Abstract
A persistent goal of vaccine development is the enhancement of the immunogenicity of antigens while maintaining safety. One strategy involves alteration of the presentation of the antigen by combining antigens with a multimeric scaffold. Multi-antigen vaccines are under development, and there are presently far more candidate antigens than antigen scaffolding strategies. This is potentially problematic, since prior immunity to a scaffold may inhibit immune responses to the antigen-scaffold combination. In this study, a series of domains from S. aureus which have been shown to crystallise into multimeric structures have been examined for their scaffolding potential. Of these domains, SAR1376, a 62 amino acid member of the 4-oxalocrotonate tautomerase (4-OT) family, was pro-immunogenic in mice when fused to a range of pathogen antigens from both S. aureus and P. falciparum, and delivered by either DNA vaccination, viral vector vaccines or as protein-in-adjuvant formulations. The adjuvant effect did not depend on enzymatic activity, but was abrogated by mutations disrupting the hexameric structure of the protein. We therefore propose that SAR1376, and perhaps other members of the 4-OT protein family, represent very small domains which can be fused to a wide range of antigens, enhancing immune responses against them.
Similar content being viewed by others
Introduction
Modern vaccinology approaches use highly purified protein antigens which often have limited innate stimulatory activity and so may be poorly immunogenic. To address this, multiple strategies have been developed, including incorporating protein antigens into oil emulsions, formulating with aluminium salts, and addition of, or fusion to, Toll-like receptor agonists1. Alternatively, by using DNA or recombinant viral vectors, innate immune stimulation can be achieved by the vector while exploiting the host’s translational machinery to provide in vivo expression of the antigen2.
A separate strategy, which has a good safety profile, involves alteration of the size and multimerisation of the antigen either by attachment of the protein to a self-assembling bacteriophage or by fusion to part of a viral capsid3,4,5 or other similar protein6. Such virus-like particles (VLPs), which are typically 20–200 nm in diameter, include commercially deployed human papilloma virus and hepatitis B vaccines4 as well as numerous products at earlier stages in development5, 7. The varied immunological mechanism(s) behind VLP induced enhanced immunogenicity include ready access to lymphatics, rapid dendritic cell uptake and activation and arrayed-antigen mediated B-cell receptor cross-linking4. In some cases, particle entry into cells mediated by specific host receptors has been demonstrated8, 9. VLP immunogenicity is likely a result of a combination of these mechanisms.
A limited number of non-viral antigen multimerisation domains have been described. One such domain, IMX313, is derived from the multimerisation domain of a vertebrate complement C4 binding protein (C4bp)2, and extensively re-engineered to minimise cross-reactivity with human C4bp. Marked improvements in immunogenicity to some antigens have been observed with this strategy10, 11. Other related technologies include fusion to ferritin or encapsulin molecules12, or fusion with the highly multimerising protein lumazine synthetase13.
Vaccine development is continuing for a range of bacterial pathogens, including S. aureus, pathogenic Neisseria species14, M. tuberculosis 15, E. coli 14, and against Apicomplexa (e.g. P. falciparum 10). Multi-antigen vaccines are under development, and there are presently far more candidate antigens than antigen scaffolding strategies. This is potentially problematic, since prior immunity to a scaffold may inhibit immune responses to the antigen-scaffold combination, as was observed with circumsporozoite protein-hepatitis B surface antigen fusions in human adults16. It is at present unclear how many molecules exist biologically which are capable of enhancing immunogenicity when fused to other antigens, what the required biophysical properties are, and whether multimerisation is necessary for the adjuvanting effect. Nevertheless, if such proteins exist, pro-immunogenic domains unrelated both to mammalian proteins and to existing viral-like particle components, including Hepatitis B surface antigen, might have utility in a range of vaccines which are currently being developed.
In this study, we have examined a series of domains from S. aureus proteins which have been shown to crystallise into multimeric structures. We show that SAR1376, a member of the 4-oxalocrotonate tautomerase (4-OT) family, is pro-immunogenic in mice when fused to a range of pathogen antigens from S. aureus and from P. falciparum, whether delivered by DNA vaccination, viral vectored vaccines or as protein-in-adjuvant formulations. We demonstrate by mutagenesis that the adjuvant effect does not depend on enzymatic activity, but is abrogated by mutations unfolding the hexameric structure of the protein. We therefore propose that 4-OT proteins represent a very small pro-immunogenic domain which can be fused to a range of antigens, enhancing immune responses against them.
Results
Selection of scaffolding tags
Study of crystal structures within the Protein Data Bank revealed a number of bacterial proteins which form self-multimers of various orders. A subset was selected based on absence of intra- or inter-chain disulphide bonding, absence of transmembrane regions and absence of toxicity and oncogenic activity, with a view to increasing the probability of efficient expression. In the first instance we chose to proceed with self-multimerising proteins from S. aureus, a pathogenic microbe causing disease that is controlled by both T cell and antibody mediated mechanisms17. Figure 1 shows the structures of the four protein chosen: Dps-like Peroxide Resistance Protein Dpr, with a structure similar to that of ferritin12, QacR, a multidrug binding transcriptional repressor, SA138818, a protein of unknown function annotated as a homologue of E. coli ygbL aldolase class 2-like gene, and SAR1376, a 4-Oxalocrotonate Tautomerase (4-OT). The size of the crystallising unit varied from more than 20 nm (for QacR) to less than 5 nm (for SAR1376) (Fig. 1).
Fusion of SAR1376 to S. aureus antigens enhances their immunogenicity in mice
We hypothesised that these four bacterial molecules might have pro-immunogenic activity similar to other multimerising scaffold proteins, and so might enhance immune responses to antigens fused to them2, 12, 13. Expression vectors producing fusions of the four proteins (Table 1) with a series of S. aureus antigens were constructed. The expression cassette was composed of a human tissue plasminogen activator leader sequence, the antigen of interest, an epitope tag (V5) used to monitor protein expression, and the scaffolding domain (Table 1) via a GSG linker (Fig. 2A).
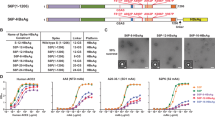
Design and immunogenicity of DNA vectors expressing the S. aureus BitC antigen fused to the novel multimerising scaffolds. (A) Design of pMono2 DNA vaccination vectors constructs. CMV: CMV IE94 promoter, TPA: human tissue plasminogen activator signal sequence, antigen: antigen being tested, V5: epitope tag and linker sequence, scaffold: multimerising domain, BGH pA: Bovine growth hormone polyadenylation sequence. (B) BitC-specific IgG response after two immunisations of groups of 4 or 5 BALB/c mice with 50 µg BitC-scaffold DNA two weeks apart, measured by LIPS assay at 3 weeks post boost. Each symbol represents a single mouse. Dotted line: threshold for background response, *p < 0.05.
We studied scaffold fusions to two S. aureus antigens: BitC19, a cell surface lipoprotein (accession NP_370379) which we have previously investigated as a vaccine candidate (unpublished data, patent 14/433565), and the extracellular domain of the Clumping factor B precursor20 (ClfB, accession YP_001333563). As comparators, constructs expressing antigen without scaffold were made (Fig. 2A). Additionally, in some experiments, the multimerising tag IMX3132 fused to the antigen C-terminus was used as a positive control.
Groups of BALB/c mice were immunised intramuscularly with a mammalian expression DNA vector expressing BitC fused to one of the four test scaffolding domains. A priming and boosting immunisation was administered, separated by two weeks. Measurement of the antibody response against BitC three weeks post-boost indicated that fusion of SAR1376 might increase immunogenicity of BitC relative to the no-scaffold comparator (p = 0.16), while the mice vaccinated with the Dps scaffold responded poorly against BitC (Fig. 2B). In this experiment, the fold change in antibody concentrations (if any) was small.
We further tested the SAR1376 and QacR scaffolds with both ClfB S. aureus antigen and with BitC, in the same 2-week prime-boost vaccination regimen. Humoral and cellular (IFN-γ ELISpot) responses were measured following the second immunisation. Comparator constructs containing the QacR domain were included in order to assess the specificity of the observed response. The immunogenicity results supported the previous experiment, in which a small increase was observed in antibody responses BitC when BitC was linked to SAR1376 but not QacR (Fig. 3A; p = 0.06 for BitC-SAR1376 when pooling data from experiments shown in Figs 2 and 3). A significantly higher antibody response was observed to ClfB was observed using ClfB-SAR1376 compared to ClfB without a scaffold (p = 0.002, Fig. 3C). This experiment allowed comparison of the fold change elicited in antibodies against two antigens, BitC (where significant changes were not observed) and ClfB, where an estimated 10 fold change (2.2 log10 units vs. 3.2 log10 units, p = 0.002) was observed. The SAR1376 scaffold did not enhance IFN-γ producing T-cell numbers with either of these two antigens (Fig. 3B and D), although the IMX313 tag, used here as a positive control, did increase the T cell responses to BitC (Fig. 3B, p = 0.01).
SAR1376 scaffold enhances immune responses to BitC and ClfB in BALB/c mice. Immunogenicity was assessed following prime-boost vaccination with DNA constructs encoding different scaffolds (as indicated on the X axes) fused to the antigen C-terminus; antigen specific IgG responses against BitC (panel A) and ClfB (panel C) measured by LIPS assay, and number of IFN-γ producing T-cells induced by BitC (panel B) and ClfB (panel D) at week 3 post boost. Each symbol represents a single mouse. Dotted line: threshold for background response. **p < 0.01, ***p < 0.001.
The 4-OT family
4-OT-like enzymes are common in bacteria21. Using a protein-based search strategy, 2780 discrete family members (modal length of 63 amino acids) were found across Eubacteria, with examples in Archaea also noted (see Supplementary Data S1). One example was chosen randomly from the 342 different genera identified (Supplementary Data S1). Extensive diversity is observed within the protein family, with only 20% identity in primary protein sequences between diverse members of the family (Fig. 4A). Examination of eleven bacterial crystal structures, however, show a very similar crystal structures in family members despite huge evolutionary distance (Fig. 4B, and also Supplementary Movie 2). Conserved motifs, including a highly conserved initial proline, do exist within the primary sequences from genera known to be pathogenic to man (Fig. 4C, see also Supplementary Data S1).
4-OT family members. (A) Family tree, 2780 discrete family members (modal length of 63 amino acids) were found across Bacteria. (B) Despite the extensive primary amino acid sequence diversity observed, crystal structures are very similar. (C) Cobalt alignment of 4-OT like sequences from selected genera which include human pathogens.
The crystal structure of SAR1376 reveals a hexamer forming an approximately spherical structure of about 5 nm diameter. It further suggests that the amino terminus of a short linker attached to the N-terminus of SAR1376 is surface accessible. Three such amino termini are present on each side of the sphere (Supplementary Movie S1). This suggests a model in which fusion of antigens to the N-terminus of SAR1376 generates a small sphere with six antigens displayed outwards (Fig. 1, Supplementary Movie S1).
Inactivated SAR1376 enzyme retains adjuvant activity
Since the mechanism of 4-OT catalysis has been heavily investigated, we mutated two critical residues involved in the active site, Proline-1 (P1) and arginine-35 (R35), a site corresponding to R39 in other crystallised family members21. P1A mutations disrupt enzymatic activity, but leave the protein structure intact, whereas R35A or Q mutations disrupt catalysis and impair protein multimerisation21. The immunogenicity of ClfB fused to these variants was compared (Fig. 5). The SAR1376 mutant P1A enhanced both the antibody and T-cell responses to ClfB significantly compared to no scaffold (p = 0.021) (Fig. 5A and B). By contrast, the immune response to ClfB with the scaffold containing the R35A mutation was not significantly different from ClfB without a scaffold (Fig. 5A and B). The effect of SAR1376 mutant P1A on the ClfB antibody response was similar in both BALB/c (Fig. 5A) and outbred CD1 mice (Fig. 5C). Enhancement on IFN-γ producing T-cell numbers by P1A was observed in one mouse strain (Fig. 5B and D). Taken together, this suggests that protein multimerisation contributes to the enhanced immunogenicity, but catalytic activity does not.
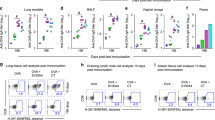
SAR1376 mutant P1A scaffold enhances immune responses to ClfB in BALB/c and CD1 mice. Mice were vaccinated twice with DNA vectors expressing the antigens shown. Antigen specific antibody responses against ClfB in BALB/c (panel A) and CD1 (panel C) mice as measured by LIPS assay, and number of IFN-γ producing T-cells induced by ClfB in BALB/c (panel B) and CD1 (panel D) mice after vaccination with mutant scaffolds fused to the c-terminus of the antigen. Sera were collected 3 weeks post second injection. Each symbol represents a single mouse. Dotted line: threshold for background response. *p < 0.05; **p < 0.01.
SAR1376 and the P1A variant are immunogenic
We next investigated the capacity of SAR1376 fusion proteins to raise a humoral response against SAR1376 itself. Antibody responses against SAR1376 fused to either BitC (Fig. 6A) or ClfB (Fig. 6B) in immunised BALB/c mice were not significantly raised, as compared to the no scaffold groups. By contrast, immunization with ClfB fusion constructs containing the SAR1376 mutant P1A scaffold raised a significant antibody response to SAR1376 in both BALB/c (Fig. 6B) and CD1 outbred (Fig. 6C) mice whereas the R35A mutation did not (Fig. 6B). In both strains, serological responses against SAR1376 in the absence of SAR1376 immunisation were indistinguishable from the assay background (Fig. 6), suggesting that SAR1376 homologues, if present in colonising murine microbiota, do not generate detectable cross reactivity with SAR1376.
SAR1376 specific antibodies measured after vaccination with DNA vectors encoding the ClfB antigen with our without scaffold fused to the C-terminus of the antigen. (A) Antibody response against SAR1376 in BALB/c mice immunised with BitC-SAR1376 fusion protein. (B) Anti-SAR1376 antibody levels after vaccination of BALB/c mice with ClfB fused to SAR1376, SAR1376-P1A or SAR1376-R35A. (C) Anti-SAR1376 antibodies in CD1 mice immunised with ClfB-SAR1376 mutant P1A scaffold, as measured by LIPS assay. Each symbol represents a single mouse. Dotted line: three s.d. above assay background. ***p < 0.001; ****p < 0.0001.
Viral Vectored vaccines expressing SAR1376-P1A fused to truncated Hla increase immunogenicity
We investigated whether the pro-immunogenic effect of SAR1376 fusion was restricted to DNA vaccination. Because the effect of SAR1376 fusion appeared most marked on antibody induction, we elected to study the S. aureus alpha toxin (AT), a haemolytic multimeric β-pore forming toxin which is a critical virulence factor in S. aureus 22 and is encoded by the Hla gene. AT can be neutralised by antibody22. We designed a truncated form of AT, designated tHla75, comprising amino acids 1–75, the portion of the molecule reported to contain the receptor ADAM10 (A disintegrin and metalloproteinase 10) binding domain23, 24. Recombinant adenoviral (AdH5) and MVA vectors expressing SAR1376 fused to tHla75 were constructed. BALB/c mice were vaccinated with AdH5-tHla75 followed eight weeks later by MVA-tHla, a prime-boost regime known to be highly immunogenic2. Analysis of the immune response against alpha toxin showed that SAR1376 fusion did not increase the immunogenicity of adenovirally expressed proteins (Fig. 7A), but two weeks post MVA boost a significantly enhanced antibody response was observed in the tHla75-P1A group relative to the tHla75-no scaffold group (Fig. 7A). However, the tHla75 construct did not induce substantial functional (neutralising) antibody titres (Fig. 7C) in either configuration. As expected, antibodies against SAR3176 were raised and boosted in tHla75-P1A vaccinated group animals (Fig. 7B).
Impact of fusion of SAR1376 mutant P1A to truncated Hla on immunogenicity. (A) Anti-AT antibody levels. (B) Anti-SAR1376 antibody levels after vaccination of BALB/c mice with tHla75-no scaffold, tHla75-P1A, or empty vectors (control) as assessed respectively by ELISA and LIPS assay. Sera were collected pre boost (day 56 post Adenovirus prime) and post MVA boost (day 70 post prime). Dotted line: threshold for background response. (C) AT neutralizing activity of the antibodies was assessed post boost by Toxin Neutralisation Assay. Assay minimum and maximum levels were determined with mAb 8B7 and recombinant AT as described in Methods. **p < 0.01.
In summary, fusion of S. aureus antigens to SAR1376-P1A significantly increased antibody responses to the antigens, when expressed from DNA vaccine vectors and viral vectors. The enhancement was observed in two different mouse strains, and was abrogated by a mutation known to disrupt multimerisation of SAR1376. Baseline immune responses against SAR1376 were not detected in the two mouse strains, but SAR1376-P1A variant is itself immunogenic.
Fusion of SAR1376-P1A to Pfs25 recombinant protein improves immunogenicity in BALB/c mice
We investigated whether the pro-immunogenic effect of SAR1376 fusion extended to recombinant protein antigens by studying the effect SAR1376 fusion on immunogenicity of a P. falciparum protein, Pfs25. Pfs25 is a candidate antigen for a transmission blocking vaccine and antibodies against Pfs25 have been shown in several studies to interfere with sexual reproduction of the parasite in the mosquito vector25. Recently, we have reported that heptamerisation of Pfs25 by fusion to IMX313 increased its immunogenicity significantly in pre-clinical studies10.
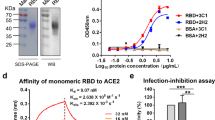
Recombinant monomeric Pfs25 and Pfs25-SAR1376-P1A proteins were produced in Pichia pastoris as secreted proteins and purified using a 6-Histidine tag (Fig. 8A and B). Mice were immunised twice with either Pfs25-SAR1376-P1A or monomeric Pfs25 protein-in-Alhydrogel formulations at 2-week intervals.
Fusion of SAR1376-P1A to Pfs25 improves functional activity. (A) Plasmid construct for expression of pfs25-SAR1376 P1A in Pichia pastoris. (B) SDS page and Western Blot of SAR1376 (anti-His mAb), and Pfs25-SAR1376-P1A (anti Pfs25 mAb). (C) Antibody response after vaccination of BALB/c mice with monomeric Pfs25 or Pfs25 fused to SAR1376-P1A. Sera were collected at day 14, 28 and day 61 post prime and IgG responses measured using a standardised ELISA. Each solid symbol represents a single mouse. (D) Pooled-purified IgG mixed with P. falciparum gametocytes was fed to Anopheles stephensi mosquitoes through a membrane. Midguts were dissected 9 days post-feed and the number of oocysts per gut were counted. Each symbol represents the count from one mosquito. AU = antibody units. *p < 0.05; ***p < 0.001; ****p < 0.0001.
Little response was seen in mice following vaccination with Pfs25. This is not unexpected as we have previously observed that monomeric protein is poorly immunogenic using Alhydrogel adjuvant10. The antibody levels at all time points were significantly higher in the group that received Pfs25-SAR1376-P1A than the mice receiving monomeric Pfs25, demonstrating that fusion of Pfs25 to SAR1376-P1A significantly improved the immune response (Fig. 8C). The pooled and purified IgG from the Pfs25-SAR1376-P1A immunised mice (day 28) were able to completely block oocyst development in the mosquito midgut in the ex vivo Standard Membrane Feeding Assay (SMFA), demonstrating that antibodies produced by immunisation with this platform were functional (Fig. 8D).
Discussion
In this study we show that fusion of the 4-OT enzyme family member SAR1376 to different antigens enhances antibody responses against the fused antigen. The effect was demonstrated using three vaccine delivery platforms: DNA, viral vector delivery systems and recombinant proteins, and several antigens: three from S. aureus and one from Plasmodium falciparum. For some proteins, the fold change in antibody levels observed was modest (such as the lipoprotein BitC, where less than 2-fold change was observed), but for others (such as Pfs25, and ClfB) more substantial enhancements were observed. In the case of P. falciparum Pfs25, we could demonstrate that this increase was associated with a functional antibody activity.
4-Oxalocrotonate Tautomerase (4-OTs), of which S. aureus enzyme SAR1376 is an example, are typically 60–80 amino acids in length, placing them among the smallest enzymes known. They have an unusual mechanism of action, involving the proline at residue 1 (after the initiator methionine) and are involved in the catalytic breakdown of polycyclic compounds into tri-carboxylic acid (Krebs’) cycle precursors in a variety of bacteria21. A range of other enzymatic activities have been described in proteins with 4-OT-like structures21, 26, but all depend on the initial proline. By contrast, the enhancement of immunogenicity observed here does not depend on the initial proline, and is in fact enhanced following P1A mutation, suggesting that enzymatic activity is not required for the adjuvant effect.
The ability of 4-OT proteins to spontaneously multimerise into hexamers was exploited to enhance the immune response to several different antigens. This ability is not compromised by the ligation of the antigen of interest via a short linker to the N-terminus of the enzyme, as judged by crystal structures in which the N-terminus of the linker remains surface-exposed in the resulting hexamer (Movie S1). The results of our mutagenesis experiments support the idea that multimerisation of SAR1376 is required for enhanced immunogenicity. The mechanism by which this multimerisation increases immunogenicity was not investigated but is most likely due to the particulate nature of the resulting protein and/or pattern recognition of the repetitive antigen arrangement on the hexamer surface, as described with other multimerising ligands4. It is possible that such multimerisation has a larger impact on the immunogenicity (such as Pfs25) than on others (such as BitC), perhaps because immune detection of the native proteins varies, with the impact of additional multimerisation more evident in poorly immunogenic proteins.
A range of other S. aureus proteins reported to multimerise were also tested for pro-immunogenic activity (Dps, QacR, SA1388) using the DNA vaccination system which detected the activity of SAR1376. None displayed a similar effect. This is surprising given the structural similarities between Dps and ferritin, a self-multimerising molecule which is successful at increasing immunogenicity to some antigens when fused to their C-terminus12. It is possible that of the proteins examined, only SAR1376 adopts a pro-immunogenic, likely multimeric, structure in vivo when produced by the expression systems we studied.
Of note, 4-OTs are widespread in pathogenic bacteria27. There are thousands of sequences known from multiple bacterial species, both Gram positive and Gram negative. Given their conserved structure, inactivating the enzymatic activity while retaining ability to multimerise and fusion of diverse bacterial 4-OT proteins may be a general strategy for enhancing antibody responses to vaccine antigens. In the development of novel vaccines the approach combining potent viral vectors with a new generation of short, multimerising approach could represent a promising alternative to virus like particles, which have recently become an attractive vaccination platform4, as it combines the inherent immunogenicity and safety of viral vectored vaccines28 with stable spherical structures surface-displaying the antigen.
Methods
Scaffolds used and their production
Chosen scaffold sequences where human codon optimised and DNA synthetized (GeneArt, Life Technologies Ltd) (Table 1). Scaffolds were fused to the C-terminus of the S. aureus antigens in a mammalian expression vector (pMono2) using a restriction enzyme based strategy. Antigens fused were S. aureus BitC, a S. aureus cell surface lipoprotein (accession NP_370379) which we have previously investigated as a vaccine candidate (unpublished data, patent 14/433565), the extracellular domain of the S. aureus Clumping factor B precursor20 (ClfB, accession YP_001333563), S. aureus α–hemolysin (accession YP_111574996, amino acids 1–75, designed tHla75 here), and P. falciparum protein Pfs25 (accession AAN35500)10. Sequences for all these were synthesised by Life Technologies Ltd.
S. aureus tHla75 was ligated into the pMono2 vector from which the construct was subcloned into shuttle vectors and transfected into replication-deficient adenovirus human serotype 5 (AdHu5) and Modified Vaccinia Ankara (MVA) as described elsewhere29, 30.
The gene coding for Plasmodium falciparum transmission-blocking antigen, Pfs25 with a 6-his tag (His6-Pfs25) was fused to the 5′ upstream of the gene coding for SAR1376-P1A and ligated into the pPinkα-HC plasmid (Fig. 8A) which puts protein expression under methanol inducible control and allows secretion of the expression product from Pichia pastoris via the α-mating factor secretion signal. Electrocompetent P. pastoris were transformed with the expression plasmid. Colonies were screened for optimal expression and the highest expressing clone selected for scale up. A one litre shake flask culture of the selected clone was grown under inducing conditions and the supernatant harvested. Supernatant containing the secreted expression product was harvested by centrifugation and the product purified by nickel-chelate affinity chromatography.
Vaccination experiments
All mouse procedures were conducted in accordance to the Animal (Scientific Procedures) Act 1986 (Project licence 30/2825) and were approved by the University of Oxford Animal Care and Ethical Review Committee. Six to eight week old female BALB/c or CD1 mice from Harlan Laboratories UK were used.
In DNA vaccination experiments, groups of 4-12 mice (BALB/c or CD1) were immunised intramuscularly with 50 μg vector DNA in 50 μl PBS (25 µl/hind leg). Immunisation was repeated 2 weeks later. On day 35, blood samples were taken from all animals under terminal anaesthesia (heart bleeds) for immune assays (IFN-gamma secreting T-cell numbers (ELISpot), and antibody levels by Luciferase ImmunoPrecipitation System (LIPS) assay)31, 32.
For viral vector immunization, groups of 6 mice were immunised intramuscularly with 109 i.u. AdHu5 in 25 μl PBS followed at least 8 weeks later by 107 pfu MVA as prime-boost sequence, a regime we refer to as AM7. Venous blood samples were taken from the tail vein of all animals pre boost and 2 weeks post boost.
For immunization with recombinant proteins, groups of 6 BALB/c mice were immunised intramuscularly with 50 µl aliquots (25 µl/hind leg) of protein-in-Alhydrogel formulations, containing 2.5 µg of either Pfs25-P1A or monomeric Pfs25 twice at 2 week intervals. Blood was collected from the tail vein on day 14 (2 weeks post prime) and day 28 (2 weeks post boost).
Assessment of immune responses against BitC and ClfB
A Luciferase ImmunoPrecipitation System (LIPS) assay was used to detect specific serum anti-S. aureus BitC and ClfB antibodies as described32. Briefly, recombinant BitC and ClfB fusion protein with Renilla luciferase were produced in 293 cells as described32. Serially diluted sera were incubated with Renilla luciferase-BitC or ClfB fusion proteins. The mix was added to filter plates loaded with A/G beads (Thermo Fisher). After incubation and subsequent washings, chemiluminescence was measured in a Luminometer (ClarioStar, BMG Labtech) after adding substrate (Renilla luciferase assay system, Promega UK Ltd.). Log transformation was applied to luminescence data prior to statistical analysis. Specific luminescence was generated by subtracting the assay background, which was considered to be the luminescence observed in the absence of any sera. The assay limit of detection was considered to be four standard deviations above the specific luminescence in the control groups.
Anti-alpha toxin immune responses
The anti-tHla antibody levels and functional activity (neutralizing activity, NA) of the antibodies in serum were assessed respectively by ELISA and Toxin Neutralisation Assay (TNA) as described by Oscherwitz and Cease33. In brief, the ability of antibody to block recombinant alpha toxin (AT) cytotoxicity in vitro was assessed using the Jurkat T cell line (TIB-152, ATCC, Manassas, VA). Mouse anti-Staphylococcal alpha hemolysin mAb (8B7) (IBT Bioservices # 0210-001) was used as standard positive to obtain minimum and maximum levels for neutralisation of AT (H9395, Sigma Biologicals).
Anti-Pfs25 immune responses
Antibody levels in serum were assessed by standardised anti-Pfs25 ELISA, as described10. A serially diluted standard reference serum with a known antibody titre was used to determine the antibody titre of individual samples. Total IgG was purified from the pooled serum of the mice immunised with Pfs25-SAR1376-P1A and assessed by functional assay by Standard Membrane Feeding Assay (SMFA). This assay involves feeding malaria infected blood mixed with purified IgG to Anopheles stephensi mosquitos through a membrane34. If the IgG has functional activity it will block development of the malaria sexual stage in the mosquito midgut and at 9 days post feed there will be a reduction in the number of oocysts observed in the gut compared to a non-functional IgG control.
Statistical analysis
Data on antibody response and IFNγ-specific spots were statistically analysed for effect of added scaffold by means of an F-test after a log10 transformation and correction for background. Log (number of IFN-γ secreting cells) was used, because of the approximate log-normal distribution of ELISpot counts in the animals (not shown). Specific antibody levels from the LIPS assay were generated by subtracting the assay luminescence background, which was considered to be the luminescence observed in the absence of any sera, from the luminescence observed with serum dilutions added. The assay limit of detection was considered to be four standard deviations above the background. Post-hoc pairwise comparisons were performed using Dunnett’s Multiple Comparison Test. Differences were considered significant when p < 0.05. The statistical packages used were R 2.15 (http://www.cran.org), and GraphPad Prism version 5.04 (GraphPad Software, Inc.).
Bioinformatic identification of 4-OT-like proteins
The NCBI RefSeq database was queried using BLASTp and delta-BLAST14 using default parameters with the S. aureus 4-OT enzyme (YP_040781.1) as a query. Further searches were performed using distant hits and results pooled, and then filtered using custom R scripts to include hits encoding proteins of 55 to 85 amino acids. Manual curation was performed, and the sequence start of predicted proteins was trimmed to begin with MP, as a proline is present in position 2 of all canonical family members21, i.e. any amino acids purported to originate from upstream initiator codons were removed. A single sequence was selected per genus; using genus-specific sequences, an alignment was prepared using the NCBI Cobalt multiple alignment engine35 with default parameters. Additionally, a tree was constructed using PhyML36 using default parameters, and visualised using Archeopterix37 software.
Presentation of crystal structures
Crystal structures of proteins of interest were downloaded from the Protein data bank. A single hexameric structure was isolated from each set of crystal data using Pymol v.1.8.2 for Windows. For comparison of multiple 4-OT crystals, structures were aligned using CEAlign (Pymol) using default parameters. Pymol was also used to render images.
References
De Gregorio, E., Caproni, E. & Ulmer, J. B. Vaccine adjuvants: mode of action. Front Immunol 4, 214, doi:10.3389/fimmu.2013.00214 (2013).
Rollier, C. S., Reyes-Sandoval, A., Cottingham, M. G., Ewer, K. & Hill, A. V. Viral vectors as vaccine platforms: deployment in sight. Curr Opin Immunol 23, 377–382, doi:10.1016/j.coi.2011.03.006 (2011).
Tissot, A. C. et al. Versatile virus-like particle carrier for epitope based vaccines. PLoS One 5, e9809, doi:10.1371/journal.pone.0009809 (2010).
Bachmann, M. F. & Jennings, G. T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nature reviews. Immunology 10, 787–796, doi:10.1038/nri2868 (2010).
Lopez-Sagaseta, J., Malito, E., Rappuoli, R. & Bottomley, M. J. Self-assembling protein nanoparticles in the design of vaccines. Comput Struct Biotechnol J 14, 58–68, doi:10.1016/j.csbj.2015.11.001 (2016).
Lee, L. A. & Wang, Q. Adaptations of nanoscale viruses and other protein cages for medical applications. Nanomedicine 2, 137–149, doi:10.1016/j.nano.2006.07.009 (2006).
Richardson, C., Bargatze, R. F., Goodwin, R. & Mendelman, P. M. Norovirus virus-like particle vaccines for the prevention of acute gastroenteritis. Expert review of vaccines 12, 155–167, doi:10.1586/erv.12.145 (2013).
Da Silva, D. M. et al. Physical interaction of human papillomavirus virus-like particles with immune cells. International Immunology 13, 633–641, doi:10.1093/intimm/13.5.633 (2001).
Al-Barwani, F., Young, S. L., Baird, M. A., Larsen, D. S. & Ward, V. K. Mannosylation of virus-like particles enhances internalization by antigen presenting cells. PLoS One 9, e104523, doi:10.1371/journal.pone.0104523 (2014).
Li, Y. et al. Enhancing immunogenicity and transmission-blocking activity of malaria vaccines by fusing Pfs25 to IMX313 multimerization technology. Sci Rep 6, 18848, doi:10.1038/srep18848 (2016).
Forbes, E. K. et al. T cell responses induced by adenoviral vectored vaccines can be adjuvanted by fusion of antigen to the oligomerization domain of C4b-binding protein. PLoS One 7, e44943, doi:10.1371/journal.pone.0044943 (2012).
Kanekiyo, M. et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 162, 1090–1100, doi:10.1016/j.cell.2015.07.043 (2015).
Jardine, J. et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716, doi:10.1126/science.1234150 (2013).
Boratyn, G. M. et al. Domain enhanced lookup time accelerated BLAST. Biology direct 7, 12, doi:10.1186/1745-6150-7-12 (2012).
Kaufmann, S. H. Tuberculosis vaccine development at a divide. Current opinion in pulmonary medicine 20, 294–300, doi:10.1097/MCP.0000000000000041 (2014).
White, M. T. et al. A combined analysis of immunogenicity, antibody kinetics and vaccine efficacy from phase 2 trials of the RTS, S malaria vaccine. BMC medicine 12, 117, doi:10.1186/PREACCEPT-5212460251240508 (2014).
Daum, R. S. & Spellberg, B. Progress toward a Staphylococcus aureus vaccine. Clin Infect Dis 54, 560–567, doi:10.1093/cid/cir828 (2012).
Saikatendu, K. S. et al. Structure of a conserved hypothetical protein SA1388 from S. aureus reveals a capped hexameric toroid with two PII domain lids and a dinuclear metal center. BMC structural biology 6, 27, doi:10.1186/1472-6807-6-27 (2006).
Dugourd, D., Martin, C., Rioux, C. R., Jacques, M. & Harel, J. Characterization of a periplasmic ATP-binding cassette iron import system of Brachyspira (Serpulina) hyodysenteriae. J Bacteriol 181, 6948–6957 (1999).
Ni Eidhin, D. et al. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Molecular microbiology 30, 245–257, doi:10.1046/j.1365-2958.1998.01050.x (1998).
Whitman, C. P. The 4-oxalocrotonate tautomerase family of enzymes: how nature makes new enzymes using a beta-alpha-beta structural motif. Archives of biochemistry and biophysics 402, 1–13, doi:10.1016/S0003-9861(02)00052-8 (2002).
Sampedro, G. R. et al. Targeting Staphylococcus aureus alpha-toxin as a novel approach to reduce severity of recurrent skin and soft-tissue infections. J Infect Dis 210, 1012–1018, doi:10.1093/infdis/jiu223 (2014).
Inoshima, I. et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med 17, 1310–1314, doi:10.1038/nm.2451 (2011).
Wilke, G. A. & Bubeck Wardenburg, J. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus alpha-hemolysin-mediated cellular injury. Proc Natl Acad Sci U S A 107, 13473–13478, doi:10.1073/pnas.1001815107 (2010).
Kaslow, D. C. et al. A vaccine candidate from the sexual stage of human malaria that contains EGF-like domains. Nature 333, 74–76, doi:10.1038/333074a0 (1988).
Huddleston, J. P., Burks, E. A. & Whitman, C. P. Identification and characterization of new family members in the tautomerase superfamily: analysis and implications. Archives of biochemistry and biophysics 564, 189–196, doi:10.1016/j.abb.2014.08.019 (2014).
4-Oxalocrotonate like proteins: Uniprot query, http://www.uniprot.org/uniprot/?query=family:%224-oxalocrotonate+tautomerase+family%22 (2014).
Ewer, K. J. et al. Viral vectors as vaccine platforms: from immunogenicity to impact. Curr Opin Immunol 41, 47–54, doi:10.1016/j.coi.2016.05.014 (2016).
Gilbert, S. C. et al. Enhanced CD8 T cell immunogenicity and protective efficacy in a mouse malaria model using a recombinant adenoviral vaccine in heterologous prime-boost immunisation regimes. Vaccine 20, 1039–1045, doi:10.1016/S0264-410X(01)00450-9 (2002).
Draper, S. J. et al. Effective induction of high-titer antibodies by viral vector vaccines. Nat Med 14, 819–821, doi:10.1038/nm.1850 (2008).
Burbelo, P. D., Goldman, R. & Mattson, T. L. A simplified immunoprecipitation method for quantitatively measuring antibody responses in clinical sera samples by using mammalian-produced Renilla luciferase-antigen fusion proteins. BMC biotechnology 5, 22, doi:10.1186/1472-6750-5-22 (2005).
van Diemen, P. M. et al. Irradiated wild-type and Spa mutant Staphylococcus aureus induce anti-S. aureus immune responses in mice which do not protect against subsequent intravenous challenge. Pathog Dis 68, 20–26, doi:10.1111/2049-632X.12042 (2013).
Oscherwitz, J. & Cease, K. B. Identification and validation of a linear protective neutralizing epitope in the beta-pore domain of alpha toxin. PLoS One 10, e0116882, doi:10.1371/journal.pone.0116882 (2015).
Miura, K. et al. Qualification of standard membrane-feeding assay with Plasmodium falciparum malaria and potential improvements for future assays. PLoS One 8, e57909, doi:10.1371/journal.pone.0057909 (2013).
Papadopoulos, J. S. & Agarwala, R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079, doi:10.1093/bioinformatics/btm076 (2007).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology 59, 307–321, doi:10.1093/sysbio/syq010 (2010).
Creech, C. B. 2nd et al. Vaccination as infection control: a pilot study to determine the impact of Staphylococcus aureus vaccination on nasal carriage. Vaccine 28, 256–260, doi:10.1016/j.vaccine.2009.09.088 (2009).
Acknowledgements
The research was supported by three sources: the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre based at Oxford University Hospitals NHS Trust and University of Oxford, by the James Martin School, University of Oxford, and received funding in part from the European Union’s Seventh Framework Programme under the grant agreement number 601783 (BELLEROPHON project). C.S.R. is a Jenner investigator. S.B. is a NDM Leadership Fellow and Junior Research Fellow of St Catherine’s College, University of Oxford. In addition, K.M. and C.A.L. were supported by the intramural program of the National Institute of Allergy and Infectious Disease/NIH and by PATH Malaria Vaccine Initiative. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors are grateful for the assistance of the Viral Vector Core Facilities, Jenner Institute, University of Oxford.
Author information
Authors and Affiliations
Contributions
D.H.W. designed and initiated the study and performed bioinformatics. P.M.v.D. designed and made the constructs, performed the DNA and viral vectored experiments, immune tests and statistical analysis. I.J.B., S.B. and D.B.L. performed the malaria experiments and statistical analysis. K.M. and C.A.L. performed the standard membrane feeding assays. S.B. and D.B.L. designed the Pfs25 constructs. D.H.W., C.S.R., A.M., S.B. and C.A.L. provided advice on study design. P.M.v.D. and D.H.W. wrote the paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
P.M.v.D, C.S.R and D.H.W are named on patent applications relating to BitC (patent 14/433565) and SAR1376 vaccines. The authors declare no other conflict of interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Diemen, P.M., Leneghan, D.B., Brian, I.J. et al. The S. aureus 4-oxalocrotonate tautomerase SAR1376 enhances immune responses when fused to several antigens. Sci Rep 7, 1745 (2017). https://doi.org/10.1038/s41598-017-01421-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-01421-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.