Ltd.). Briefly, reagent was added into wells and the plate was incubated at 37 °C for 4 h. The optical density of formazan was detected at 450 nm by GloMax® (Promega, Madison, WI, USA) for calculating cell viability. The wavelength of 600 nm was used as reference.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 10 min. And cells were permeabilized with 0.1% tritonX/PBS for 30 min at RT. The 2% goat serum was used for blocking reaction for 60 min. Samples were incubated with primary antibody (rabbit anti-SOD1 antibody, 1:150, Enzo Life Sciences, Inc, Farmingdale, NY, USA) at 4 °C overnight. And we incubated cells with second antibody (goat anti-mouse antibody Alexia 555, (Thermo Fisher Scientific)) at RT for 30 min. Samples were observed by using confocal imaging system (Zeiss LSM 700).
ROS production
To detect mutant SOD1 induced ROS production, we used a Red mitochondrial superoxide indicator, MitoSOX® Red (Thermo Fisher Scientific). N2a cells were prepared in uncoated glass-bottomed microwells. After transfection of each plasmid, MitoSOX® Red was added to the cell culture to a final concentration of 5 μM to the cells and incubate for 30 min at 37 °C following the manufacturer’s instructions (Thermo Fisher Scientific). Fluorescent microscopy images were acquired using a confocal fluorescence microscope (LSM700, Carl Zeiss) and the images were analyzed using ZEN software (Carl Zeiss).
ESR analysis
The measurement condition for ESR (JES-FA 200, JEOL) was applied as following: center field 330 mT; sweep width, 1.5 × 10 mT; sweep time, 4 min; field modulation width, 2 × 0.1 mT; amplitude, 5.0 × 100; time constant, 0.3 s; microwave power, 4.0 mW. The •OH scavenging activity was calculated from the integration values of the ESR spectra corrected with a Mn2+ marker. The •OH was generated by the Fenton reaction. Sample solution (50 μL), 72 mM DMPO (50 μL), 2 mM H2O2 (50 μL), and 0.2 mM FeSO4 (50 μL) were mixed and transferred to an ESR spectrometry cell. Exactly 1 min after the addition of FeSO4, ESR spectrum of DMPO-OH spin adducts was recorded.
Statistical analysis
Data are given as the mean ± standard error of the mean (SEM). The significance of differences was determined by Student’s t test or an analysis of variance (ANOVA). Further statistical analysis for post hoc comparisons was performed using the Bonferroni/Dunn test (StatView, Abacus, Baltimore, MD, USA). A p-value of less than 0.05 was considered to be statistically significant.
References
Julien, J. P. Amyotrophic lateral sclerosis. unfolding the toxicity of the misfolded. Cell 104, 581–591, doi:10.1016/S0092-8674(01)00244-6 (2001).
Banerjee, R., Beal, M. F. & Thomas, B. Autophagy in neurodegenerative disorders: pathogenic roles and therapeutic implications. Trends Neurosci 33, 541–549, doi:10.1016/j.tins.2010.09.001 (2010).
Cheroni, C. et al. Functional alterations of the ubiquitin-proteasome system in motor neurons of a mouse model of familial amyotrophic lateral sclerosis. Hum Mol Genet 18, 82–96, doi:10.1093/hmg/ddn319 (2009).
Allen, S. et al. Analysis of the cytosolic proteome in a cell culture model of familial amyotrophic lateral sclerosis reveals alterations to the proteasome, antioxidant defenses, and nitric oxide synthetic pathways. J Biol Chem 278, 6371–6383, doi:10.1074/jbc.M209915200 (2003).
Urushitani, M., Kurisu, J., Tsukita, K. & Takahashi, R. Proteasomal inhibition by misfolded mutant superoxide dismutase 1 induces selective motor neuron death in familial amyotrophic lateral sclerosis. J Neurochem 83, 1030–1042, doi:10.1046/j.1471-4159.2002.01211.x (2002).
Kikuchi, H. et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc Natl Acad Sci USA 103, 6025–6030, doi:10.1073/pnas.0509227103 (2006).
Wong, E. & Cuervo, A. M. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 13, 805–811, doi:10.1038/nn.2575 (2010).
Chhangani, D. et al. Mahogunin ring finger 1 confers cytoprotection against mutant SOD1 aggresomes and is defective in an ALS mouse model. Neurobiol Dis 86, 16–28, doi:10.1016/j.nbd.2015.11.017 (2016).
Drago, L. et al. In vitro antimicrobial activity of propolis dry extract. J Chemother 12, 390–395, doi:10.1179/joc.2000.12.5.390 (2000).
Amoros, M. et al. Comparison of the anti-herpes simplex virus activities of propolis and 3-methyl-but-2-enyl caffeate. J Nat Prod 57, 644–647, doi:10.1021/np50107a013 (1994).
Paulino, N. et al. Evaluation of the analgesic and anti-inflammatory effects of a Brazilian green propolis. Planta Med 72, 899–906, doi:10.1055/s-2006-947185 (2006).
Xuan, H. et al. Antitumor Activity of Chinese Propolis in Human Breast Cancer MCF-7 and MDA-MB-231 Cells. Evid. Based Complement Alternat Med 2014, 280120–11, doi:10.1155/2014/280120 (2014).
Kudo, D. et al. Conditioned medium of dental pulp cells stimulated by Chinese propolis show neuroprotection and neurite extension in vitro. Neurosci Lett 589, 92–97, doi:10.1016/j.neulet.2015.01.035 (2015).
Maruyama, H. et al. Antihypertensive effects of flavonoids isolated from brazilian green propolis in spontaneously hypertensive rats. Biol Pharm Bull 32, 1244–1250, doi:10.1248/bpb.32.1244 (2009).
Abel, O., Powell, J. F., Andersen, P. M. & Al-Chalabi, A. ALSoD: A user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum Mutat 33, 1345–1351, doi:10.1002/humu.v33.9 (2012).
Cao, X. et al. Structures of the G85R variant of SOD1 in familial amyotrophic lateral sclerosis. J Biol Chem 283, 16169–16177, doi:10.1074/jbc.M801522200 (2008).
Watanabe, S., Hayakawa, T., Wakasugi, K. & Yamanaka, K. Cystatin C protects neuronal cells against mutant copper-zinc superoxide dismutase-mediated toxicity. Cell Death Dis 5, e1497, doi:10.1038/cddis.2014.459 (2014).
Watanabe, S. et al. SIRT1 overexpression ameliorates a mouse model of SOD1-linked amyotrophic lateral sclerosis via HSF1/HSP70i chaperone system. Mol Brain 7, 62, doi:10.1186/s13041-014-0062-1 (2014).
Oh, Y. K., Shin, K. S., Yuan, J. & Kang, S. J. Superoxide dismutase 1 mutants related to amyotrophic lateral sclerosis induce endoplasmic stress in neuro2a cells. J Neurochem 104, 993–1005, doi:10.1111/jnc.2008.104.issue-4 (2008).
Naramoto, K., Kato, M. & Ichihara, K. Effects of an ethanol extract of Brazilian green propolis on human cytochrome P450 enzyme activities in vitro. J Agric Food Chem 62, 11296–11302, doi:10.1021/jf504034u (2014).
Huang, W. W. et al. Kaempferol induces autophagy through AMPK and AKT signaling molecules and causes G2/M arrest via downregulation of CDK1/cyclin B in SK-HEP-1 human hepatic cancer cells. Int J Oncol 42, 2069–2077, doi:10.3892/ijo.2013.1909 (2013).
Filomeni, G. et al. Neuroprotection of kaempferol by autophagy in models of rotenone-mediated acute toxicity: possible implications for Parkinson’s disease. Neurobiol Aging 33, 767–785, doi:10.1016/j.neurobiolaging.2010.05.021 (2012).
Jeong, J. M. et al. Antioxidant and chemosensitizing effects of flavonoids with hydroxy and/or methoxy groups and structure-activity relationship. J Pharm Pharm Sci 10, 537–546, doi:10.18433/J3KW2Z (2007).
Zimmerman, M. C., Oberley, L. W. & Flanagan, S. W. Mutant SOD1-induced neuronal toxicity is mediated by increased mitochondrial superoxide levels. J Neurochem 102, 609–618, doi:10.1111/jnc.2007.102.issue-3 (2007).
Bosco, D. A. et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci 13, 1396–1403, doi:10.1038/nn.2660 (2010).
Forsberg, K. et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS One 5, e11552, doi:10.1371/journal.pone.0011552 (2010).
Hata, T. et al. Artepillin C, a major ingredient of Brazilian propolis, induces a pungent taste by activating TRPA1 channels. PLoS One 7, e48072, doi:10.1371/journal.pone.0048072 (2012).
Basso, M. et al. Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J Biol Chem 281, 33325–33335, doi:10.1074/jbc.M603489200 (2006).
Cheroni, C. et al. Accumulation of human SOD1 and ubiquitinated deposits in the spinal cord of SOD1G93A mice during motor neuron disease progression correlates with a decrease of proteasome. Neurobiol Dis 18, 509–522, doi:10.1016/j.nbd.2004.12.007 (2005).
Tan, W., Pasinelli, P. & Trotti, D. Role of mitochondria in mutant SOD1 linked amyotrophic lateral sclerosis. Biochim Biophys Acta 1842, 1295–1301, doi:10.1016/j.bbadis.2014.02.009 (2014).
Tafuri, F. et al. SOD1 misplacing and mitochondrial dysfunction in amyotrophic lateral sclerosis pathogenesis. Front Cell Neurosci 9, 336, doi:10.3389/fncel.2015.00336 (2015).
Mancuso, R. et al. Resveratrol improves motoneuron function and extends survival in SOD1(G93A) ALS mice. Neurotherapeutics 11, 419–32, doi:10.1007/s13311-013-0253-y (2014).
Daniel, B., Green, O., Viskind, O. & Gruzman, A. Riluzole increases the rate of glucose transport in L6 myotubes and NSC-34 motor neuron-like cells via AMPK pathway activation. Amyotroph. Lateral Scler. Frontotemporal Degener 14, 434–443, doi:10.3109/21678421.2013.808226 (2013).
Lim, M. A. et al. Reduced activity of AMP-activated protein kinase protects against genetic models of motor neuron disease. J Neurosci 32, 1123–1141, doi:10.1523/JNEUROSCI.6554-10.2012 (2012).
Acknowledgements
We thank Mr. Daichi Kudo for technical assistance. This work was supported by grants from JSPS KAKENHI, 17H04198 (I.H.), 17K08311 (M.I.) and 17K18001 (H.K.).
Author information
Authors and Affiliations
Contributions
T.U., M.I., and I.H. designed research; T.U., M.I., K.S., S.S., Y.M., and H.K. performed research; T.U., M.I., K.S., S.S., Y.M., H.K., K.I., T.I., and H.I. analyzed data; T.U., M.I., and I.H. wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ueda, T., Inden, M., Shirai, K. et al. The effects of Brazilian green propolis that contains flavonols against mutant copper-zinc superoxide dismutase-mediated toxicity. Sci Rep 7, 2882 (2017). https://doi.org/10.1038/s41598-017-03115-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-03115-y
This article is cited by
-
2(3H)-Dihydrofranolactone metabolites from Pleosporales sp. NUH322 as anti-amyotrophic lateral sclerosis drugs
Journal of Natural Medicines (2024)
-
Impact of Plant-Derived Compounds on Amyotrophic Lateral Sclerosis
Neurotoxicity Research (2023)
-
Propolis: An update on its chemistry and pharmacological applications
Chinese Medicine (2022)
-
The neuroprotective effects of activated α7 nicotinic acetylcholine receptor against mutant copper–zinc superoxide dismutase 1-mediated toxicity
Scientific Reports (2020)
-
Comparison of aqueous, polyethylene glycol-aqueous and ethanolic propolis extracts: antioxidant and mitochondria modulating properties
BMC Complementary and Alternative Medicine (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.