Abstract
Guanine nucleotide-binding protein/α-subunit (GNAS) mutations are involved in fibrous dysplasia (FD) pathogenesis. Here, we analyzed GNAS mutations in FD which were performed by pyrosequencing DNA isolated from formalin-fixed paraffin-embedded (FFPE) tissue. The mutation detection rate was determined in FD specimens with and without decalcification. GNAS mutation was identified in 28 cases out of 87 FDs (32.18%) [p.R201C (N = 14) and p.R201H (N = 14)]. GNAS mutation was more likely to occur in polyostotic FD (7/28, 25.0%); FD without GNAS mutation was mostly monostotic form (56/59, 94.9%, P = 0.011). The G > A (R201H) mutation was more frequent in polyostotic FD (6/14 patients, 42.9%) than the C > T (R201C) mutation (1/14, 7.1%) (P = 0.077). We divided the FD cases into two subgroups: tissue specimens that were not decalcified (N = 35, 40.2%), and tissue specimens that were decalcified (N = 52, 59.8%). GNAS mutation was more frequently identified in FD specimens that were not subjected to decalcification (23/35, 65.7%) than in FD specimens that were decalcified (5/52, 9.6%) (P = 0.001). In conclusion, mutation analysis of GNAS by pyrosequencing has diagnostic value in FFPE tissue of patients with FD, especially in specimens that were not decalcified. The R201H substitution mutation of GNAS may be involved in the pathogenesis of polyostotic FD.
Similar content being viewed by others
Introduction
Fibrous dysplasia (FD) is a frequently encountered benign fibro-osseous lesion. FD is histologically characterized by fibrous tissue with bland-looking fibroblasts, irregularly shaped woven bone, and no osteoblastic rimming. FD can be involved in one site (monostotic disease) or several sites (polyostotic disease) of any bone. In approximately 5% of all cases, polyostotic disease may be associated with McCune-Albright syndrome1. Monostotic and polyostotic FD, McCune-Albright syndrome, and soft tissue myxoma with FD all result from somatic mutation of the same guanine nucleotide-binding protein/a-subunit (GNAS) gene2,3,4.
GNAS is located on chromosome 20q13.3, which encodes the α-subunit of the heterotrimeric G (Gsα) protein complex4,5,6. GNAS mutations induce the activation of G-protein a-subunit and cause FD, also known as McCune-Albright syndrome5,6,7. FD is thought to be predominantly caused by two mutations in exon 8 of the GNAS gene8. These mutations are substitutions in codon 201, which lead to the replacement of arginine 201 with histidine (R201H) or cysteine (R201C). Substitution of arginine 201 with serine (R201S)9, leucine (R201L)10, 11, and glycine (R201G) have been reported in rare cases12. One study reported a point mutation at codon 227 (glutamine) in exon 9 of GNAS, which led to replacement with leucine (Q227L)13.
Several studies showed that no GNAS mutations were detected in all other benign fibro-osseous lesions, and suggested that mutation analysis of GNAS is a reliable and valuable diagnostic tool for FD. A meta-analysis of 203 patients with FD reported the positive detection of GNAS mutation in 71.9% of the patients14. However, the detection rate in subsequent studies varied from 45–95% due to tissue mosaicism and sensitivity of the technique15,16,17,18,19.
Several methods can be used to detect GNAS mutations in paraffin-embedded tissues or frozen samples, including Sanger direct sequencing14, 17, allele-specific PCR20, PCR with mutation-specific restriction enzyme digestion13, and coamplification at lower denaturation temperature PCR (COLD-PCR)4. Recently, Liang et al. reported a detection sensitivity of GNAS mutation as high as 95% using next-generation pyrosequencing19. They isolated DNA from undercalcified, formalin-fixed, methylmethacrylate-embedded tissue. However, most other studies used tissue specimens that had been subjected to various decalcification methods before preparing formalin-fixed paraffin-embedded (FFPE) FD tissue samples. Calcified tissues were treated with strong acid for decalcification before fixation and embedding. This harsh treatment may affect DNA integrity, and reduce the quantity and quality of DNA for amplification and sequencing21.
In this study, we evaluated the effect of tissue decalcification on the diagnosis of FD by pyrosequencing for GNAS mutations. We selected FFPE specimens of patients with FD according to whether they had undergone decalcification or not, and used these samples to detect GNAS mutations. We also analyzed clinicopathological features according to GNAS mutation status.
Results
Clinical and pathological features
The clinical features of 87 patients with FD are presented in Supplementary Table 1. The male-to-female ratio of patients with FD was approximately 1:1 (43 males and 44 females). The median onset age was 31.02 years (range, 1–85 years). A total of 34 patients had FD located at craniofacial sites (34/87, 39.1%), and the remainder were located at extracraniofacial sites (53/87, 60.9%). Ten patients had multiple bone lesions at FD diagnosis, which radiologically suggested polyostotic FD. One of these 10 patients was diagnosed with Albright-McCune syndrome. Tissue samples were not decalcified before formalin fixation for 35 patients; the tissues samples of the remaining 52 patients were decalcified before formalin fixation.
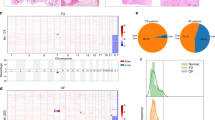
Histological examination indicated that most FD lesions contained varying proportions of fibrous tissue with irregular, curvilinear, and trabeculae of woven bone without osteoblastic rimming (Fig. 1). Some samples displayed aggregation of foamy histiocytes and cystic change. Benign cartilaginous differentiation was observed in one sample.
Histological features of fibrous dysplasia, which contains fibrous tissue with irregular, curvilinear, and trabeculae of woven bone in varying proportions. (A) Low fibrous tissue. (B) Abundant fibrous tissue. (C) Typical irregularly shape woven bones without osteoblastic rimming were observed in most cases. Some cases showed (D) aggregation of foamy histiocytes and (E) cystic changes. (F) Benign cartilaginous differentiation was observed in one case.
GNAS mutation in fibrous dysplasia
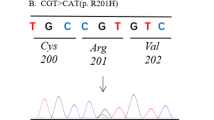
We performed pyrosequencing and detected GNAS mutations in 28 out of 87 patients with FD (32.2%, Table 1). Fourteen cases had the R201H mutation, and 14 cases had the R201C mutation (Fig. 2). We divided the FD patients into two groups, FD with GNAS mutation and FD without GNAS mutation, and compared clinical features in the two groups (Table 1). The results showed that polyostotic disease was observed more frequently in FD cases with GNAS mutation (7/28, 25.0%) than in FD without GNAS mutation (3/59, 5.1%, P = 0.011). However, there were no significant differences between the two groups with respect to gender, age at diagnosis, or lesion site.
GNAS mutation rate in fibrous dysplasia specimens with or without decalcification
The observed GNAS mutation rate in tissue samples from patients with FD (32.2%) was remarkably lower than that reported by a previous study (95%)19. Both studies used the same next-generation sequencing method, a pyrosequencing. Therefore, we hypothesized that mutation rates are affected by decalcification, and compared mutation rates according to whether or not the FD tissue samples were decalcified (Table 2). The observed GNAS mutation rate was much lower in FD tissue specimens that had been subjected to decalcification (5/52, 9.6%) than in FD specimens without decalcification (23/35, 65.7%, P = 0.001).
To verify the effect of tissue decalcification on the detection rate of GNAS mutations by pyrosequencing, we divided tissue specimens from 15 FD patients into two parts: one part of each specimen was decalcified, and the other part was not decalcified. Subsequent tissue processing, PCR, and pyrosequencing reactions were the same for both tissue samples. Table 2 compares the observed GNAS mutation rates in these two tissues (decalcified or not) for the 15 FD patients. Consistent with the previous experiment, the observed GNAS mutation rates in decalcified tissue specimens were lower (2/15, 13.3%) that those in undercalcified tissue specimens (9/15, 60.0%, P = 0.021). These combined results indicate that GNAS mutation was not detected in 7 FD patients when the source of DNA for diagnostic pyrosequencing reactions was derived from decalcified tissue specimens.
Clinical features of fibrous dysplasia according to GNAS mutation type
We detected GNAS mutation in 28 out of 87 FD lesions (Table 1). Fourteen of these patients had C > T substitution mutations (R201C) and the other 14 patients had G > A substitution mutations (R201H). Other rare mutations were not identified.
The R201H mutation occurred more frequently in polyostotic FD, whereas the R201C mutation occurred more frequently in monostotic FD (P = 0.077). There were no significant differences in gender, age at diagnosis, or lesion site between patients with R201H and R201C mutations in GNAS leading to FD.
Discussion
GNAS encodes a subunit of the stimulatory cAMP pathway-associated G-protein, Gsα. Activating or gain-of-function GNAS mutations in osteoblastic cells result in adenylate cyclase activation and constitutive elevation of intracellular cyclic adenosine monophosphate (cAMP). Consequently, mutated osteoblastic cells undergo increased cell proliferation and reduced cell differentiation, resulting in overproduction of disorganized fibrotic bone matrix. This process is responsible for the development of FD, which may also present as a feature of McCune-Albright syndrome (endocrine diseases or Café au lait macules occurs in addition to FD)22,23,24,25.
The diagnosis of FD usually requires clinical information, radiographical images, and histological findings, but sometimes it can be challenging. In many cases, it can be difficult or impossible to distinguish FD from other fibro-osseous lesions only based on histological findings. Several studies reported that GNAS mutation was absent in other fibro-osseous disorders such as low-grade osteosarcoma, ossifying fibroma, and osteofibrous dysplasia, and therefore GNAS gene analysis can aid the differntiation of FD from such disease entities15, 16, 26.
A previous meta-analysis reported that the detection rate of GNAS mutation varied from 45–95%15,16,17,18,19, and the overall positive detection rate of GNAS mutation was 71.9% in 203 patients with FD14. In our study, pyrosequencing results detected GNAS mutations in only 32.2% of FD cases, regardless of decalcification. This result might be explained by differences in methodologies used to detect GNAS mutations, which result in different sensitivities. Several methods for detecting mutations have been published, including direct sequencing7, mutation-specific restriction enzyme digestion13, 26, PCR coupled with allele-specific oligonucleotide hybridization6, PCR–restriction fragment length polymorphism27, PCR with peptide nucleic acid28, and PCR with pyrosequencing19. A previous study performed pyrosequencing and reported the detection of GNAS mutations in 95% of FD patients, which is a much higher detection rate than that observed in the present study.
Next, we evaluated the effect of tissue decalcification on the detection of GNAS mutations in patients with FD. Many standard protocols for preparing bony tissue samples include treatment with strong acid to decalcify the tissue before fixation and embedding. This harsh treatment may affect the integrity of DNA isolated from the tissue, and thereby reduce the quantity and quality of DNA for PCR amplification and sequencing21. In this study, specimens were decalcified with Calci-Clear Rapid solution for 24 hours. The detection rate of GNAS mutations was 9.6% (5/52) in decalcified specimens and 65.7% (23/35) in non-decalcified specimens. A comparison of the detection rates of GNAS mutations in FD samples from 15 FD patients that were either decalcified or undercalcified also indicated that the detection rate of GNAS mutation was much higher in undercalcified tissues (9/15, 60.0%) than in decalcified tissues (2/15, 13.3%, p = 0.021).
The present study showed that patients with polyostotic FD more frequently displayed p.R201H mutation in GNAS than patients with monostotic FD (p = 0.077). This result was not consistent with a previous report that patients with polyostotic FD had R201C mutation in GNAS 29. We identified p.R201C mutation in one of seven patients with polyostotic FD. The lack of statistical significance in our result may be due to the small number of cases in our study.
In this study, we demonstrated that GNAS mutations were more frequently detected in FD tissue samples without decalcification than in decalcified FD tissue samples. And, most of the polyostotic FD patients displayed R201H substitution mutation in GNAS. Therefore, GNAS mutation test should be conducted without performing decalcification. Also, if sequencing analysis detects an R201H mutation in GNAS, we suggest that the clinician should consider the possibility of polyostotic FD.
Materials and Methods
Patients and samples
We retrospectively recruited 93 patients who were diagnosed with FD at Severance Hospital, Yonsei University College of Medicine, from 2006 to 2016. A total of 107 tissue samples were collected. The specimens included 72 decalcified FFPE specimens and 35 non-decalcified FFPE specimens. The DNA isolated from six patients was not suitable for molecular studies; therefore, we analyzed the DNA of 87 patients with FD. Ten patients presented with multiple bone lesions on radiological studies when FD was initially diagnosed, suggesting polyostotic FD. Among the ten polyostotic patients, only two patients underwent multiple biopsies and/or curettages from different locations, whereas the other patients received only one biopsy or curettage from a representative lesion. In 15 cases, only part of the specimen was subjected to decalcification and the rest was not decalcified, which enabled a direct comparison of the effect of decalcification on the detection of mutations in a tissue specimen. For decalcification, the tissue samples were decalcified with Calci-Clear Rapid solution containing EDTA and hydrochloric acid (National Diagnostics, Georgia, USA) for 24 hours, fixed in 10% formalin, and then embedded in paraffin.
To select the most representative FFPE tissues for molecular studies, samples were mounted on slides, stained with hematoxylin and eosin (H&E), and reviewed by two pathologists (SJS and SKK). Medical records were reviewed for clinical features, and radiological and pathological findings.
All methods and experimental protocols using human tissue (FFPE tissue) were carried out in accordance with relevant guidelines and regulations approved by the Institutional Review Board of Severance Hospital, Yonsei University Health System (4-2016-0276). The informed consent was waved because the IRB decided that this retrospective study had a minimal risk to the patients (risk level I).
DNA extraction and PCR amplification
Genomic DNA was extracted from 10 μm sections cut from FFPE tissue blocks using the Maxwell® CSC DNA FFPE extraction kit (Promega, Wisconsin, USA) and the Maxwell® CSC instrument. The genomic DNA was subjected to PCR amplification optimized for pyrosequencing analysis; reactions contained 9 μl (10 ng/μl) of DNA, 12.5 μl of 2 × PyroMark PCR master mix (QIAGEN, Hilden, Germany), 2.5 μl of 10x CoralLoad PCR loading buffer (QIAGEN), and 1 μl each of the forward and reverse primers, in a total reaction volume of 25 μl (Supplementary Table 2). The following program was used for PCR amplification: 95 °C for 15 minutes; followed by 45 cycles of 94 °C for 30 seconds, 59 °C for 30 seconds, and 72 °C for 30 seconds; and a final extension step of 72 °C for 10 minutes.
Pyrosequencing
After PCR analysis, single-stranded DNA suitable for pyrosequencing was prepared using the PyroMark Q24 MDx Vacuum Workstation (QIAGEN, Hilden, Germany). In workstation, the biotinylated PCR products were bound to streptavidin-coated sepharose beads. The beads were released into a PyroMark Q24 Plate containing 25 μl of annealing buffer and 0.3 μM of sequencing primer. The sample was heated to 80 °C for 2 minutes, and then cooled to room temperature. Pyrosequencing was performed in a PyroMark Q24 MDx (QIAGEN) according to the manufacturer’s instructions. Sequencing data were analyzed using PyroMark Q24 MDx Software 2.0 (QIAGEN).
Statistical analysis
All statistical analyses were performed using SPSS version 18.0 (IBM, Chicago, IL, USA). The relationships between groups were compared using the χ2 test, Fisher’s exact test, or the Mann-Whitney U-test. Two-sided P values < 0.05 were considered as statistically significant.
References
Gorham, L. W., Campbell, E. H., Howard, W. C., Donhauser, J. L. & Rust, N. H. Albright’s Syndrome-A Group of Cases Characterized by Osteitis Fibrosa Disseminata, Areas of Pigmentation and a Gonadal Dysfunction. Trans Am Clin Climatol Assoc 57, 179–187 (1941).
Okamoto, S. et al. Activating Gs(alpha) mutation in intramuscular myxomas with and without fibrous dysplasia of bone. Virchows Arch 437, 133–137, doi:10.1007/s004280000217 (2000).
Marie, P. [Cellular and molecular biology of fibrous dysplasia]. Ann Pathol 21, 489–498 (2001).
Delaney, D. et al. GNAS1 mutations occur more commonly than previously thought in intramuscular myxoma. Mod Pathol 22, 718–724, doi:10.1038/modpathol.2009.32 (2009).
Weinstein, L. S. et al. Activating mutations of the stimulatory G protein in the McCune-Albright syndrome. N Engl J Med 325, 1688–1695, doi:10.1056/NEJM199112123252403 (1991).
Shenker, A., Weinstein, L. S., Sweet, D. E. & Spiegel, A. M. An activating Gs alpha mutation is present in fibrous dysplasia of bone in the McCune-Albright syndrome. J Clin Endocrinol Metab 79, 750–755, doi:10.1210/jcem.79.3.8077356 (1994).
Bianco, P. et al. Mutations of the GNAS1 gene, stromal cell dysfunction, and osteomalacic changes in non-McCune-Albright fibrous dysplasia of bone. J Bone Miner Res 15, 120–128, doi:10.1359/jbmr.2000.15.1.120 (2000).
Alman, B. A., Greel, D. A. & Wolfe, H. J. Activating mutations of Gs protein in monostotic fibrous lesions of bone. J Orthop Res 14, 311–315, doi:10.1002/jor.1100140221 (1996).
Candeliere, G. A., Roughley, P. J. & Glorieux, F. H. Polymerase chain reaction-based technique for the selective enrichment and analysis of mosaic arg201 mutations in G alpha s from patients with fibrous dysplasia of bone. Bone 21, 201–206, doi:10.1016/S8756-3282(97)00107-5 (1997).
Gorelov, V. N. et al. Overexpression of Gs alpha subunit in thyroid tumors bearing a mutated Gs alpha gene. J Cancer Res Clin Oncol 121, 219–224, doi:10.1007/BF01366965 (1995).
Lietman, S. A., Ding, C. & Levine, M. A. A highly sensitive polymerase chain reaction method detects activating mutations of the GNAS gene in peripheral blood cells in McCune-Albright syndrome or isolated fibrous dysplasia. J Bone Joint Surg Am 87, 2489–2494, doi:10.2106/JBJS.E.00160 (2005).
Riminucci, M. et al. A novel GNAS1 mutation, R201G, in McCune-albright syndrome. J Bone Miner Res 14, 1987–1989, doi:10.1359/jbmr.1999.14.11.1987 (1999).
Idowu, B. D. et al. A sensitive mutation-specific screening technique for GNAS1 mutations in cases of fibrous dysplasia: the first report of a codon 227 mutation in bone. Histopathology 50, 691–704, doi:10.1111/j.1365-2559.2007.02676.x (2007).
Lee, S. E. et al. The diagnostic utility of the GNAS mutation in patients with fibrous dysplasia: meta-analysis of 168 sporadic cases. Hum Pathol 43, 1234–1242, doi:10.1016/j.humpath.2011.09.012 (2012).
Tabareau-Delalande, F. et al. Diagnostic value of investigating GNAS mutations in fibro-osseous lesions: a retrospective study of 91 cases of fibrous dysplasia and 40 other fibro-osseous lesions. Mod Pathol 26, 911–921, doi:10.1038/modpathol.2012.223 (2013).
Shi, R. R., Li, X. F., Zhang, R., Chen, Y. & Li, T. J. GNAS mutational analysis in differentiating fibrous dysplasia and ossifying fibroma of the jaw. Mod Pathol 26, 1023–1031, doi:10.1038/modpathol.2013.31 (2013).
Walther, I., Walther, B. M., Chen, Y. & Petersen, I. Analysis of GNAS1 mutations in myxoid soft tissue and bone tumors. Pathol Res Pract 210, 1–4, doi:10.1016/j.prp.2013.09.003 (2014).
Salinas-Souza, C. et al. GNAS mutations are not detected in parosteal and low-grade central osteosarcomas. Mod Pathol 28, 1336–1342, doi:10.1038/modpathol.2015.91 (2015).
Liang, Q. et al. Quantitative analysis of activating alpha subunit of the G protein (Gsalpha) mutation by pyrosequencing in fibrous dysplasia and other bone lesions. J Mol Diagn 13, 137–142, doi:10.1016/j.jmoldx.2010.10.003 (2011).
Schwindinger, W. F., Francomano, C. A. & Levine, M. A. Identification of a mutation in the gene encoding the alpha subunit of the stimulatory G protein of adenylyl cyclase in McCune-Albright syndrome. Proc Natl Acad Sci USA 89, 5152–5156, doi:10.1073/pnas.89.11.5152 (1992).
Wickham, C. L. et al. Formic acid decalcification of bone marrow trephines degrades DNA: alternative use of EDTA allows the amplification and sequencing of relatively long PCR products. Mol Pathol 53, 336–336, doi:10.1136/mp.53.6.336 (2000).
Ringel, M. D., Schwindinger, W. F. & Levine, M. A. Clinical implications of genetic defects in G proteins. The molecular basis of McCune-Albright syndrome and Albright hereditary osteodystrophy. Medicine (Baltimore) 75, 171–184, doi:10.1097/00005792-199607000-00001 (1996).
Spiegel, A. M. Inborn errors of signal transduction: mutations in G proteins and G protein-coupled receptors as a cause of disease. J Inherit Metab Dis 20, 113–121, doi:10.1023/A:1005393501786 (1997).
Spiegel, A. M. The molecular basis of disorders caused by defects in G proteins. Horm Res 47, 89–96, doi:10.1159/000185441 (1997).
Marie, P. J., de Pollak, C., Chanson, P. & Lomri, A. Increased proliferation of osteoblastic cells expressing the activating Gs alpha mutation in monostotic and polyostotic fibrous dysplasia. Am J Pathol 150, 1059–1069 (1997).
Pollandt, K., Engels, C., Kaiser, E., Werner, M. & Delling, G. Gsalpha gene mutations in monostotic fibrous dysplasia of bone and fibrous dysplasia-like low-grade central osteosarcoma. Virchows Arch 439, 170–175, doi:10.1007/s004280100453 (2001).
Sakamoto, A., Oda, Y., Iwamoto, Y. & Tsuneyoshi, M. A comparative study of fibrous dysplasia and osteofibrous dysplasia with regard to Gsalpha mutation at the Arg201 codon: polymerase chain reaction-restriction fragment length polymorphism analysis of paraffin-embedded tissues. J Mol Diagn 2, 67–72, doi:10.1016/S1525-1578(10)60618-6 (2000).
Toyosawa, S. et al. Ossifying fibroma vs fibrous dysplasia of the jaw: molecular and immunological characterization. Mod Pathol 20, 389–396, doi:10.1038/modpathol.3800753 (2007).
Cohen, M. M. Jr. & Howell, R. E. Etiology of fibrous dysplasia and McCune-Albright syndrome. Int J Oral Maxillofac Surg 28, 366–371, doi:10.1016/S0901-5027(99)80085-X (1999).
Acknowledgements
This work was supported by a faculty research grant of Yonsei University College of Medicine (6-2015-0087 and 6-2016-0034).
Author information
Authors and Affiliations
Contributions
S.K.K. designed the study. S.K.K. and S.J.S. conducted the study. S.J.S. and S.J.L. collected data. S.J.S. and S.J.L. analyzed data. S.K.K., S.J.S., and S.J.L. interpreted data. S.K.K. and S.J.S. drafted the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shin, SJ., Lee, S.J. & Kim, S.K. Frequency of GNAS R201H substitution mutation in polyostotic fibrous dysplasia: Pyrosequencing analysis in tissue samples with or without decalcification. Sci Rep 7, 2836 (2017). https://doi.org/10.1038/s41598-017-03093-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-03093-1
This article is cited by
-
GNAS mutation analysis assists in differentiating chronic diffuse sclerosing osteomyelitis from fibrous dysplasia in the jaw
Modern Pathology (2022)
-
Effect of decalcification protocols on immunohistochemistry and molecular analyses of bone samples
Modern Pathology (2020)
-
Head and neck manifestations of an undiagnosed McCune-Albright syndrome: clinicopathological description and literature review
Virchows Archiv (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.