Abstract
The chloroplast genome originated from photosynthetic organisms and has retained the core genes that mainly encode components of photosynthesis. However, the causes of variations in chloroplast genome size in seed plants have only been thoroughly analyzed within small subsets of spermatophytes. In this study, we conducted the first comparative analysis on a large scale to examine the relationship between sequence characteristics and genome size in 272 seed plants based on cross-species and phylogenetic signal analysis. Our results showed that inverted repeat regions, large or small single copies, intergenic regions, and gene number can be attributed to the variations in chloroplast genome size among closely related species. However, chloroplast gene length underwent evolution affecting chloroplast genome size in seed plants irrespective of whether phylogenetic information was incorporated. Among chloroplast genes, atpA, accD and ycf1 account for 13% of the variation in genome size, and the average Ka/Ks values of homologous pairs of the three genes are larger than 1. The relationship between chloroplast genome size and gene length might be affected by selection during the evolution of spermatophytes. The variation in chloroplast genome size may influence energy generation and ecological strategy in seed plants.
Similar content being viewed by others
Introduction
The variation in genome size, which simultaneously reflects genotype and phenotype, has been a puzzle for researchers for almost half a century1,2,3. Previous studies have reported the significant associations between the variation in genome size and life history4, 5, taxonomy6, evolutionary affiliation7 and geographical distribution8. These associations were suggested to be determined by selective force1, 3, 9. Genome size change has also been linked to remarkable changes in non-coding sequences, and random drift is regarded as a strong evolutionary force that affects genome size variation10, 11. However, these associations between DNA composition and genome size2, 9 have not been clarified in species over a broad range of evolutionary time. Currently, the development of genome sequence technology and population genetics methods has enabled researchers to identify the signatures of selection or genetic drift of genome size variation12, 13.
Chloroplasts originated from endosymbiotic photosynthetic organisms and retain their own unique DNA encoding multiple genes, including components of light reactions in the photosynthesis process to convert light energy into chemical energy14, 15, and photosynthesis is strictly controlled by the genes in chloroplasts16. Most plant chloroplast genomes have been examined, and they have a very constrained size that ranges from 120 kb to 160 kb17. The limited size change in chloroplast genomes in nearly all of the main lineages in plants indicates the possibility that the chloroplast genome is maintained by natural selection, especially when compared to the random and large-scale size variations in both mitochondrial18 and nuclear genomes19. In seed plants, the chloroplast genome exhibits a conserved genome structure17 that includes two inverted repeats (IRs), through which a long single-copy section (LSC) and a short single-copy section (SSC) are separated. In addition, compared to nuclear and certain plant mitochondrial genomes, chloroplast genomes are small and less prone to recombination, which provides distinct information for studying genome size variation and evolutionary status20, 21. These described features are advantageous for comparative studies because they enable researchers to investigate genome divergences over a broad range of evolutionary time, from early land plants22 to recently domesticated plants, and to detect selection signals of genome size evolution23.
Three important factors have been proposed to drive the variation in chloroplast genome size in previous studies of seed plants: (a) intergenic region variation, which mainly affects the variation in chloroplast genome size within a genus24,25,26,27; (b) variation of an IR region, which is an important characteristic of specific groups, such as gymnosperms, Poaceae and Leguminosae28,29,30,31,32,33,34,35; and (c) gene loss, which is an important reason for the shrinking of chloroplast genome size in some parasitic plants28, 35. However, previous studies of chloroplast genome size that have used limited taxon sampling or comparisons among very distant relatives have yielded results of uncertain generality, and there is a lack of systemic and comprehensive phylogenetic studies. It remains unclear which of the three factors has a greater influence on genome size, and the recontribution of natural selection to genome variation is still unknown.
In this study, we collected and annotated 272 complete chloroplast genomes of seed plants, and phylogenies were constructed as a basis to infer the evolutionary mechanism of chloroplast genome size. We first analyzed the general structures of the 272 chloroplast genomes with phylogenetic information incorporated; then, we compared the general structures of chloroplast genomes among monocots, eudicots, basal angiosperms and gymnosperms. Second, we assessed the influence of different sequence characteristics on the variations in chloroplast genome size through a conventional analysis of variance based on cross-species and phylogenetic signal analyses. The analyses suggested that variations in genome size originate from lineage-specific differences in intergenic region variation, and the generality of the genome size and gene length relationship was confirmed by a cross-species and independent contrasts analysis. Finally, the variation patterns and the results of principal component analyses of 126 chloroplast genes were compared among the 272 species. It was demonstrated that atpA, accD and ycf1 may influence photosynthesis for plant adaptation. The variations in chloroplast genome size may play an important genetic role in influencing the energy generation and ecological strategy of a species.
Results
General variation of the chloroplast genome in seed plants
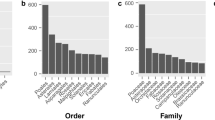
Two-hundred and seventy-two complete or nearly complete chloroplast genomes were collected from 67 families of 45 orders, including 32 genomes from gymnosperms, 15 from basal angiosperms, 50 from monocots and 175 from eudicots respectively. The 32 gymnosperm chloroplast genomes were derived from 10 families, which included all the major basal lineages of gymnosperms except Araucariaceae and Taxodiaceae. The 240 angiosperm chloroplast genomes were collected from 57 families, including all eight orders of basal angiosperms, six (out of 12) orders of monocots and 22 (out of 43) orders of eudicots, which covered the four major branches fabids, malvids, lamiids and campanulids. General variations in the chloroplast genome were analyzed in these species for the total length of the chloroplast genome (TL), the length of the inverted repeat region (IRL), large single copy (LSCL), small single copy (SSCL), gene region (GRL), intergenic region (IGRL), GC content (GCC), and gene number (GN) (see Supplementary Table S1 and Fig. 1).
Variations in chloroplast genome size and the sequence characteristics of chloroplast genomes within seed plants. The box plots of chloroplast genome size and sequence characteristics of chloroplast genome are shown for each order. The complete order names are in Supplementary Table S1. The box plots represent the median (central line), first and third quartiles (black box), and outliers (black circles). TL, IRL, LSCL, SSCL, GRL, IGRL, and GN indicate the total length of the chloroplast genome, length of the inverted repeat region, large single copy, small single copy, gene region, intergenic region, and gene number, respectively. Red triangles in TL indicate there were no inverted repeat regions in these species. The numbers below order names are the number of species collected in each order. The number in the brackets indicates the number of species without an inverted repeat region. The three lines from the top to bottom represent the first quartiles, the median, and the third quartiles of the sequence characteristics of all seed plants examined in this study, respectively.
The TL ranged from 70,028 bp (Epifagus virginiana, eudicot) to 217,942 bp (Pelargonium x hortorum, eudicot). The median and the first and the third quartiles of TL were 155,621 bp (Fragaria virginiana, eudicot), 160,076 bp (Eucalyptus marginata, eudicot) and 143,164 bp (Erycina pusilla, monocot), respectively. Trachelium caeruleum (eudicot), Capsicum annuum (eudicot) and Pelargonium x hortorum (eudicot) had the longest LSCL (100,114), SSCL (25,783) and IRL (75,741), respectively. The LSCL (19,799) and SSCL (4759) of Epifagus virginiana (eudicot) were smaller than those of other species, and the IRL (15,114) of Illicium oligandrum (basal angiosperms) was the smallest among all the existing chloroplast genome sequences. The coefficients of variation (standard deviation/mean) of SSCL (0.20) and IRL (0.17) were nearly twice as high as those of LSCL (0.08), IGRL (0.11) and GRL (0.10), which indicated that more samples deviated from the average in the distributions of SSCL and IRL than in those of LSCL, IGRL and GRL. The coefficient of variation of TL (0.09) was close to that of LSCL and GRL. The variation of GCC in chloroplast genomes was small and ranged from 33.80% (Typha latifolia, monocot) to 39.60% (Pelargonium x hortorum, eudicot). The coefficient of variation for GCC was only 0.03. The GNs ranged from 56 (Epifagus virginiana, eudicot) to 165 (Oryza nivara, monocot), and the GNs of 87% species ranged from 110 to 140. The coefficient of variation of GN (0.09) was the same as TL.
We further compared the general variations in chloroplast genomes among monocots, eudicots, basal angiosperms and gymnosperms using a t-test (Fig. 2). LSCL and GRL showed a similar distribution to TL in gymnosperms, basal angiosperms, monocots, and eudicots. The median values of TL (p = 0.001), LSCL (p = 0.08) and GRL (p = 0.001) of gymnosperms were significantly lower than those of angiosperms according to a t-test. In angiosperms, the monocots had a significantly smaller chloroplast genome size compared to eudicots and basal angiosperms (p = 0.003 for TL; p = 0.001 for LSCL; p = 0.003 for GRL). Although the median values of TL, LSCL and GRL were similar in eudicots and basal angiosperms, basal angiosperms (13 for TL; 6 for LSCL; 13 for GRL) had more outliers than eudicots (5 for TL; 2 for LSCL; 1 for GRL). For SSCL and IRL, monocots, eudicots and basal angiosperms had similar ranges and variations (p = 0.23), but gymnosperms had higher ranges and variances than angiosperms (p = 0.003). The distribution of IGRL (p = 0.16) and GCC (p = 0.53) was not significantly different among monocots, eudicots, basal angiosperms and gymnosperms. The GN (p = 0.02) of gymnosperms was significantly smaller than that of angiosperms, whereas monocots, eudicots and basal angiosperms had similar median GN values (p = 0.51).
Variations in chloroplast genome size and the sequence characteristics of chloroplast genome in gymnosperms (pink), basal angiosperms (blue), monocots (yellow), and eudicots (green). The box plots represent the median (central line), first and third quartiles (black box), and outliers (black circles).
Factors influencing chloroplast genome size based on cross-species and phylogenetic signal analyses
We found that inverted repeat region variation and gene loss occurred many times independently. To further explore the relationship between genome size and other characteristics (see Supplementary Table S1), a conventional analyses of variance (ANOVA) was performed based on cross-species and phylogenetic signal analysis (Table 1). For cross-species analyses, large amounts of data related to chloroplast genome size and chloroplast sequence characteristics among species were collected (Fig. 1), and analyses across all species showed that chloroplast genome size was significantly and positively associated with IRL, LSCL, SCCL, GRL, IGRL and GN but not GCC (Table 1). The variations in chloroplast genome size could explain the lower variations in IRL (r 2 = 23%), LSCL (r 2 = 24%) and SCCL (r 2 = 25%) than in GRL (r 2 = 81%), IGRL (r 2 = 69%) and GN (r 2 = 76%). The estimated slopes for chloroplast genome size and IRL (0.05), LSCL (0.04) or SCCL (0.05) were similar and were significantly smaller than those for GRL (1.11), IGRL (0.83) and GN (0.74) (Table 1).
The genome character and sequence variation are significantly associated with phylogenetic signal36. Therefore, closely related species are more likely to have similar genome sizes and characteristics. We first compared the data from the above comparative analysis with the values obtained from 1000 Monte Carlo simulations that randomized the data from the phylogeny tree. We used K to describe the degree of the difference between the F-statistic of simulated data and observed F-statistic distributions. The descriptive statistics (K) of LSCL (K = 0.93), IGRL (K = 0.95), GN (K = 0.96) and IR (K = 0.92) were close to 1 and larger than GRL (K = 0.59), indicating that LSCL, IGRL, GN and IR were more strongly affected by the phylogenetic signal than was GRL. Therefore, we performed a conventional analysis of variance with phylogeny being taken into account and compared the chloroplast genome size with genome characteristics standardized by branch lengths. The slope estimate between chloroplast genome size and GRL obtained from the comparison based on phylogeny analyses (0.91) was significantly smaller when compared to the cross-species results (slope = 1.11). However, the inclusion or exclusion of phylogeny in the analyses for chloroplast genome size and IRL, LSCL, SCCL, IGRL or GN did not lead to any differences in r 2 (Table 1).
IRL, LSCL, SCCL, GRL, IGRL and GN had the same distribution in four phylogenetic groups as TL (Fig. 2). However, the critical values (C) based on a phylogenetically corrected ANOVA, which were obtained from a distribution of 1000 Monte Carlo simulated F-statistics assuming a gradual model of Brownian motion, suggested that IRL (C = 70.52, p = 0.25), LSCL (C = 76.41, p = 0.35), SCCL (C = 71.46, p = 0.55), IGRL (C = 34.51, p = 0.15) and GN (C = 26.42, p = 0.07) were not significantly associated with chloroplast genome size. In other words, the associated relationship observed among IRL, LSCL, SCCL, IGRL and GN could be attributed to phylogenetic signaling. However, after incorporating both chance and phylogeny into the ANOVA, the variations in chloroplast genome size were significantly associated with the variations in GRL (C = 18.26, p = 0.00002), which were higher than the values predicted by the Brownian motion model. The above results indicate that the variation in gene length plays an important role in the variation in chloroplast genome size.
Comparison of chloroplast genes among seed plants
So we explored the variation patterns of chloroplast genes and we standardized gene annotation and gene length. All collected chloroplast genomes were re-annotated using DOGMA with default settings. Two or more successive genes with the same name were annotated as one gene, such as clpP and rpl2, which had more than one exon. Multiple genes with overlapping regions were manually adjusted into one gene, such as orf188 and ndhA. The genes annotated in only one species, including orf221, orf332, orf365 and orf574, were not used in subsequent analyses. In addition, we standardized gene length based on its average and variance. We obtained standardized contrasts (SCs) for further analyses by dividing the difference between the gene length of each species and the average total gene length by the standard deviations.
A total of 126 chloroplast genes were annotated, which were divided into three broad categories and 13 subcategories. The first category (I) comprised genes for the photosynthetic apparatus, including 6 photosystem I, 15 photosystem II, 7 cytochrome b6f, 6 ATP synthase, 1 RuBisCo and 11 NAD(P)H dehydrogenase genes; the second category (II) comprised RNA genes and genes for the genetic apparatus, including 31 transfer RNA, 4 ribosomal RNA, 4 RNA polymerase and 21 ribosomal subunit genes; and the third category (III) consisted of potential genes, including 8 conserved hypothetical chloroplast open reading frames (ycfs), 2 open reading frames (ORFs) and 10 potential protein-coding genes. A total of 106 genes (81% of the total) were found in more than 90% (245) of the species, and 13 genes were found in less than 10% of the species (27). Among these low-frequency genes, three were photosynthetic apparatus genes, one was a tRNA gene and nine were ORFs or ycfs. All of these rare genes were found in gymnosperms and a small proportion of angiosperms (such as Fabaceae, Cucurbitaceae, Araceae and Geraniaceae).
The SC of each gene is shown in Fig. 3a. The coefficient of variation for the SC of each gene ranged from 0.052 (trns-UGA) to 16.49 (orf574) in all genes, and the average was 1.02. The variation in SC of the genes in the first category was smaller than that of the genes in the second and third categories. Principal component analysis of SC indicated that atpA, accD and ycf1 accounted for 13% of the variation in plant chloroplast genome size. The gene atpA, which codes for a small photosystem II polypeptide, accD, which affects leaf development, and ycf1, which is associated with plant survival37, may influence photosynthesis and are associated with plant adaptation. Therefore, variations in these genes during plant evolution may play an important genetic role in determining the energy generation and ecological strategy of a species. Thus, we detected the selection signal of these genes by calculating the Ka/Ks of their homologous gene pairs with the most recent common ancestor of the plants, and the SC of these gene pairs was larger than 1. In addition, we selected five genes (atpI, ndhE, rbcL, rps8, and matK) that had smaller effects on the length of genome than atpA, accD and ycf1 according to principal component analysis. Strength of selection is commonly measured by calculating the ratio of nonsynonymous (change in amino acid) substitution over synonymous (silent) substitutions (Ka/Ks). We calculated the Ka/Ks values associated with terminal branches to measure the strength of selection during the most recent divergence of each species. We investigated the Ka/Ks values for the 8 protein-coding genes from the chloroplast genome, including atpA, accD, ycf1, atpI, ndhE, rbcL, rps8, and matK (Fig. 3b). The average Ka/Ks values of atpA, accD and ycf1 in homologous genes of plants were greater than 1, which was larger than that of other genes.
Relationship between chloroplast genome size and chloroplast gene length. (a) Heat maps of standardized contrast (SC) values for each gene. SCs were obtained by dividing the raw contrasts between gene length and the average gene length by the standard deviation. Green indicates that the gene was lost in the species. White indicates that SC is zero, which indicates that the sequence characteristics of this species were equal to the average of the sequence characteristics of all seed plants collected for this study. Red stands for an SC larger than zero, blue stands for an SC smaller than zero, and larger absolute values of SC are indicated by darker colors. I indicates genes for the photosynthetic apparatus, II comprises RNA genes and genes for the genetic apparatus, and III represents potential genes. (b) The box plots of Ka/Ks values of atpA, accD, ycf1 atpI, ndhE, rbcL, rps8, and matK in the homologous genes of plants.
Discussion
The main purpose of this study was to re-examine the relationship between chloroplast genome size and chloroplast sequence characteristics within seed plants using large datasets of species and two comparative approaches. Across 272 species of 67 families, we found that the variations in the chloroplast genome in closely related species were affected by intergenic region length. The log-scale linearity in the relationship between chloroplast genome size and chloroplast gene length was revealed by both cross-species and phylogenic analyses (Table 1). Moreover, we found that across all species, IGRL or GN accounted for more than 60% of the total variation in chloroplast genome size. However, tests of phylogenetic signal indicated that this pattern of GN was not independent of ancestry. The variations in IRL, LSCL and SCCL were related to the variation in genome size, but they only explained nearly 20% of the total variation in chloroplast genome size based on cross-species analyses (Table 1). Therefore, our results support the assumption that chloroplast gene length is a predictor of plant chloroplast genome size across a long evolutionary timespan (Fig. 3a and Table 1). Additional factors, such as IGRL, GN, IRL, LSCL and SCCL, may play a role in determining variations in the chloroplast genome among closely related species (Table 1), and IRL, LSCL and SCCL may only modulate the chloroplast genome size of a few groups of species (Fig. 1). The variations in IGRL, GN, IRL, LSCL and SCCL are an important reason for variations in chloroplast genomes in closely related species.
Previous studies have reported that gene loss28, 35, inverted repeat region variation29, 32,33,34 and intergenic region variation24,25,26,27 are three important factors driving the variation in chloroplast genome size in plants. Chloroplast gene loss is an important reason for the reduction of chloroplast genomes in some parasitic plants38,39,40,41,42. These parasitic plants are not closely related in phylogeny, but all have undergone similar functional changes in photosynthesis40,41,42. However, both intergenic region variation and inverted repeat region variation associated with the chloroplast genome length diversity were observed in previous study based on comparisons within a genus or a family. For example, the loss of the inverted repeat region often occurs in some species groups, such as Fabaceae43, 44, Pinaceae45 and Geraniaceae46, and this loss is very rare in most other plant families (Fig. 1). Intergenic region variation was the result of comparisons of chloroplast genome length among species within a genus, such as Poaceae10 and Orchidaceae47, 48. All these previous research studies also supported our conclusion.
Chloroplast gene length is an important factor affecting the variations in chloroplast genome size based on phylogenetic signals (Table 1). This result contrasts with the results for nuclear genome size, which was primarily affected by the non-protein-encoding fraction of the genome49,50,51,52,53. Three reasons may explain this outcome: (1) the mutation and recombination rate of the chloroplast genome is much lower compared to the nuclear genome, which results in fewer repeat sequences and transposons54, 55; (2) the chloroplast genome originated from endosymbiotic photosynthetic organisms and retained core genes, which led the length of gene region of the chloroplast genome to be significantly larger than the intergenic region in most plants56,57,58; and (3) the chloroplast genome originated from prokaryotes, whose GN and genome size were strongly correlated because prokaryotes generally exhibit a paucity of non-coding DNA59,60,61.
In our study we considered the phylogenetic factors, which is a big step forward. Our results demonstrate that chloroplast gene length is significantly associated with chloroplast genome size based on both cross-species and phylogenic signal analyses across 272 species. Moreover, we found that among all chloroplast genes, atpA, accD and ycf1 accounted for 13% of the variation in plant chloroplast genome size through principal component analysis (Fig. 3b). AtpA, accD and ycf1 may influence photosynthesis and may be useful for predicting plant responses to environment variation62,63,64,65. AtpA, accD and ycf1 have been completely or partially lost in the plastid genome multiple times during evolution. In the grass chloroplast genome, the degradation of accD and ycf1 occurred in the ancestors of grass. In addition, the accD reading frame underwent a length expansion in cupressophytes66. We also found that the average Ka/Ks values of the homologous gene pairs of the three genes in plants were higher than 1 (Fig. 3b). These findings indicate that these genes, which have a considerable effect on the variations in chloroplast genome size, have undergone strong selection67. Generally, nonsynonymous substitutions are more likely to cause functional changes than synonymous substitutions because the mutation accumulation of atpA, accD and ycf1 can cause variations in photosynthesis efficiency. The relationship between chloroplast genome size and functional gene content variation suggested that the variation in chloroplast genome size may influence photosynthesis, which may cause a higher level of ecological diversity for organisms. These results are important for understanding the processes underlying the complexity of chloroplast genomes and highlight the interdependence between chloroplast genome size and environmental complexity.
Materials and Methods
Plant materials and genome annotation
A total of 272 complete or nearly complete chloroplast genomes were collected from NCBI (National Center for Biotechnology Information), including the genomes of five gymnosperm groups, four clades of eudicots (fabids, malvids, lamiids, and campanulids), one major clade of monocots (commelinids), and basal angiosperms (magnoliids). The details (species name, family names, and accession numbers) of 272 chloroplast genomes are listed in Supplementary Table S1.
In 1986, for the first time, the complete chloroplast genomes of tobacco (Nicotiana tabacum 68) and liverwort (Marchantia polymorpha 69) were obtained and the chloroplast genes were annotated by gene expression. With the expansion of the NCBI database, homology searches by Blastx and Blastn against the GenBank database have been used to annotate chloroplast genes70 for several years. Consequently, the gene names and data annotation information are inconsistent among different studies15, 70. In addition, it is possible that some hypothetical chloroplast open reading frames (ycfs) or open reading frames (ORFs), whose functions and features have been identified, were not updated in previous studies70, 71. DOGMA (Dual Organellar GenoMe Annotator) is a web-based annotation package that solves some of these problems, including typos, incorrect sequences and gene names in GenBank70. Therefore, protein-coding, ribosomal RNA (rRNA) and transfer RNA (tRNA) genes of all the collected chloroplast genomes were re-annotated using DOGMA with the default settings. However, because BLAST cannot provide a precise search for start and stop codons for the protein coding genes and those genes with more than one intron were annotated as two genes70, the start and stop codons must be chosen by manual operation. Thus, we further modified the annotation information using our own Perl scripts.
Phylogenetic analysis
Chloroplast genomes were analyzed at the order and species level. We collected 45 orders, and the phylogenetic relationship of these orders was an integration of previously published phylogenies established by Jansen et al.72, Moore et al.73 and APG III74. For the species tree, maximum likelihood (ML) analyses were performed on datasets of 40 genes to ensure sufficient information for the calculation of branch length75, 76. An individual gene matrix was aligned using T-Coffee77 and then manually adjusted. We used group-to-group profile alignments78, 79 by taking advantage of previously recognized phylogenetic relationships72,73,74, which yielded data matrices with fewer missing data compared to other methods79. We then identified and concatenated alignment clusters of homologous gene regions. ML analysis was conducted using RAxML version 7.0.480 using the PROTGAMMAJTT substitution model and default settings. Support for each node for ML analysis was tested with 1000 bootstrap replicates. These trees were viewed and edited with the TreeExploter program in MEGA 5.081.
Statistical tests based on cross-species and phylogenetic signal analysis
To identify the relationship between chloroplast genome size and all the other characteristics of chloroplast genome sequences shown in Supplementary Table S1, we conducted a conventional analysis of variance (ANOVA) to test the differences between genome size and all sequence characteristics based on cross-species and phylogenetic signal analyses82. In cross-species analysis, the relationship between each pair-wise characteristic and chloroplast genome size was described using their standardized major axes without taking phylogeny into account (SMA; model II regression). We computed the common slope using SMA analyses among species with a likelihood ratio procedure83. The smatr package84 of R85 was used to perform the SMA analyses.
The ANOVAs were carried out using the PDAP package to test whether there was significant cross-species association between sequence characteristics and genome size that could also be a small-probability event based on a random model of Brownian motion evolution86. We first used Pdsimul to generate 1000 Monte Carlo simulated data by taking the tree topology and branch length information into account (see the phylogenetic analysis section)86. The F-statistic of ANOVA of the simulated data was analyzed by pdanova, and the obtained values were compared against the observed F-statistic from the cross-species analysis. If the observed F-statistic was greater than 95% of the simulated data, the relationship between chloroplast genome size and other characteristics was not random and was affected by phylogenetic signals. This analysis was implemented separately for each characteristic. K was the descriptive statistical parameter to describe the degree of the difference between the F-statistic of simulated data and observed F-statistic distributions87. In brief, the K statistic was the ratio of the observed mean square error derived from a phylogenetically corrected mean and the expected mean square error obtained from the analysis by considering tree topology and branch length information based on a Brownian motion evolution model82. K = 1 would denote that the species had a close relationship with the same characteristic values as those obtained from a Brownian motion evolution model, whereas K < 1 would indicate that the relationship of the characteristic values was not affected by phylogenetic signals. Slope estimates and r 2 from SMA analyses86 were obtained from the results of our standardized contrasts utilizing pdtree and the R package smatr85. In addition, we performed the same likelihood ratio procedure as described earlier in this section to test the common slope for within-group SMA analyses84.
Ratio of nonsynonymous to synonymous nucleotide substitutions (Ka/Ks)
The ratio of nonsynonymous to synonymous substitutions (Ka/Ks) of all individual datasets was estimated for each branch of the phylogenetic tree using PAML88, 89. A free-ratio model was implemented in PAML, and an independent Ka/Ks value was assumed separately for each branch. Because independent estimation of the Ka/Ks ratio for each branch of the tree was extremely time-consuming, the phylogenetic tree of angiosperms was divided into six monophyletic sub-trees, while the phylogenetic tree of gymnosperms was divided into three sub-trees, and each of the sub-trees was evaluated independently. A free-ratio model was implemented in PAML, and an independent Ka/Ks value was separately assumed for each branch90, 91. Only the Ka/Ks values between modern species and their most recent reconstructed ancestors were used in subsequent analyses. Thus, we focused only on the rate of accumulation of mutations between homologous gene pairs with the most recent common ancestors.
References
Grime, J. P. & Mowforth, M. A. Variation in genome size-an ecological interpretation. Nature 299, 151–153, doi:10.1038/299151a0 (1982).
Petrov, D. A., Sangster, T. A., Johnston, J. S., Hartl, D. L. & Shaw, K. L. Evidence for DNA loss as a determinant of genome size. Science 287, 1060–1062, doi:10.1126/science.287.5455.1060 (2000).
Feng, P., Zhao, H. B. & Lu, X. Evolution of mitochondrial DNA and its relation to basal metabolic rate. Mitochondrial DNA 26, 566–571, doi:10.3109/19401736.2013.873895 (2015).
Bennett, D. R., Gorzinski, S. J. & LeBeau, J. E. Structure-activity relationships of oral organosiloxanes on the male reproductive system. Toxicology and Applied Pharmacology 21, 55–67, doi:10.1016/0041-008X(72)90027-0 (1972).
Lane, N. & Martin, W. The energetics of genome complexity. Nature 467, 929–934, doi:10.1038/nature09486 (2010).
Jeremy, E. et al. Development and evaluation of a high-throughput, low-cost genotyping platform based on oligonucleotide microarrays in rice. Plant Methods 4, 13, doi:10.1186/1746-4811-4-13 (2008).
Leitch, I. J., Chase, M. W. & Bennett, M. D. Phylogenetic analysis of dna c-values provides evidence for a small ancestral genome size in flowering plants. Annals of botany 82, 85–94, doi:10.1006/anbo.1998.0783 (1998).
Vesely, P., Bures, P., Smarda, P. & Pavlicek, T. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Annual of Botany 109, 65–75, doi:10.1093/aob/mcr267 (2011).
Bennetzen, J. L., Ma, J. X. & Devos, K. M. Mechanisms of recent genome size variation in flowering plants. Annals of Botany 95, 127–132, doi:10.1093/aob/mci008 (2005).
Wu, Z. Q. & Ge, S. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Molecular Phylogenetics and Evolution 62, 573–578, doi:10.1016/j.ympev.2011.10.019 (2012).
Lynch, M. & Marinov, G. K. The bioenergetic costs of a gene. Proc. Natl. Acad. Sci. USA 112, 15690–15695, doi:10.1073/pnas.1514974112 (2015).
Sabeti, P. C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918, doi:10.1038/nature06250 (2007).
Bigham, A. et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genetics 6, e1001116, doi:10.1371/journal.pgen.1001116 (2010).
Schimper, A. F. W. Über die Entwicklung der Chlorophyllkörner und Farbkörper. Botanische Zeitung 41, 105–120, 126–131, 137–160 (1883).
Martin, T., Oswald, O. & Graham, I. A. Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon: nitrogen availability. Plant Physiology 128, 472–481, doi:10.1104/pp.010475 (2002).
Soll, J. & Schleiff, E. Protein import into chloroplasts. Nature Reviews. Molecular Cell Biology 5, 198–208, doi:10.1038/nrm1333 (2004).
Palmer, J. D. Comparative organization of chloroplast genomes. Annual Review of Genetics 19, 325–354, doi:10.1146/annurev.ge.19.120185.001545 (1985).
Alexeyev, M. F., Ledoux, S. P. & Wilson, G. L. Mitochondrial DNA and aging. Clinical Science 107, 355–364, doi:10.1042/CS20040148 (2004).
Greilhuber, J., Doležel, J., Lysák, M. & Bennett, M. D. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Annals of Botany 95, 255–260, doi:10.1093/aob/mci019 (2005).
Ravi, V., Khurana, J. P., Tyagi, A. K. & Khurana, P. An update on chloroplast genomes. Plant Systematics and Evolution 271, 101–122, doi:10.1007/s00606-007-0608-0 (2008).
Fleischmann, A. et al. Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibulariaceae), with a new estimate of the minimum genome size in angiosperms. Annals of Botany 114, 1651–1663, doi:10.1093/aob/mcu189 (2014).
Kugita, M. et al. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: insight into the earlier land plants. Nucleic Acids Research 31, 716–721, doi:10.1093/nar/gkg155 (2003).
Yamane, H., Ikeda, K., Ushijima, K., Sassa, H. & Tao, R. A pollen expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiology 44, 764–769, doi:10.1093/pcp/pcg088 (2003).
Shahid Masood, M. et al. The complete nucleotide sequence of wild rice (Oryza nivara) chloroplast genome: first genome wide comparative sequence analysis of wild and cultivated rice. Gene 340, 133–139, doi:10.1016/j.gene.2004.06.008 (2004).
Tang, J. et al. A comparision of rice chloroplast genomes. Plant Physiology 135, 412–420, doi:10.1104/pp.103.031245 (2004).
Wu, C. S., Wang, Y. N., Liu, S. M. & Chaw, S. M. Comparative chloroplast genomes of Pinaceae: insight into the mechanism of diversified genomic organizations. Genome Biology and Evolution 3, 309–319, doi:10.1093/molbev/msm059 (2011).
Greiner, S. et al. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Research 36, 2366–2378, doi:10.1093/nar/gkn081 (2008).
Wakasugi, T. et al. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 91, 9794–9798, doi:10.1073/pnas.91.21.9794 (1994).
Parks, M., Cronn, R. & Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology 7, 84, doi:10.1186/1741-7007-7-84 (2009).
Palmer, J. D., Nugent, J. M. & Herbon, L. A. Unusual structure of geranium choroplast DNA: A triplesized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc. Natl. Acad. Sci. USA 84, 769–773, doi:10.1073/pnas.84.3.769 (1987).
Chumley, T. W. et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Molecular Biology and Evolution 23, 2175–2190, doi:10.1093/molbev/msl089 (2006).
Lin, T. P., Chuang, W. J., Huang, S. S. & Hwang, S. Y. Evidence for the existence of some dissociation in an otherwise strong linkage disequilibrium between mitochondrial and chloroplastic genomes in Cyclobalanopsis glauca. Molecular Ecology 12, 2661–2668, doi:10.1046/j.1365-294X.2003.01912.x (2003).
Glöckner, G., Rosenthal, A. & Valentin, K. The structure and gene repertoire of an ancient red algal plastid genome. Journal of Molecular Evolution 51, 382–390, doi:10.1007/s002390010101 (2000).
Gockel, G. & Hachte, L. W. Complete gene map of the plastid genome of the nonphotosynthetic euglenoid flagellate Astasia longa. Protist 151, 347–351, doi:10.1078/S1434-4610(04)70033-4 (2000).
Wolfe, K. H., Morden, C. W. & Palmer, J. D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 89, 10648–10652, doi:10.1073/pnas.89.22.10648 (1992).
Lanfear, R., Kokko, H. & Eyre-Walker, A. Population size and the rate of evolution. Trends in Ecology and Evolution 29, 33–41, doi:10.1016/j.tree.2013.09.009 (2014).
Drescher, A., Ruf, S., Calsa, T. Jr., Carrer, H. & Bock, R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. The Plant Journal 22, 97–104, doi:10.1046/j.1365-313x.2000.00722.x (2000).
Selossea, M., Albertb, B. & Godellec, B. Reducing the genome size of organelles favours gene transfer to the nucleus. Trends in Ecology & Evolution 16, 135–141 (2001).
Bungard, R. A. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays 26, 235–247, doi:10.1002/bies.10405 (2004).
Delannoy, E., Fujii, S., Francs-Small, C. C., Brundrett, M. & Small, I. Rampant gene loss in the underground Orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol 28, 2077–2086, doi:10.1093/molbev/msr028 (2011).
Sanderson, M. J. et al. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. American Journal of Botany 102, 1115–1127, doi:10.3732/ajb.1500184 (2015).
Lam, V. K., Gomez, M. S. & Graham, S. W. The highly reduced plastome of Mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biol Evol 7, 2220–2236, doi:10.1093/gbe/evv134 (2015).
Jansen, R. K., Wojciechowski, M. F., Sanniyasi, E., Lee, S. B. & Daniell, H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Molecular Phylogenetics and Evolution 48, 1204–1217, doi:10.1016/j.ympev.2008.06.013 (2008).
Magee, A. M. et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Research 20, 1700–1710, doi:10.1101/gr.111955.110 (2010).
Lin, C. P., Huang, J. P., Wu, C. S., Hsu, C. Y. & Chaw, S. M. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biology and Evolution 2, 504–517, doi:10.1093/gbe/evq036 (2010).
Chris Blazier, J., Guisinger, M. M. & Jansen, R. K. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Molecular Biology 76, 263–272, doi:10.1007/s11103-011-9753-5 (2011).
Pan, I. C. et al. Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding. PLoS One 7, e34738, doi:10.1371/journal.pone.0034738 (2012).
Yang, J. B., Tang, M., Li, H. T., Zhang, Z. R. & Li, D. Z. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evolutionary Biology 13, 84, doi:10.1186/1471-2148-13-84 (2013).
Mirsky, A. E. & Ris, H. The desoxyribonucleic acid content of animal cells and its evolutionary significance. The Journal of General Physiology 34, 451–462, doi:10.1085/jgp.34.4.451 (1951).
Piegu, B. et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Research 16, 1262–1269, doi:10.1101/gr.5290206 (2006).
Hawkins, R. et al. Accounting for English article interpretation by L2 speakers. EUROSLA Yearbook 6, 7–25, doi:10.1075/eurosla.6 (2006).
Grover, C. E., Yu, Y., Wing, R. A., Paterson, A. H. & Wendel, J. F. A phylogenetic analysis of indel dynamics in the cotton genus. Molecular Biology and Evolution 25, 1415–1428, doi:10.1093/molbev/msn085 (2008).
Hawkins, J. D., Kosterman, R., Catalano, R. F., Hill, K. G. & Abbott, R. D. Effects of social development intervention in childhood 15 years later. Archives of Pediatrics and Adolescent Medicine 162, 1133–1141, doi:10.1001/archpedi.162.12.1133 (2008).
Neiman, M. & Taylor, D. R. The causes of mutation accumulation in mitochondrial genomes. Proceeding Biological Sciences 276, 1201–1209, doi:10.1098/rspb.2008.1758 (2009).
Smith, D. R. Mutation rates in plastid genomes: They are lower than you might think. Genome Biol Evol 7, 1227–1234, doi:10.1093/gbe/evv069 (2015).
Stiller, J. W. Plastid endosymbiosis, genome evolution and the origin of green plants. Trends in plant science 12, 391–396, doi:10.1016/j.tplants.2007.08.002 (2007).
Maier, U. et al. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol Evol 5, 2318–2329, doi:10.1093/gbe/evt181 (2013).
McFadden, G. I. Origin and evolution of plastids and photosynthesis in Eukaryotes. Cold Spring Harb Perspect Biol 6, a016105–a016105, doi:10.1101/cshperspect.a016105 (2014).
Lynch, M. Streamlining and simplification of microbial genome architecture. Annual Review of Microbiology 60, 327–349, doi:10.1146/annurev.micro.60.080805.142300 (2006).
Lynch, M. The Origins of Genome Architecture. Sinauer Associates Sunderland, MA (2007).
Gregory, T. R. & DeSalle, R. Comparative Genomics in Prokaryotes. The Evolution of the Genome (pp. 585–675. Elsevier, San Diego: CA, 2005).
Leu, S., Schlesinger, J., Michaels, A. & Shavit, N. Complete DNA sequence of the Chlamydomonas reinhardtii chloroplast atpA gene. Plant molecular biology 18, 613–616, doi:10.1007/BF00040681 (1992).
Drapier, D. et al. The chloroplast atpA gene cluster in Chlamydomonas reinhardtii. Plant Physiology 117, 629–641 (1998).
Kode, V., Mudd, E. A., Iamtham, S. & Day, A. The tobacco plastid accD gene is essential and is required for leaf development. The plant journal 44, 237–244, doi:10.1111/j.1365-313X.2005.02533.x (2005).
Wicke, S., Schneeweiss, G. M., Pamphilis, C. W., Müller, K. F. & Quandt, D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Molecular Biology 76, 273–297, doi:10.1105/tpc.113.113373 (2011).
Robbins, J. C., Heller, W. P. & Hanson, M. R. A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15, 1142–1153, doi:10.1261/rna.1533909 (2009).
Sabeti, P. C. Positive natural selection in the human lineage. Science 312, 1614–1620, doi:10.1126/science.1124309 (2006).
Shinozaki, K. et al. The complete nucleotide sequence of the tobacoo chloroplast genome: its gene organization and expression. The EMBO Journal 5, 2043–2049 (1986).
Ohyama, K. et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322, 572–574, doi:10.1038/322572a0 (1986).
Wyman, S. K., Jansen, R. K. & Boore, J. L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255, doi:10.1093/bioinformatics/bth352 (2004).
Cosner, M. E., Jansen, R. K., Palmer, J. D. & Downie, S. R. The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Current Genetics 31, 419–429, doi:10.1007/s002940050225 (1997).
Jansen, R. K. et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 104, 19369–19374, doi:10.1073/pnas.0709121104 (2007).
Moore, M. J., Soltis, P. S., Bell, C. D., Burleigh, J. G. & Soltis, D. E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 107, 4623–4628, doi:10.1073/pnas.0907801107 (2010).
The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society 161, 105–121, doi:10.1111/(ISSN)1095-8339 (2009).
Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. Journal of Molecular Evolution 39, 306–314, doi:10.1007/BF00160154 (1994).
Parks, S. L. & Goldman, N. Maximum likelihood inference of small trees in the presence of long branches. Systematic Biology 63, 798–811, doi:10.1093/sysbio/syu044 (2014).
Notredame, C., Higgins, D. G. & Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology 302, 205–217, doi:10.1006/jmbi.2000.4042 (2000).
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 19, 113, doi:10.1186/1471-2105-5-113 (2004).
Smith, S. A. & Donoghue, M. J. Rates of molecular evolution are linked to life history in flowering plants. Science 322, 86–89, doi:10.1126/science.1163197 (2008).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690, doi:10.1093/bioinformatics/btl446 (2006).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739, doi:10.1093/molbev/msr121 (2011).
Beaulieu, J. M. et al. A β-arrestin 2 signaling complex mediates lithium action on behavior. Cell 132, 125–136, doi:10.1016/j.cell.2007.11.041 (2008).
Warton, D. I. & Weber, N. C. Common slope tests for Bivariate Errors-in-Variables models. Biometrical Journal 44, 161–174, doi:10.1002/(ISSN)1521-4036 (2002).
Warton, D. I. & Ormerod, J. smatr: (Standardised) major axis estimation and testing routines. R package version 2.1. http://web.maths.unsw-edu.au/dwarton (2007).
R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna (2007).
Garland, N. L., Medhurst, L. J. & Nelson, H. H. Potential chlorofluorocarbon replacements: OH reaction rate constants between 250 and 315 K and infrared absorption spectra. Journal of Geophysical Research 98, 0148–0227, doi:10.1029/93JD02550 (1993).
Blomberg, S. P., Garland, T. J. & Ives, A. R. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745, doi:10.1111/evo.2003.57.issue-4 (2003).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in the Biosciences 13, 555–556 (1997).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24, 1586–1591, doi:10.1093/molbev/mst179 (2007).
Yang, Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15, 568–573, doi:10.1093/oxfordjournals.molbev.a025957 (1998).
Pond, S. L. & Frost, S. D. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol Biol Evol 22, 478–485, doi:10.1093/molbev/msi031 (2005).
Acknowledgements
We thank Zhang Qiang and Wang Wei for their helpful suggestions regarding plant phylogeny. This research was supported by the National Key Research and Development Program of China (2016YD0100301) and the National Natural Science Foundation of China (31670211, 31100176).
Author information
Authors and Affiliations
Contributions
Z.X.M. and Y.Q. designed the study, W.J. analyzed the data, and Z.X.M. interpreted the results and wrote the paper. F.L. and L.S. drew and modified the figures. P.H. edited the paper. Q.L., L.J., S.Y., Q.W., Z.L. and C.Y. helped collect the data and edited the paper. All authors are aware of the content of this paper and have read and edited the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xiao-Ming, Z., Junrui, W., Li, F. et al. Inferring the evolutionary mechanism of the chloroplast genome size by comparing whole-chloroplast genome sequences in seed plants. Sci Rep 7, 1555 (2017). https://doi.org/10.1038/s41598-017-01518-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-01518-5
This article is cited by
-
Complete chloroplast genomes of 13 species of sect. Tuberculata Chang (Camellia L.): genomic features, comparative analysis, and phylogenetic relationships
BMC Genomics (2024)
-
Global exact optimisations for chloroplast structural haplotype scaffolding
Algorithms for Molecular Biology (2024)
-
Regardless of having identical photosynthetic pathways, chloroplast genomes vary depending on whether the host plant is monocotyledonous or dicotyledonous
Genetic Resources and Crop Evolution (2024)
-
Complete chloroplast genome structural characterization of two Phalaenopsis (Orchidaceae) species and comparative analysis with their alliance
BMC Genomics (2023)
-
Chloroplast genome characterization of Uncaria guianensis and Uncaria tomentosa and evolutive dynamics of the Cinchonoideae subfamily
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.