Abstract
The COP9 signalosome (CSN) is an evolutionary conserved multiprotein complex that regulates many aspects of plant development by controlling the activity of CULLIN-RING E3 ubiquitin ligases (CRLs). CRLs ubiquitinate and target for proteasomal degradation a vast number of specific substrate proteins involved in many developmental and physiological processes, including light and hormone signaling and cell division. As a consequence of CSN pleiotropic function, complete loss of CSN activity results in seedling lethality. Therefore, a detailed analysis of CSN physiological functions in adult Arabidopsis plants has been hampered by the early seedling lethality of csn null mutants. Here we report the identification and characterization of a viable allele of the Arabidopsis COP9 signalosome subunit 4 (CSN4). The allele, designated csn4-2035, suppresses the adventitious root (AR) phenotype of the Arabidopsis superroot2-1 mutant, potentially by altering its auxin signaling. Furthermore, we show that although the csn4-2035 mutation affects primary and lateral root (LR) formation in the 2035 suppressor mutant, CSN4 and other subunits of the COP9 complex seem to differentially control AR and LR development.
Similar content being viewed by others
Introduction
The CSN was first discovered in Arabidopsis, during a screening for mutants exhibiting constitutive photomorphogenic development in darkness, and was subsequently shown to be evolutionary conserved across eukaryotes (reviewed in ref. 1). The complex is composed of eight subunits, CSN1-CSN8. Six (CSN1-CSN4, CSN7 and CSN8) contain a PCI (Proteasome, COP9 signalosome and eukaryotic initiation factor 3, eIF3) domain, and two (CSN5 and CSN6) contain a MPN (Mpr1p-Pad1p-N-terminal) domain2. In Arabidopsis, the PCI domain-containing subunits are encoded by single copy genes, while the MPN domain-containing subunits are each encoded by two highly homologous genes. The two genes encoding CSN5 (CSN5A and CSN5B) play unequal roles in the regulation of plant development, while CSN6A and CSN6B, the genes encoding the CSN6 subunit, act largely redundantly3. The PCI and MPN subunits are structurally interdependent during the formation of the COP9 complex, thus explaining why loss of any of the eight CSN subunits leads to an apparently identical seedling lethal phenotype in Arabidopsis3. Therefore, a detailed analysis of CSN physiological functions in adult Arabidopsis plants has been hampered by the early seedling lethality of the null csn mutants.
Before the availability of T-DNA insertion lines, the only known Arabidopsis csn mutants were the pleiotropic seedling lethal mutants, now collectively known as the cop (constitutively photomorphogenic)/det (de-etiolated)/fus (fusca), which were identified through genetic screenings (reviewed in refs 1 and 4). Recent identification of alleles with partial loss of CSN function has shed light on some aspects of CSN functions beyond the seedling stage. However, to our knowledge, viable csn mutants are available only for five of the eight CSN subunits, including the double encoded MPN domain-containing subunits CSN55 and CSN63, and only for three out of the six single copy gene-encoded PCI domain-containing subunits, CSN16, CSN27, and CSN38. It has been suggested that one potential reason for the lack of viable known csn mutants is that CSN-independent functions of CSN subunits can only be uncovered under specific conditions4, and probably in particular types of screening. In this report, we introduce a viable allele of Arabidopsis CSN4, identified in a screening for mutants suppressing the adventitious root formation of the auxin overproducer superroot2 9.
Results and Discussion
Isolation of the 2035 mutant and identification of the csn4-2035 mutation
Aiming to identify new Arabidopsis genes involved in the control of adventitious root (AR) formation, we screened for suppressors of the superroot2-1 (sur2-1) mutant10. We isolated, mapped and characterized a number of suppressors, and, for a subset of mutants, we identified the causal mutations in the corresponding genes9. Using the combined advantages of classical map-based-cloning11 and whole genome re-sequencing12, we identified a point mutation in the locus At5g42970, which encodes the subunit 4 of the COP9 signalosome complex, as the potential causal mutation for the phenotype of one of the sur2-1 suppressors, designated 2035. The mutant carries a G-to-A mutation at position 2592 in the tenth exon of the CSN4/COP8/FUS4 gene, which results in an Ala-302-to-Val amino acid substitution (Fig. 1a,b). The Ala302 is part of a putative helix-loop-helix domain centered around amino acids 294 and 30213. A comparison of the CSN4 protein with homologs from other organisms reveals that the Ala302 mutated in 2035 is highly conserved even in more divergent proteins (Fig. 1b), being located in the PCI domain of the protein, that has previously been identified to be critical for the stability of the complex13, and recently shown to act as the scaffold for CSN4-6-7 interaction in Arabidopsis2. More recently, the crystal structure of the human COP9 signalosome has highlighted the important role of the PCI domain CSN4 subunit in sensing the binding of the neddylated Cullin–RINGE3 ubiquitin ligases to CSN, which is subsequently communicated to CSN5 and CSN6 for de-neddylation14,15,16. Therefore, the mutation of the Ala302 could induce a destabilization of the CSN and/or affect the de-neddylation process.
The csn4 alleles used in this study. (a) Structure of the Arabidopsis CSN4 subunit gene, with the position of the csn4-2035 point mutation and of the two T-DNA insertion lines. Exons are indicated by black boxes, introns by lines. (b) A comparison of a fragment from the Arabidopsis CSN4 protein with homologs from other organisms. The position of highly conserved Ala302, mutated in csn4-2035, is highlighted. The position and nature of the amino acid substitution found in CSN4-2035 mutant protein is shown. Below, an alignment of CSN4 proteins from Arabidopsis thaliana (NP_199111.1; residues 286–345), Populus tricocharpa (XP_002320585.1; residues 286–345), Brachypodium distachyon (XP_003558584.1; residues 289–348), Oriza sativa (NP_001049272.1; residues 289–348), Drosophila melanogaster (NP_477444.1; residues 294–353) and Homo sapiens (NP_057213.2; residues 287–346), around Arabidopsis Ala302 is shown.
Segregation analysis of the F2 progeny from a sur2-1gl1 × 2035 cross showed a 3:1 ratio of superroot:suppressor phenotype consistent with a single recessive mutation9. The mutant is viable and fertile as a homozygote, both in the sur2-1 mutant and in the Ws-4 wild-type backgrounds and does not exhibit the characteristic csn mutant phenotype, in contrast to seedling lethal csn4-1 (Salk_043720) and csn4-2 (Salk_053839) T-DNA insertion alleles, both isolated in the Col-0 background (Fig. 2a and ref. 17). To demonstrate unambiguously that the mutation in the suppressor 2035 affects CSN4, we conducted an allelism test using both insertion alleles. Due to early seedling lethality of the homozygous T-DNA insertion alleles, we crossed the homozygous suppressor 2035 with heterozygote csn4-1/+ and csn4-2/+ plants. A 1:1 mutant to wild-type phenotype segregation ratio was observed in the F1 generation of both crosses, as shown in Fig. 2b for the cross with csn4-1. The individuals that were potential csn4 trans-heterozygote mutants had flat cotyledons, shorter hypocotyls than wild-type like sur2-1/+ heterozygote plants, and no AR, except for a few individuals developing only one AR, as compared to sur2-1/+ wild-type like plants which on average, developed 1.8 AR/hypocotyl. We genotyped all the F1 individuals and confirmed that in the progeny of both combinations of crosses, all seedlings with a short hypocotyl and flat cotyledons were trans-heterozygotes, carrying either csn4-1/csn4-2035 or csn4-2/csn4-2035 hetero-allelic combination, while seedlings with a wild-type phenotype did not carry any T-DNA insertion in the CSN4 gene and were heterozygote for what we called the csn4-2035 allele. All trans-heterozygous mutants were viable and grew in soil, indicating that the mutation in the suppressor 2035 was responsible for the observed phenotypes.
Phenotype and characterization of the csn4 alleles. (a) The phenotype of in vitro grown 2035 suppressor mutant, together with Ws-4, Col-0, sur2-1gl1, csn4-2035, the two trans-heterozygotes double mutants, sur2-1csn4-1/csn4-2035 and sur2-1csn4-2/csn4-2035, respectively, is shown, as compared to the non-viable csn4-1 and csn4-2 alleles. Seedlings were first etiolated in the dark, until their hypocotyls were 6 mm long, and then transferred to light for seven days to induce AR formation on the etiolated hypocotyls. Arrowheads indicate the root-hypocotyl junction; arrows indicate ARs. Bar, 5 mm. (b) Allelism test. A cross between the homozygous 2035 and the heterozygote csn4-1 gives a 1:1 wild-type to mutant phenotype segregation ratio in the F1 generation. Arrowheads indicate the root-hypocotyl junction; arrows indicate ARs. Bar, 5 mm. (c) Numbers of AR were counted on the hypocotyls treated as in (a), and averaged. (d) Numbers of emergent lateral roots were counted on seedlings grown in vitro directly in light conditions for 10 d and averaged. (e) Primary root length was measured on the same seedling as in (d) and averaged. (f) Lateral root density was estimated by dividing the average number of lateral roots (d) by the average root length (e). (g) Phenotypes of 3-week-old wild-type, sur2-1gl1, 2035 and csn4-2035 mutant plants. Seeds were sown in vitro in light conditions and were subsequently transferred to soil where the plants were grown for 3 weeks in a controlled environment, as described in the Methods. Bar, 20 mm. For (c to e) at least 50 seedlings of each line were analyzed and the experiments were repeated three times and the data pooled. Error bars indicate standard error. One-way ANOVA combined with Tukey’s multiple-comparison post-test revealed that the values indicated by different letters were significantly different from each other (P < 0.05; n > 150). (h) Free IAA content was quantified on apical parts (cotyledons+hypocotyls) and on roots of seedlings grown in vitro under light for nine days; samples were extracted, purified, and analyzed by GC-MRM-MS as described by55. Three biological replicates of at least 50 seedlings from each line were analyzed and averaged. Error bars indicate standard error. One-way ANOVA combined with Tukey’s multiple-comparison post-test revealed that the values indicated by different letters were significantly different from each other (P < 0.05; n = 3). (i) csn4-2035 is slightly resistant to exogenously applied auxin. Percentage inhibition was calculated by dividing the average growth on 2,4-D containing medium by the average growth on control medium and subtracting this ratio from 100% (n > 25).
Phenotypic characterization of the suppressor 2035
When grown in vitro on vertical plates, sur2-1gl1 developed numerous AR on the etiolated hypocotyls9. The suppressor 2035, which carries the mutation csn4-2035 in a sur2-1gl1 background, developed significantly fewer AR compared to sur2-1gl1 (Fig. 2a,c). In addition, the AR formation was almost completely abolished in the csn4-2035 single mutant compared to the Ws-4 wild-type, since the majority of the seedlings had no initiated AR on the hypocotyl seven days after being transferred to the light, and only a limited number of individuals developed one AR (Fig. 2a,c). We had previously shown that the main root system of suppressor 2035 was also significantly reduced compared to sur2-1gl1 (Fig. 2d,e and ref. 9). Here we show that the main root system is also reduced in the csn4-2035 single mutant as compared to the Ws-4 wild-type. Both the primary root (PR) length and the lateral root (LR) number were significantly reduced (Fig. 2d,e,f), confirming that the csn4-2035 mutation also affects PR and LR formation.
Interestingly, the AR number vs LR number and PR length were differentially affected by the csn4-1/csn4-2035 and csn4-2/csn4-2035 heteroallelic combinations in the two trans-heterozygote double mutants, sur2-1csn4-1/2035 and sur2-1csn4-2/2035. While the average number of AR in the trans-heterozygote double mutants, sur2-1csn4-1/2035 and sur2-1csn4-2/2035 was not affected compared to 2035 (Fig. 2c), the PR length and the LR number and density were significantly decreased (Fig. 2d,e,f). As shown in Fig. 2c, the number of AR was not significantly different in the two populations derived from sur2-1csn4-1/2035 and sur2-1csn4-2/2035 compared to the 2035 suppressor. This was surprising, since it has been reported that the phenotype of a hypomorph allele had a more severe effect in trans to a deletion allele than when homozygous18. Indeed, a more extreme phenotype in the populations containing 50% trans mutants was observed for PR and LR development, since the LR density was decreased in the two populations to 32% and 34%, respectively as compared to 2035 (Fig. 2d,e,f). It should be noted that in Fig. 2c–f, the data corresponding to the trans-heterozygotes sur2-1csn4-1/2035 and sur2-1csn4-2/2035 were collected from segregating populations including viable sur2-1csn4-1/2035 (50%) and 2035 (25%), and sur2-1csn4-2/2035 (50%) and 2035 (25%), respectively. These were generated by self-pollinating the sur2-1csn4-1/2035 and sur2-1csn4-2/2035 parents, respectively. In both situations, the segregating sur2-1csn4-1 −/− (25%) and sur2-1csn4-2 −/− (25%) had the characteristic fusca phenotype and were not considered for the characterization. Apparently, losing one copy of the csn4-2035 in 50% of the individuals from both populations did not have an impact on the AR number compared to the suppressor 2035, but this lack could not be compensated for in the case of PR and LR formation. This suggests that CSN4 and/or the COP9 complex differentially control AR and LR development.
When grown in soil, in LD conditions, both 2035 and csn4-2035 develop smaller rosettes than the wild type (Fig. 2g), and the adult plants show a mild dwarfism and loss of apical dominance phenotype as observed for other hypomorphic csn mutants7.
The seedling and rosette phenotype of 2035 (short petioles, round leaves) is the result of the suppression of the sur2-1 phenotype. The apical part (not only the roots) loses the auxin related phenotype. Since the sur2-1 mutant phenotype is the result of auxin overproduction, its suppression may be due to either altered auxin homeostasis or auxin perception in the suppressor mutant. It has been reported that the viable CSN alleles csn1-10, csn2-5, csn3-3, csn5a and csn5b have altered auxin perception and auxin-resistant root growth5,6,7,8, 19. To test whether the csn4-2035 mutation suppresses the sur2-1 phenotype by disturbing the auxin signaling or the auxin homeostasis in the 2035 mutant, we measured the free IAA content both in the apical parts and the roots of in vitro grown seedlings. As we previously described in ref. 9 and show here in Fig. 2h, the 2035 mutant retains the high auxin level of sur2-1gl1, which indicates that the reduction of the AR number in 2035 is unlikely to be due to a reduced auxin content, suggesting a possible alteration in auxin signaling.
Auxin signaling is perturbed in 2035
It is well-known that the CSN plays a role in auxin signaling, acting as a de-neddylase to regulate SCFTIR1/AFB activity, and mutants altered in any CSN subunit are more or less resistant to inhibition of root growth by exogenously applied auxin and have reduced lateral root formation5,6,7, 20. Therefore, we performed a dose-response assay and measured the auxin inhibition of root elongation and found that csn4-2035 was resistant to 2,4-D compared to Ws-4 (Fig. 2i and Supplementary Fig. S1). After transfer to the medium supplemented with 0.1 μM 2,4-D, the wild type Ws-4 displayed inhibition of root elongation by 65% whereas csn4-2035 seedlings displayed a 20% inhibition only (Fig. 2i) and did not develop LR (Supplementary Fig. S1).
In previous work, we have demonstrated that a regulatory module, composed of three AUXIN RESPONSE FACTOR genes (ARF6, ARF8, and ARF17) and three auxin-responsive genes (GH3.3, GH3.5, and GH3.6) controls AR initiation in Arabidopsis hypocotyls by modulating JA homeostasis21, 22. In addition, we recently showed that selected suppressor mutations of the sur2-1 phenotype differentially affected the expression of the ARF/GH3 regulatory module9. In the suppressors analyzed, the reduced number of AR was independent of the endogenous auxin content, but positively correlated with the transcript amount of the three GH3 genes9. In the present work we show that despite the endogenous content of free IAA in the suppressor 2035, which is still as high as that of the sur2-1gl1 mutant (Fig. 2h and ref. 9), the relative transcript amount of the GH3 genes is significantly reduced compared to sur2-1gl1 (Fig. 3a), probably explaining the reduced number of AR. Interestingly, this could be explained neither by a down-regulation of the expression of the positive regulators ARF6 or/and ARF8 nor an up-regulation of the negative regulator ARF17 (Fig. 3a). ARF transcriptional activity is negatively regulated by transcriptional repressors of the Aux/IAA family, which are unstable proteins rapidly degraded through the SCFTIR1/AFB1-3 dependent ubiquitin-proteasome system in the presence of a high auxin concentration. It was also shown that mutants altered in the expression of CSN3, CSN4, CSN5 or CSN8 subunits were partially impaired in the SCFTIR1/AFB1-3-mediated protein degradation, resulting in a partially altered auxin response5, 17.
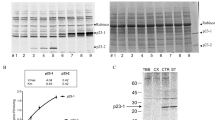
Auxin signaling is perturbed in 2035. (a) Quantification by quantitative real-time PCR of ARF6, ARF8, ARF17, GH3.3, GH3.5 and GH3.6 transcript abundance in hypocotyls of sur2-1gl1 and 2035 seedlings, which were etiolated until their hypocotyl had reached 6 mm (T0) and then transferred to the light for 72 h (T0+72 h light), or kept in dark for an additional 72 h (T0+72 h dark). Gene expression values are relative to the expression in the wild type, for which the value is set to 1. Error bars indicate standard error obtained from three independent biological replicates; (*) indicates values that were significantly different from sur2-1gl1 values according to one-way ANOVA combined with Bonferroni’s comparison post-test; (ns) indicates values not significantly different; (P < 0.05; n = 3). (b) csn4-2035 is a weak allele mutant that produces as much protein as the wild type. CSN4 western blot analysis of protein extracts prepared from WS-4, csn4-2035 and the null allele csn4-1. (c) csn4-2035 mutation affects the de-neddylation activity of CSN4 protein. CUL1 western blot analysis of protein extracts prepared from Ws-4 and csn mutant seedlings. The upper band indicates the modified (neddylated) CUL1. Ponceau stained polyvinylidene difluoride (PVDF) membrane is shown as a loading control. (d) Quantification by quantitative real-time PCR of GH3.3, GH3.5 and GH3.6 transcript abundance in hypocotyls of csn4-2035 seedlings. Gene expression values are relative to the expression in the wild type, for which the value is set to 1. Error bars indicate standard error obtained from three independent replicates; (*) indicate values that were significantly different from wild-type values (P < 0.05; n = 3).
Because the CSN cleaves the RUB/NEDD8 peptide from the cullin subunit of CRL ubiquitin-ligases, we checked whether or not the csn4-2035 mutation affected CUL1 modification. We first showed that the csn4-2035 mutant produced as much CSN4 protein as the wild type, while the null allele mutant retained no protein as expected (Fig. 3b). These results suggested that the phenotype was not the result of a reduced amount of CSN4 protein but most likely due to a modification of its activity or to a defect in the CSN assembly or structure. Therefore, we analyzed de-neddylation activity (Fig. 3c). We included csn1-10 and csn3-3 mutants as controls because they were shown to exhibit similar auxin resistance but had different de-neddylation activity8. Figure 3c shows that there was no de-neddylation of CUL1-NEDD8 in the csn4-1 null allele mutant as expected and a clear accumulation of de-neddylated CUL1 in the wild type and csn3-3 mutant as reported in previous studies8. In contrast, we observed an accumulation of CUL1-NEDD8 in csn4-2035, as is the case in csn10-1, indicating that the csn4-2035 mutation partially affects the de-neddylation activity of the CSN and, as a consequence, decreases auxin sensitivity. This hypothesis is supported by the slight but significant down-regulation of the auxin inducible GH3 genes in the csn4-2035 mutant (Fig. 3d).
Therefore, the down-regulation of the auxin inducible GH3 genes in the 2035 suppressor mutant, resulting in the reduced number of AR, can reasonably be explained by an inefficient degradation of AUX/IAA proteins and, as a consequence, the down-regulation of the auxin signaling pathway which in turn would induce the expression of the activating ARFs such as ARF6 and ARF8 in a regulatory feedback loop. This would explain why, despite high endogenous auxin content and a combined increase in the relative transcript amount of the ARF6 and ARF8 positive regulators and decrease in the transcript amount of the ARF17 negative regulator, compared to sur2-1gl1 (Fig. 3a), the mutant 2035 develops fewer AR.
csn4-2035 mutation affects blue and red light perception in a sur2-1 background
No obvious csn phenotypes - cop/det (open cotyledons, short hypocotyl and absence of apical hook of the dark-grown seedling) and fusca (accumulation of anthocyanins) - typical of the other reported Arabidopsis mutants of all six PCI-domain subunits, including the two CSN4 T-DNA alleles csn4-1 and csn4-2, were observed, either in 2035 mutant or in the csn4-2035 single mutant (Fig. 2a). Grown in vitro, in LD conditions, the hypocotyl of 2035 had an intermediate length between that of the wild-types and sur2-1gl1 9. Early observations have shown that light-grown CSN null mutants have very short hypocotyls23. Shorter hypocotyls than the wild-type controls have also been observed in week CSN alleles5, 19, and the role of CSN in hypocotyl elongation has recently been demonstrated24. In a screening for novel CSN interactors, the authors identified CFK1 (COP9 INTERACTING F-BOX KELCH 1), a new plant-specific CSN-interacting F-box protein that is regulated by the CSN and the proteasome-dependent proteolysis. This physical interaction suggests that CFK1 function might be required for CSN-mediated hypocotyl inhibition, as demonstrated by enhanced hypocotyl length in the double mutant csn5a-2 CFKRNAi compared to either parents24. The authors have shown that light induces accumulation of the CFK1 transcript in the hypocotyl, and that CFK1 promotes hypocotyl elongation by increasing cell size. Moreover, while reduction of CSN levels enhances the short hypocotyl phenotype of CFK1RNAi seedlings, complete loss of CSN activity suppresses the long-hypocotyl phenotype of CFK1-overexpressing seedlings. In light of these findings, we can speculate that suppression of the hypocotyl growth in the 2035 suppressor mutant compared to sur2-1gl1 could be explained by a reduced activity of the CSN due to the mutation in the CSN4 subunit.
In Arabidopsis, normal hypocotyl elongation is controlled both by cryptochromes, which respond to blue/UV-A light, and phytochromes, which sense red/far-red light. An allelic mutant of sur2-1, red1 (r ed e longate d1), has been identified during a screening for mutants that display enhanced etiolation in continuous red light (cR), suggesting a defect in red light perception25. In addition, the CSN was originally discovered as an essential complex that regulates light-induced development in Arabidopsis26,27,28,29, and increased photomorphogenic responses were observed in different light conditions for viable alleles of the CSN subunit genes, csn5a, csn5b 5 and csn6a-1 and csn6a-2, respectively3. To investigate further whether the csn4-2035 mutation alters light perception, we measured the hypocotyl length of 2035, sur2-1gl1 and Ws-4 under five fluence rates of continuous red (cR) and continuous blue (cB) light. In the dark, 2035 and sur2-1gl1 had the same hypocotyl length and were shorter than the wild type, as previously described for sur2-1 (Fig. 4a,b and ref. 10). When grown under cR at 40 or 210 μmol m−2 s−1 irradiance, both sur2-1gl1 and 2035 were longer than the wild type. Nevertheless the 2035 mutant had a significantly longer hypocotyl compared to sur2-1gl1 when grown under 1, 3 or 11 μmol m−2 s−1 irradiance (Fig. 4a), suggesting that a mutation in the CSN4 subunit affects red light perception. Similarly, a significant difference between the suppressor mutant 2035 and sur2-1gl1 was observed when seedlings were grown under cB light at 2 and 10 μmol m−2 s−1 irradiance (Fig. 4b). From these data, we can conclude that a mutation in the CSN4 subunit affects red and blue light perception to some extent, but we cannot say whether the AR phenotype and the light related phenotype are connected, and additional experiments are required. Nevertheless, we have previously shown that AR initiation is also controlled by light21, 30, as well as by a signaling cross-talk between auxin and jasmonate involving MYC2 transcription factor22, therefore the csn4-2035 mutation could also highlight cross-talk between light and these signaling pathways in the control of AR initiation. Indeed, it was recently suggested that inhibition of hypocotyl elongation by jasmonates is enhanced under red light in phyB-dependent manner31, and that JA-related transcription factors MYC2, MYC3, and MYC4 are short-lived proteins degraded by the proteasome, and stabilized by JA and light, in Arabidopsis thaliana. Darkness and CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) destabilize MYC2, MYC3, and MYC4 proteins, whereas red and blue lights stabilize them through the activation of the corresponding photoreceptors32. Therefore, the csn4-2035 mutation, by perturbing red and blue light perception, may also affect the stabilization of MYC proteins and thereby have an impact on adventitious root formation.
The csn4-2035 mutation affects red and blue light perception in a sur2-1gl1 background. All measurements were performed on three independent biological replicates with a minimum of 40 seedlings each, as described in the Methods. Error bars indicate standard error. One-way ANOVA combined with Tukey’s multiple-comparison post-test revealed that the values indicated by (*) were significantly different from Ws-4 values; (#) indicates values significantly different from sur2-1gl1 values; while (o) and (◻) indicate values not significantly different from Ws-4 and sur2-1gl1 values, respectively; (P < 0.05; n > 120).
CSN subunits play differential roles in AR and LR formation
The CSN regulates multiple plant hormone signaling and developmental processes through SCF-type CRLs, known as SCF complexes, which act in many pathways such as SCFTIR1 in auxin7, 20, SCFCOI1 in jasmonate33, 34 and SCFSLY1 in gibberellic acid signaling35, SCFUFO in flower development36, and SCFCFK1 in hypocotyl elongation24. The CSN also controls photomorphogenic development and abscisic acid signaling through a CUL4-CDD (COP10, DDB1 and DET137), and a CRL4-DDA1 complex38, respectively. The isolation of viable csn mutants provides an opportunity to study CSN functions beyond seedling stages by elucidating the role CSN and its individual subunits play in these processes. Interestingly, the viable CSN mutant alleles described so far, both for the double encoded MPN subunits and single encoded PCI subunits, either display pleiotropic developmental defects or have subtler phenotypes. The first viable csn mutants described were the loss-of-function T-DNA insertion mutants in the double encoded MPN domain-containing subunit CSN5A and CSN5B, csn5a-1 (null), csn5a-2 (hypomorphic), and csn5b-1 5. The csn5a mutant alleles display increased photomorphogenic responses and pleiotropic developmental defects both at the seedling and adult stage, affecting lateral root and root hair formation, and flower size. In contrast, the csn5b mutant shows comparably subtle phenotypes5. However, both csn5a-1csn5-b and csn5a-2csn5b double mutants mimic the phenotype of the previously described cop/det/fus mutants, indicating that the two CSN5 genes have redundant functions, with CSN5A having a stronger relative contribution to the respective phenotype than CSN5B. On the other hand, except for a very mild photomorphogenic phenotype in the dark and under blue light, the T-DNA insertion mutants in the CSN6A and CSN6B genes, coding for the second MPN subunit, do not display any obvious morphological defects in white light and after the seedling stage, while loss of function for both CSN6 proteins leads to seedling lethality and a fusca phenotype in the double mutant csn6a-1csn6b-1 3.
Of the three single gene encoded PCI subunits for which viable alleles have been identified, csn1-10 has been described to have severe pleiotropic developmental defects associated with altered auxin responses6, while csn2-5, except for a mild dwarfism of adult plants7 and a “curly hypocotyl” phenotype when seedlings were grown in darkness19 and csn3-3 8, although impaired in auxin responses, retain a wild-type like phenotype, under the conditions investigated and for the analyzed parameters.
In order to test whether the viable csn mutants show differences in AR formation and whether they differentially interact with sur2-1, we counted the AR number in the single mutants csn1-10, csn2-5, csn3-3, csn5a-1, csn5a-2, csn5b and in the double mutants with sur2-1. As shown in Fig. 5a, the AR number varied considerably among the tested csn mutants. The csn1-10 and csn3-3 mutants developed significantly fewer AR as compared to Col-0, and csn5a-1 and csn5a-2 did not form any AR, while csn2-5 was not significantly different from its corresponding wild-type Ler (Fig. 5a). Interestingly, in contrast to csn5a-1 and csn5a-2, the csn5b mutant developed an increased number of AR compared to the wild-type Col-0 (Fig. 5a), and although all csn mutants suppress the AR phenotype of sur2-1 to various degrees, csn5b enhanced the sur2-1 phenotype (Fig. 5b). This indicates that the two CSN5 subunits differentially contribute to the regulation of AR formation.
CSN subunits play differential roles in AR and LR formation. (a,b) Adventitious roots were counted on seedlings first etiolated in the dark, until their hypocotyls were 6 mm long, then transferred to the light for 7 d, and averaged. (c,d) Numbers of emergent lateral roots were counted on seedlings grown in vitro directly in light conditions for 10 d and averaged as described in the Methods. At least 30 seedlings of each line were analyzed, the experiments were repeated three times and the data pooled. Error bars indicate standard error; A one-way ANOVA combined with Dunnett’s comparison post-test was used to compare single mutant lines with their respective wild type, and double mutant lines with sur2-1gl1; (*) indicates values significantly different from Col-0 values; (#) indicates values significantly different from Ler values; (o) indicate values that were significantly different from sur2-1gl1 values; ns indicates values not significantly different from Ler (c) or sur2-1gl1 (d) values (P < 0.05; n > 90).
As shown in Fig. 2c,d, the csn4-2035 mutation differentially impacted on AR vs. LR development. We wondered whether this was also true for mutations in other CSN subunits. We counted the average number of LR on the seedlings of the csn1-10, csn2-5, csn3-3, csn5a-1, csn5a-2 and csn5b single mutants, and in the double mutants with sur2-1 grown for 10 days under light (Fig. 5c,d). Interestingly, all single and double mutants (except sur2-1csn5b double mutant) showed a reduced number of LR compared to the wild-type controls (Fig. 5c) or sur2-1 (Fig. 5d). Interestingly the number of LR in the sur2-1csn5b double mutant was not significantly different from sur2-1gl1. Analyzing the impact that the individual csn mutations have on AR vs. LR development, we can see that the number of both AR and LR is significantly reduced in csn1-10, csn3-3, csn5a-1, csn5a-2 single mutants compared to Col-0, as well as in their double mutants with sur2-1, compared to sur2-1gl1. In contrast, the csn2-5 mutant is affected with respect to LR number, but shows no significant difference in AR number compared to the Ler wild type, although the csn2-5 mutation strongly suppresses the sur2-1 phenotype in the double mutant sur2-1csn2-5 (Fig. 5a). The csn5b mutant, although showing a significant reduction of the number of LR, developed significantly more AR on the hypocotyl compared to Col-0. Moreover, it enhanced the AR phenotype in the double mutant sur2-1csn5b, while the LR number was not significantly different from sur2-1gl1.
Mutations in CSN subunits have previously been shown to impact LR formation in the viable identified single csn mutants or generated double mutants. While LR numbers in the single mutants csn1-10 and csn3-3 8 were comparable to wild-type control, single csn5a-2, csn5b-1 5, as well as double mutants csn3-3 csn1-10 8, developed significantly fewer LR. This reduction was attributed to the impaired auxin response of the mutants5, 8, or to a defect in the cell cycle progression39. Nevertheless we have previously shown that jasmonate, through the CORONATIN INSENSITIVE 1 (COI1) signaling pathway, negatively regulates AR initiation in Arabidopsis22 while others have shown that it promotes LR formation40, 41. Therefore, considering the fact that the COP9 signalosome modulates JA responses by physically interacting with the SCFCOI1 complex on one hand33 and that it modulates JA biosynthesis on the other hand41, we can speculate that the different CSN subunits may differentially control AR and LR formation by differentially modulating the JA signaling pathway or JA biosynthesis.
Alternatively, several lines of evidence suggest that light-auxin cross-talk takes place during AR development in Arabidopsis. First, in the ago1 mutant, the defect in AR formation correlates with an alteration of auxin homeostasis and a hypersensitivity to light30. Second, a model for genetic control of AR formation in Arabidopsis hypocotyls that integrates light and auxin signaling has been published21. In addition, previous studies suggest that shoot-localized phytochromes regulate lateral root development42, and root-localized phytochromes and cryptochromes regulate phototropic responses and growth of primary roots43,44,45. Also, it has been shown that light regulates developmental processes at least in part through cross-talk with the phytohormone auxin45. Light signaling involves nucleo-cytoplasmic partitioning of phytochromes46 and negative regulators such as CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), an E3 ubiquitin ligase involved in 26S proteasome-mediated protein degradation47. In darkness, COP1 accumulates in the nucleus to degrade transcription factors including HY5, HYH, HFR and LAF, therefore, suppressing the expression of light-regulated genes48,49,50. On the other hand, light triggers the degradation of COP1 in the nucleus, thereby, activating the expression of light-regulated genes, and promoting photomorphogenesis. The cytoplasmic-nuclear partitioning of COP1 is regulated by the multisubunit COP9 complex (COP9 signalosome or CSN). Total loss-of-function mutations in any CSN components exclude COP1 from nuclear accumulation in the dark29 and lead to the extreme seedling-lethal phenotype, whereas partial loss of function mutations in the same subunits highlight differential roles in the regulation of light perception. Therefore, the differential role of the CSN in the regulation of AR and LR formation could also be due to differential regulation of light/auxin signaling cross-talk.
Based on these data, we cannot say whether suppression of the AR phenotype in the 2035 mutant is due to defects in auxin signaling, jasmonate signaling, light perception or light regulatory pathways, cell cycle defects in roots, or a combination of these. However, given the documented function that the COP9 signalosome has in light, auxin and jasmonate signaling, the csn4-2035 allele could serve for the elucidation of auxin-jasmonate-light cross-talk in AR development. For more general interest, it will provide a tool for a further in-depth study of CSN-independent functions of the CSN4 subunit in Arabidopsis.
Methods
Plant material and growth conditions
The 2035 mutant was identified in a previously described screening9. In order to avoid selecting wild-type seedlings due to potential contamination of the mutagenized population with wild-type seeds, the glabra1 mutation was introgressed in the sur2-1 mutant background. The glabra1 mutant was identified in the Versailles collection of T-DNA insertion lines and was therefore in the same genetic background as sur2-1. Homozygote seeds from the double mutant superroot2-1gl1 (sur2-1gl1) (ecotype Wassilewskija, Ws-4) were mutagenized with ethyl methanesulphonate, as described in ref. 51.
The csn4-1 (SALK_043720) and csn4-2 (Salk_053839) segregating lines were described in ref. 17; csn5a-1 (SALK_063436), csn5a-2 (SALK_027705) and csn5b-1 (SALK_007134) were described in ref. 5; csn2-5 was described in ref. 7; csn1-10 was described in ref. 6, and csn3-3 was described in ref. 8. The 2035 mutant is in Wassilewskija (Ws-4) background, the csn2-5 mutant is in a Landsberg erecta (Ler) background while the rest are in a Columbia (Col-0) background. In vitro characterization and auxin quantification were conducted as described previously9. For the phenotypic evaluation of soil-grown plants, the seeds were first germinated in vitro and subsequently the seedlings transferred into pots. The plants were then placed in growth chambers in long day (16 h light/8 h darkness) conditions, at 22 °C/18 °C (light/dark), 130 μE m−2 s−1 irradiance on average and 60% relative humidity.
For root growth assays, 6-day-old seedlings grown in the light as described in ref. 30 were transferred to the same medium supplemented with various concentrations of 2,4-D, and root growth was measured after an additional 5 days. The conditions in the controlled environment chambers were as follows: 130 μE m−2 s−1 irradiance on average, 16/8 light/dark cycle, 22/15 °C, 60% relative humidity. Plates were scanned before and after transfer of the seedling on auxin containing medium. Root length was measured using the ImageJ software package (http://rsb.info.nih.gov/ij/index.html) and ref. 52. Percentage inhibition was calculated by dividing the average growth on 2,4-D containing medium by the average growth on control medium and subtracting this ratio from 100% (n > 25).
Complementation test and genotyping the csn4 alleles
For complementation analyses, homozygous 2035 plants were crossed with heterozygous csn4-1 and csn4-2, respectively. F1 progeny was phenotyped and subsequently genotyped, using allele specific PCR primers, as follows. The csn4-1 mutation was genotyped using CSN4-FW1 and CSN4-RV1.1 to test for the presence of the wild-type gene, and LBb1.3 and CSN4-RV1 to test for the presence of the T-DNA. The csn4-2 mutation was genotyped using CSN4-FW2 and CSN4-RV2 to test for the presence of the wild-type gene and LBb1.3 and CSN4-RV2 to test for the presence of the T-DNA. To genotype the csn4-3 point mutation, newly derived cleaved-amplified polymorphic sequence primers were designed using the dCAPS Finder 2.0 software (ref. 53, http://helix.wustl.edu/dcaps/dcaps.html). Two mismatches (underlined) were introduced in the F primer to incorporate a restriction site in the PCR product of one allele. After amplification, the PCR products were digested with HpaI (Fermentas Fast Digest) following the manufacturer’s recommendations and separated on a 4% agarose gel. The csn4-3 allele yielded two fragments of 150 and 19 bp, while the wild type gave one band of 169 bp. All primers are listed in Supplementary Table S1.
Hypocotyl growth under different light conditions
Seeds were surface sterilized, sown in rows in vitro on media without sugar, stratified for 48 h at 4 °C, then transferred to a plant growth chamber under white light conditions for 4 h to activate germination. Subsequently, five Petri dishes of each light condition/replicate were placed on top of each other, with one chromatography paper layer between them, and wrapped with black plastic leaving the top uncovered (see Supplementary Fig. S2). They were transferred to plant growth cabinets at 20°C with constant blue (cB) or constant red (cR) light, respectively. The light intensity measured on each layer, bottom to top, was as follows: 1; 3; 11; 41 and 210 μmol m−2 s−1 irradiance of cR light, and 0.1; 0.4; 2; 10 and 52 μmol m−2 s−1 irradiance of cB light, respectively. For dark growth conditions, five plates/chamber/replicate were wrapped in three layers of aluminum foil and placed vertically. After 7 d, the seedlings were bended on the surface of the medium, the plates were photographed and hypocotyls measured using ImageJ software52 (http://rsb.info.nih.gov/ij/index.html). All measurements were performed on three independent biological replicates with a minimum of 40 seedlings each.
Real-time PCR experiments and data analysis
Transcript abundance was assessed by quantitative real-time (RT) PCR as previously described22. All quantifications were repeated with three independent biological replicates, using the following standard thermal profile: 10 min at 95 °C, followed by 40 cycles at 95 °C for 10 s, 60 °C for 15 s (except for GH3.5 for which the annealing temperature was 65 °C), and 72 °C for 15 s. The sequences of primers used are presented in Supplementary Table S2. Using the RefFinder: http://www.leonxie.com/referencegene.php, EF1A (At5g60390) has been validated as the most stably expressed gene, of the four tested, and was used to normalize the RT-PCR data. The expression levels were calculated as previously described9. Gene expression values are relative to the expression in the wild type, for which the value is set to 1. All RT-PCR results presented are means from three independent biological replicates. For each independent biological replicate, the relative transcript amount was calculated as the mean of three technical replicates, using the method for calculation of standard errors in relative quantification recommended by54.
Immunoblot analysis
Total proteins were extracted from 10-day-old light grown seedlings in protein extraction buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% NP40, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), and Protease Inhibitor cocktail tablets (Roche)8. A aliquote of 40 μg of total protein for each genotype was loaded and separated by 10% SDS-PAGE gel. Samples were then blotted and used for immunodetection. anti-CSN4 (1:1000) (COP9 signalosome subunits polyclonal antibody) and anti-CUL1 (1:1000) (CUL1 (Arabidopsis thaliana) polyclonal antibody) were purchased at Enzo Life Sciences (http://www.enzolifesciences.com). Goat anti-Rabbit IgG (1:20000) (Agrisera http://www.agrisera.com/en/info/about-agrisera-.html) was used as a secondary antibody.
Change history
20 July 2017
A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
References
Wei, N. & Deng, X. W. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19, 261–286 (2003).
Kotiguda, G. G. et al. The organization of a CSN5-containing subcomplex of the COP9 signalosome. J. Biol. Chem. 287, 42031–42041 (2012).
Gusmaroli, G., Figueroa, P., Serino, G. & Deng, X. W. Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant cell 19, 564–581 (2007).
Stratmann, J. W. & Gusmaroli, G. Many jobs for one good cop - the COP9 signalosome guards development and defense. Plant Sci. 185–186, 50–64 (2012).
Dohmann, E. M., Kuhnle, C. & Schwechheimer, C. Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant cell 17, 1967–1978 (2005).
Zhang, W. et al. Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc. Natl. Acad. Sci. USA 105, 8470–8475 (2008).
Stuttmann, J. et al. COP9 signalosome- and 26S proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J. Biol. Chem. 284, 7920–7930, doi:10.1074/jbc.M809069200 (2009).
Huang, H., Quint, M. & Gray, W. M. The eta7/csn3-3 auxin response mutant of Arabidopsis defines a novel function for the CSN3 subunit of the COP9 signalosome. Plos one 8, e66578, doi:10.1371/journal.pone.0066578 (2013).
Pacurar, D. I. et al. Identification of new adventitious rooting mutants amongst suppressors of the Arabidopsis thaliana superroot2 mutation. J. Exp. Bot. 65, 1605–1618, doi:10.1093/jxb/eru026 (2014).
Delarue, M., Prinsen, E., Onckelen, H. V., Caboche, M. & Bellini, C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14, 603–611 (1998).
Pacurar, D. I. et al. A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J. Exp. Bot. 63, 2491–2501 (2012).
Pacurar, D. I., Pacurar, M. L., Pacurar, A. M., Gutierrez, L. & Bellini, C. A novel viable allele of Arabidopsis CULLIN1 identified in a screen for superroot2 suppressors by next generation sequencing-assisted mapping. Plos one 9, e100846, doi:10.1371/journal.pone.0100846 (2014).
Serino, G. et al. Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant cell 11, 1967–1980 (1999).
Lingaraju, G. M. et al. Crystal structure of the human COP9 signalosome. Nature 512, 161–165, doi:10.1038/nature13566 (2014).
Cavadini, S. et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 531, 598–603, doi:10.1038/nature17416 (2016).
Mosadeghi, R. et al. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. eLife 5, doi:10.7554/eLife.12102 (2016).
Dohmann, E. M., Levesque, M. P., Isono, E., Schmid, M. & Schwechheimer, C. Auxin responses in mutants of the Arabidopsis CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome. Plant Phys. 147, 1369–1379 (2008).
Wilkie, A. O. The molecular basis of genetic dominance. J. Med. Genet. 31, 89–98 (1994).
Stuttmann, J., Parker, J. E. & Noel, L. D. Novel aspects of COP9 signalosome functions revealed through analysis of hypomorphic csn mutants. Plant Signal. Behav 4, 896–898 (2009).
Schwechheimer, C. et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science (New York, N.Y.) 292, 1379–1382 (2001).
Gutierrez, L. et al. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant cell 21, 3119–3132 (2009).
Gutierrez, L. et al. Auxin Controls Arabidopsis Adventitious Root Initiation by Regulating Jasmonic Acid Homeostasis. Plant cell 24, 2515–2527 (2012).
Kwok, S. F., Piekos, B., Misera, S. & Deng, X. W. A complement of ten essential and pleiotropic arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Phys. 110, 731–742 (1996).
Franciosini, A. et al. The Arabidopsis COP9 SIGNALOSOME INTERACTING F-BOX KELCH 1 protein forms an SCF ubiquitin ligase and regulates hypocotyl elongation. Mol. Plant 6, 1616–1629, doi:10.1093/mp/sst045 (2013).
Wagner, D., Hoecker, U. & Quail, P. H. RED1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant cell 9, 731–743, doi:10.1105/tpc.9.5.731 (1997).
Wei, N. & Deng, X. W. COP9: a new genetic locus involved in light-regulated development and gene expression in arabidopsis. Plant cell 4, 1507–1518 (1992).
Wei, N., Chamovitz, D. A. & Deng, X. W. Arabidopsis Cop9 Is a Component of a Novel Signaling Complex Mediating Light Control of Development. Cell 78, 117–124, doi:10.1016/0092-8674(94)90578-9 (1994).
Wei, N. et al. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8, 919–922, doi:10.1016/S0960-9822(07)00372-7 (1998).
Chamovitz, D. A. et al. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86, 115–121 (1996).
Sorin, C. et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant cell 17, 1343–1359 (2005).
Chen, J. et al. Inhibition of arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner. J. Plant Res. 126, 161–168, doi:10.1007/s10265-012-0509-3 (2013).
Chico, J. M. et al. Repression of Jasmonate-Dependent Defenses by Shade Involves Differential Regulation of Protein Stability of MYC Transcription Factors and Their JAZ Repressors in Arabidopsis. Plant cell 26, 1967–1980, doi:10.1105/tpc.114.125047 (2014).
Feng, S. H. et al. The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. Plant cell 15, 1083–1094, doi:10.1105/Tpc.010207 (2003).
Lozano-Duran, R. et al. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant cell 23, 1014–1032, doi:10.1105/tpc.110.080267 (2011).
Dohmann, E. M., Nill, C. & Schwechheimer, C. DELLA proteins restrain germination and elongation growth in Arabidopsis thaliana COP9 signalosome mutants. Eur. J. Cell Biol. 89, 163–168, doi:10.1016/j.ejcb.2009.12.001 (2010).
Wang, X. P. et al. The COP9 signalosome interacts with SCFUFO and participates in Arabidopsis flower development. Plant cell 15, 1071–1082, doi:10.1105/Tpc.009936 (2003).
Chen, H. et al. Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant cell 18, 1991–2004, doi:10.1105/tpc.106.043224 (2006).
Irigoyen, M. L. et al. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant cell 26, 712–728, doi:10.1105/tpc.113.122234 (2014).
Dohmann, E. M. et al. The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135, 2013–2022, doi:10.1242/dev.020743 (2008).
Grunewald, W. et al. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO reports 10, 923–928, doi:10.1038/embor.2009.103 (2009).
Hind, S. R. et al. The COP9 signalosome controls jasmonic acid synthesis and plant responses to herbivory and pathogens. Plant J 65, 480–491, doi:10.1111/j.1365-313X.2010.04437.x (2011).
Salisbury, F. J., Hall, A., Grierson, C. S. & Halliday, K. J. Phytochrome coordinates Arabidopsis shoot and root development. Plant J 50, 429–438, doi:10.1111/j.1365-313X.2007.03059.x (2007).
Vitha, S., Zhao, L. & Sack, F. D. Interaction of root gravitropism and phototropism in Arabidopsis wild-type and starchless mutants. Plant Phys. 122, 453–462 (2000).
Correll, M. J. et al. Phytochromes play a role in phototropism and gravitropism in Arabidopsis roots. Advances in space research: the official journal of the Committee on Space Research 31, 2203–2210 (2003).
Laxmi, A., Pan, J., Morsy, M. & Chen, R. Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PloS one 3, e1510, doi:10.1371/journal.pone.0001510 (2008).
Kircher, S. et al. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant cell 11, 1445–1456 (1999).
von Arnim, A. G. & Deng, X. W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045 (1994).
Ang, L. H. et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222 (1998).
Duek, P. D., Elmer, M. V., van Oosten, V. R. & Fankhauser, C. The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14, 2296–2301, doi:10.1016/j.cub.2004.12.026 (2004).
Seo, H. S. et al. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995–999, doi:10.1038/nature01696 (2003).
Santoni, V., Bellini, C. & Caboche, M. Use Of 2-Dimensional Protein-Pattern Analysis For The Characterization Of Arabidopsis thaliana Mutants. Planta 192, 557–566 (1994).
Abramoff, M. D., Magalhaes, P. J. & Ram, S. J. Image processing with ImageJ. Biophotonics International 11, 36–43 (2004).
Neff, M. M., Turk, E. & Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18, 613–615 (2002).
Rieu, I. & Powers, S. J. Real-time quantitative RT-PCR: design, calculations, and statistics. Plant cell 21, 1031–1033, doi:10.1105/tpc.109.066001 (2009).
Andersen, S. U. et al. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant cell 20, 88–100, doi:10.1105/tpc.107.054676 (2008).
Acknowledgements
The authors would like to thank Dr. Claus Schwechheimer (Technische Universität München TUM, School of Life Sciences, Weihenstephan/Munich, Germany) for providing the seeds of csn4-1 (SALK_043720), csn4-2 (SALK_053839) segregating lines, csn5a-1 (SALK_063436), csn5a-2 (SALK_027705) and csn5b-1 (SALK_007134); Dr. Johannes Stuttmann (Institute of Biology, Martin Luther University, Halle-Wittenberg, Germany) for providing the seeds of csn2-5; and Dr. William M. Gray (College of Biological Sciences, University of Minnesota, USA) for providing the seeds of csn1-10 and csn3-3. This work was supported by the Swedish Natural Sciences Research Council (VR), the Swedish Research Council for Research and Innovation for Sustainable Growth (VINNOVA), the Wallenberg foundation, the Carl Trygger foundation (CTS08: 298) to CB, and the Erasmus Programme to AMP.
Author information
Authors and Affiliations
Contributions
D.I.P., M.L.P., A.L., A.M.P. and A.R. performed the experiments; D.I.P. and C.B. designed the experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Change History: A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
An erratum to this article is available at https://doi.org/10.1038/s41598-017-04861-9.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pacurar, D.I., Pacurar, M.L., Lakehal, A. et al. The Arabidopsis Cop9 signalosome subunit 4 (CSN4) is involved in adventitious root formation. Sci Rep 7, 628 (2017). https://doi.org/10.1038/s41598-017-00744-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-00744-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.