Abstract
Myelin elaborated by oligodendrocytes (OLs) in the central nervous system (CNS) is required for saltatory conduction of action potentials along neuronal axons. We found that TMEFF2, a transmembrane protein with EGF-like and two follistatin-like domains, is selectively expressed in differentiating/myelinating OLs. Previous studies showed that TMEFF2 is capable of binding to PDGFA, which plays important roles in the proliferation, migration and differentiation of oligodendrocyte progenitor cells (OPCs). However, molecular and genetic analysis revealed that Tmeff2 is a weak binder of PDGFA, and not required for OL differentiation and myelin gene expression in vivo. Together, our data suggested that Tmeff2 is specifically upregulated in OLs, but dispensable for OL differentiation and maturation.
Similar content being viewed by others
Introduction
Myelin sheaths formed by OLs play crucial roles in the functioning of CNS, as they provide electrical insulation to promote conduction of axonal impulse1. Recent studies demonstrated that myelin sheaths provide energy and trophic support to the wrapped axons2,3,4. During embryonic development, OPCs are generated from restricted domains of neuroepithelium, such as the pMN domain in the spinal cords. Once generated, they are dispersed to occupy the entire CNS and continue to proliferate5,6,7. At later stages, OPCs especially those in the white matter regions begin to differentiate into mature OLs and form myelin sheaths8.
Previous studies have demonstrated that PDGFA and its receptor PDGFRA are required for OPC proliferation and prevent their differentiation at early stages9,10,11,12,13,14. The deficiency of PDGFRA signaling leads to premature OL differentiation, and causes severe hypomyelination at later stages due to the reduced number of OPCs12. On the contrary, forced expression of PDGFA in vivo induced hyperproliferation9, 14. Our recent studies have also suggested that Nkx2.2 regulates the timing of OL differentiation by inhibiting the transcription of Pdgfra 12, 15. However, the delay of OL differentiation is overcome in Nkx2.2 conditional mutants at later postnatal stages, suggesting the involvement of additional intracellular or extracellular factors in regulating PDGFA/PDGFRA pathway.
TMEFF2 (also known as tomoregulin and TENB2) is a transmembrane protein with EGF-like and two follistatin-like domains, but lacks an obvious functional intracellular domain16,17,18. A previous study reported that TMEFF2 specifically binds to PDGFA and suppresses fibroblast proliferation stimulated by PDGFA19. In this study, we found that Tmeff2 is specifically up-regulated in OLs at the onset of cell differentiation. Given the importance of PDGFA/PDGFRA signaling in OL development, the selective expression of Tmeff2 in differentiating OLs prompted us to hypothesize that TMEFF2 competitively binds to PDGFA and antagonizes PDGFRA signaling. However, the lack of apparent phenotypes in OL differentiation and myelin gene expression in Tmeff2 mutant mice suggested that Tmeff2 is dispensable for oligodendrocyte development.
Results
Tmeff2 is selectively expressed in differentiating OLs
To investigate whether Tmeff2 is expressed in the developing CNS, we first examined its expression at several developmental stages by in situ hybridization (ISH). It was found that Tmeff2 is primarily expressed in the gray matter of mouse spinal cords before embryonic day 18.5 (E18.5), but rapidly down-regulated after birth (Fig. 1). At the same time, Tmeff2 expression starts to be up-regulated at E18.5 in the white matter, reaches its peak from P7 to P15, and down-regulated thereafter (Fig. 1). Similarly, in the developing cortex, Tmeff2 transcription is first detected in cortical neurons at P7 (Fig. 2a), but later up-regulated in the white matter (corpus callosum) at P15 (Fig. 2b). In addition, Tmeff2 is also expressed in the white matter of cerebellum (Fig. 2c). Together, these data suggested that Tmeff2 is specifically up-regulated in cells of oligodendrocyte lineage during differentiation stage.
Tmeff2-expressing cells are immunoreactive to CC1 in different regions of the CNS. (a–c) Tmeff2 expression was detected in cortex at P7 (a), corpus callosum (b) and white matter of cerebellum (c) at P15. Bar, 250 μm. (d–g) Tmeff2 co-expresses with CC1 in spinal cords (d), cerebellum white matter (e) and corpus callosum (f,g) at P15. Bar, 50 μm.
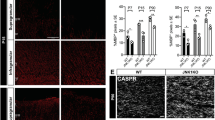
To confirm the expression of Tmeff2 in differentiating OLs, we carried out double staining with Tmeff2 ISH followed by immunohistochemistry against CC1, a specific marker for differentiating/differentiated OLs20. As expected, all Tmeff2+ cells in P15 spinal cord, corpus callosum and cerebellum are CC1+ (Fig. 2d–g). In addition, we examined Tmeff2 expression in Nkx2.2 and Myrf conditional knock-out (CKO) mice in which OL differentiation is significantly delayed and impaired12, 21, 22. The results showed that the numbers of Tmeff2+ cells in the white matter are markedly reduced in Nk2.2-CKO mutants and completely lost in Myrf-CKO mutants (Fig. 3). Taken together, these results indicated that Tmeff2 is expressed in differentiating or newly differentiated OLs in all regions of CNS.
Disruption of Tmeff2 in mice has no effect on OL differentiation
Since TMEFF2 is initially expressed in gray matter at embryonic stages (Fig. 1) and binds to PDGFA, deletion of Tmeff2 may affect the proliferation and migration of OPCs by affecting the availability of PDGFA. This possibility was investigated by examining the expression of three OPC markers, Pdgfra, Sox10 and OLIG2, in early postnatal spinal cords of Tmeff2-KO mutant mice. The phenotypes of mutants were confirmed by their smaller body size and lethality around weaning age23 (Fig. 4c), and the lack of Tmeff2 mRNA transcription in the spinal cord tissue (Fig. 4e). Unexpectedly, there is no significant difference in the number of Pdgfra+ OPCs between wild type and mutant mice at P4 and P8 stages (n = 3, p > 0.05). Similarly, the numbers of OLIG2+ and Sox10+ cells in P4 and P8 mutants are similar to those in control tissues (n = 3, p > 0.05) (Fig. 5, and Table S1). These results indicated that Tmeff2 is not necessary for OPC proliferation during early OL development.
Tmeff2 gene and Tmeff2-KO mice. (a) Structure of TMEFF2. TMEFF2 has a signal peptide (SP), two follistatin-like domains (FS I, FS II), an EGF-like domain (EGF), a shedding domain (S) and a transmembrane domain (TM). (b) A schematic of Tmeff2-KO mice. A human PLAP cassette was inserted after the start codon in the first exon. (c) The Tmeff2-KO mutant mice are smaller than their wild type littermates. (d) Genotyping results of Tmeff2-KO mice. (e) Tmeff2 mRNA transcription could not be detected by ISH in mutant mice.
The number of OPCs is not affected in Tmeff2-KO mutants. (a–f) Expression of Pdgfra (a,b), Sox10 (c,d) and OLIG2 (e,f) as detected by ISH or immunofluorescence in wild type and mutant spinal cords at P4. Bar, 100 μm. (g,h) Statistical analysis of OLIG2+, Sox10+ and Pdgfra+ OPCs in P4 (g) and P8 (h) spinal cords. n = 3, p > 0.05.
Differentiation of OLs in Tmeff2-KO mutant spinal cords was examined by ISH with two mature OL markers, Mbp and Plp. To our surprise, expression of both Mbp and Plp is normal in Tmeff2-KO mutants at various postnatal stages (Fig. 6). As Plp mRNA is mainly restricted to the cell bodies and can be visualized individually, the number of Plp+ cells per section is counted and there is no discernible difference between control and mutant tissues (Fig. 6k and Table S2). Expression of MBP and MAG myelin proteins as detected by immunofluorescence is also similar in both genotypes (Fig. 6). These results showed that Tmeff2 is not essential for OL differentiation in the spinal cord.
Normal differentiation of OLs in Tmeff2-KO mutant spinal cords. (a–p) Spinal cord sections of wild type and Tmeff2-KO mice were subjected to ISH with Mbp (a–h) and Plp (i–p) riboprobe at different stages. (q–t) Immunofluorescent staining with anti-MBP (q,r) and anti-MAG (s–t) in P8 wild type (q,s) and mutant (r,t) spinal tissues. Bar, 200 μm. (k) Statistical analysis of the number of Plp+ OLs in spinal cords from P4 to P21 stages. n = 3, p > 0.05.
In addition, we examined OL differentiation and myelin gene expression in mutant forebrains. As shown in Fig. 7, the timing of the onset of Mbp and Plp expression is not affected in the mutants, and their subsequent expression is also normal from P4 to P21 stages. Together, all these results indicated that Tmeff2 is not required for the terminal differentiation of OLs in the entire CNS.
TMEFF2 is a weak PDGFA-binding protein
Previous studies suggested that TMEFF2 regulates PDGF signaling in cancer cells by binding to PDGFA with its extracellular domain19. However, disruption of Tmeff2 does not cause any alterations in OPC proliferation and differentiation (Figs 5–7), raising the question whether TMEFF2 can effectively compete with PDGFRA for binding to PDGFA.
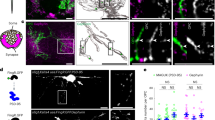
To address this question, we carried out in vitro protein binding assays by co-immunoprecipitation (co-IP) experiments. FLAG-tagged TMEFF2 and HA-tagged PDGFA were co-expressed in HEK293T cells, and FLAG-tagged PDGFRA served as a positive control. The co-IP results showed that TMEFF2 binds to PDGFA in vitro weakly, as compared to the strong binding between PDGFA and its natural receptor PDGFRA (Fig. 8a). The effects of PDGFA on PDGFRA and TMEFF2 signaling were also analyzed by the activation of pERK1/2. When 10 ng/mL PDGFA is added into the media, ERK1/2 is activated rapidly and continuously in PDGFRA overexpressing cells, but not in TMEFF2 overexpressing cells (Fig. 8b). It was previously reported that TMEFF2 acts as a receptor or co-receptor for several growth factors including PDGFA and EGF, and promotes ERK phosphorylation in vitro 24. Consistent with this finding, the level of pERK1/2 is slightly up-regulated in TMEFF2 overexpression cells, and is further increased in TMEFF2/PDGFRA co-expressing cells than in PDGFRA overexpressing cells (Fig. 8b).
TMEFF2 is a weak PDGFA-binding protein. (a) Co-IP showed that the affinity between PDGFA and TMEFF2 is lower than that between PDGFA and its natural receptor PDGFRA. (b) TMEFF2 has a weaker effect in stimulating ERK1/2 phosphorylation, compared to PDGFRA, in DMEM media containing 10% FBS or 10 ng/mL PDGFA.
Taken together, these data showed that TMEFF2 is a weak receptor/co-receptor for PDGFA-binding and ERK1/2 activation, and does not appear to function as a competitive inhibitor of PDGFRA. In this case, the lack of detectable phenotypic changes in OL differentiation in Tmeff2-KO mice may be due to the small effects of TMEFF2 on PDGFA binding and downstream signaling. Thus, the in vivo function of TMEFF2 in organ development and animal survival remains unknown.
Discussion
Oligodendrocytes arise from specific regions of neural epithelium and subsequently migrate to the entire CNS before they differentiate and form myelin around neuronal axons. These progressive developmental events are precisely regulated by a variety of intracellular and extracellular factors25,26,27,28,29. PDGFA is a well-known extracellular factor that plays a critical role in OL development30, 31. The interaction between PDGFA and its receptor PDGFRA expressed in OPCs stimulates OPC proliferation and represses their differentiation until a proper cell number is achieved9, 10. Immediately before differentiation, Pdgfra is down-regulated to allow the up-regulation or activation of other critical factors, such as Sox10 and Myrf, that promote terminal differentiation of OLs21, 32. Here, we report that TMEFF2, a type I transmembrane protein with short intracellular domain, is highly expressed by differentiating OLs (Figs 1–3). Based on the previous finding of TMEFF2 binding to PDGFA19 (Fig. 4), we initially hypothesized that TMEFF2 helps to promote and solidify OL differentiation by interfering with PDGFA/PDGFRA signaling. To our surprise, Tmeff2-KO mice do not display apparent defects in OL proliferation and differentiation (Figs 5–7). These findings have raised the possibility that TMEFF2 may not bind to PDGFA effectively and therefore does not regulate PDGFRA signaling during oligodendroglial development. Consistent with this notion, co-IP experiments showed that TMEFF2 has a much weaker binding to PDGFA than PDGFRA, and addition of PDGFA to TMEFF2 overexpressing cells only causes a weak activation of ERK1/2 (Fig. 8). These results implied that TMEFF2 may function as a low-affinity receptor/co-receptor for PDGFA, and does not appear to function as a strong inhibitor or activator of PDGFA/PDGFRA signaling under those conditions.
Although our data showed that Tmeff2 is dispensable for OL development in vivo, it is not clear whether Tmeff2 could promote OL differentiation and myelin repair after injury-induced demyelination. Considering that TMEFF2 stimulates ERK1/2 phosphorylation (Fig. 8b) and MAPK-ERK1/2 activation promotes OL differentiation and axonal myelination33,34,35,36, it remains plausible that Tmeff2 can enhance OL differentiation during development and remyelination in a redundant manner. In addition, it is possible that TMEFF2 plays an unknown function in oligodendrocyte differentiation and myelin formation. Since Tmeff2-KO mice die before adulthood, generation of conditional mutants are necessary for dissecting out the in vivo function of Tmeff2 in myelin development and repair in the future.
Methods
Immunohistochemistry and ISH
Tissues were fix in 4% PFA overnight at 4 °C, and transferred into 20% sucrose in PBS overnight at 4 °C. Tissues were then embedded in OCT and sectioned into 14 μm on a cryostat. Experimental procedures for immunohistochemistry and ISH were described previously37. For Tmeff2 probes, 750 bp fragments corresponding to 900 nt–1650 nt of mouse Tmeff2 mRNA (NCBI Reference Sequence: NM_019790.4) were cloned into pT7T3D-PacI vectors for transcription in vitro. Antibodies were used as follows: anti-OLIG2 (Millipore, AB9610, 1:1000), anti-CC1 (Abcam, ab16794, 1:500), anti-MAG (Millipore, MAB1567, 1:500), anti-MBP (Abcam, ab7349, 1:500).
Tmeff2-KO mice
All animal experiments were performed in accordance with the institutional guidelines drafted by the Laboratory Animal Center, Hangzhou Normal University, and were approved by the Animal Ethics Committee of Hangzhou Normal University, China. The generation and phenotypes of Tmeff2-KO mice were previously described by TR Chen et al.23. The primers for genotyping are as follows: Tmeff2-F (5′-TCATGCTCTCCTTTGGTCGCAG-3′), Tmeff2-WT-R (5′-AAACATCTATGGTTCCCCACACC-3′) and Tmeff2-KO-R (5′-GAGCCTCATTACCTGGGATGATG-3′). Wild-type allele resulted in a 537 bp band, while mutant allele produced a 272 bp band. Myrf flox mice were obtained from Jackson Laboratory (Stock Number: 010607). Other strains such as Nkx2.2 flox mice38, Olig1-Cre mice39 and Cnp-Cre mice40 were described previously.
Construction of expression vectors and Western blot
For TMEFF2, PDGFRA and PDGFA over-expression in vitro, ORFs of mouse Tmeff2 (NM_019790.4), Pdgfra (NM_001083316.2) and Pdgfa (NM_008808.3) were cloned into the pCDH-MCS-EF1-CopGFP vectors, with FLAG, MYC and HA tagged at the C-terminal. HEK293T cells in 12-well plates were transfected with these vectors and cultured in 10% FBS/DMEM or free DMEM for 24 hours. Cells in free DMEM media were treated with 10 ng/mL PDGFA for 10 min or 4 hours before lysed. All cells were then lysed in 300 μL lysis buffer containing 25 mM pH 7.4 Tris, 150 mM NaCl, 1% NP40, 0.25% sodium deoxycholate, 1 mM EDTA, protease and phosphatase inhibitors. 20 μL samples were used for western blot. Antibodies for western blot were used as follows: anti-FLAG (Sigma, F7425, 1:10000), anti-MYC (CST, 2276S, 1:10000), anti-GAPDH (Epitomics, 2251–1, 1:2000), anti-ERK1/2 (Epitomics, 1171–1, 1:2000), anti-pERK1/2 (Epitomics, 2219–1, 1:2000).
Co-immunoprecipitation
Empty vectors and TMEFF2-FLAG, PDGFRA-FLAG expressing vectors were co-transfected with PDGFA-HA expressing vectors into HEK293T cells. 48 hours after transfection, cells in per 3.5 cm dish were treated with 1 ml lysis buffer containing 25 mM pH 7.4 Tris, 150 mM NaCl, 1% NP40, 0.25% sodium deoxycholate, 1 mM EDTA. 0.2 mL cell lysate was used for input, and 30 μL anti-FLAG Magnetic Beads (Sigma, Catalog Number M8823) were added into the left samples and incubated at 4 °C overnight. Beads were then washed with lysis buffer, and the FLAG-tagged proteins were eluted with 120 μL lysis buffer containing 10 μg 3xFlag peptide. Both input and IP samples were subjected to western bolt with anti-FLAG (Sigma, F1804, 1:10000), anti-HA (Abcam, ab9110, 1:10000).
Statistical analysis
The experimental data were analyzed using a two-tailed unpaired Student's t-test. Statistical significance was considered to be at p < 0.05, and n = 3.
References
Tasaki, I. Conduction of impulses in the myelinated nerve fiber. Cold Spring Harb Symp Quant Biol 17, 37–41 (1952).
Funfschilling, U. et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521 (2012).
Lee, Y. et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 487, 443–448 (2012).
Barres, B. A. & Barde, Y. Neuronal and glial cell biology. Curr Opin Neurobiol 10, 642–648 (2000).
Rowitch, D. H. Glial specification in the vertebrate neural tube. Nat Rev Neurosci 5, 409–419 (2004).
Richardson, W. D., Kessaris, N. & Pringle, N. Oligodendrocyte wars. Nat Rev Neurosci 7, 11–18 (2006).
Rowitch, D. H. & Kriegstein, A. R. Developmental genetics of vertebrate glial-cell specification. Nature 468, 214–222 (2010).
Baumann, N. & Pham-Dinh, D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiological reviews 81, 871–927 (2001).
Calver, A. R. et al. Oligodendrocyte population dynamics and the role of PDGF in vivo. Neuron 20, 869–882 (1998).
Fruttiger, M. et al. Defective oligodendrocyte development and severe hypomyelination in PDGF-A knockout mice. Development 126, 457–467 (1999).
Fu, H. et al. Dual origin of spinal oligodendrocyte progenitors and evidence for the cooperative role of Olig2 and Nkx2.2 in the control of oligodendrocyte differentiation. Development 129, 681–693 (2002).
Zhu, Q. et al. Genetic evidence that Nkx2.2 and Pdgfra are major determinants of the timing of oligodendrocyte differentiation in the developing CNS. Development 141, 548–555 (2014).
Pringle, N. P. & Richardson, W. D. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development 117, 525–533 (1993).
van Heyningen, P., Calver, A. R. & Richardson, W. D. Control of progenitor cell number by mitogen supply and demand. Curr Biol 11, 232–241 (2001).
Huang, H., Zhao, X. F., Zheng, K. & Qiu, M. Regulation of the timing of oligodendrocyte differentiation: mechanisms and perspectives. Neurosci Bull 29, 155–164 (2013).
Quayle, S. N. & Sadar, M. D. A truncated isoform of TMEFF2 encodes a secreted protein in prostate cancer cells. Genomics 87, 633–637 (2006).
Uchida, T. et al. A novel epidermal growth factor-like molecule containing two follistatin modules stimulates tyrosine phosphorylation of erbB-4 in MKN28 gastric cancer cells. Biochem Biophys Res Commun 266, 593–602 (1999).
Horie, M. et al. Identification and characterization of TMEFF2, a novel survival factor for hippocampal and mesencephalic neurons. Genomics 67, 146–152 (2000).
Lin, K. et al. TMEFF2 is a PDGF-AA binding protein with methylation-associated gene silencing in multiple cancer types including glioma. PLoS One 6, e18608 (2011).
Redwine, J. M. & Evans, C. F. Markers of central nervous system glia and neurons in vivo during normal and pathological conditions. Current topics in microbiology and immunology 265, 119–140 (2002).
Emery, B. et al. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138, 172–185 (2009).
Koenning, M. et al. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci 32, 12528–12542 (2012).
Chen, T. R. et al. Generation and characterization of Tmeff2 mutant mice. Biochem Biophys Res Commun 425, 189–194 (2012).
Chen, X. & Ruiz-Echevarria, M. J. TMEFF2 modulates the AKT and ERK signaling pathways. International journal of biochemistry and molecular biology 4, 83–94 (2013).
Hornig, J. et al. The transcription factors Sox10 and Myrf define an essential regulatory network module in differentiating oligodendrocytes. PLoS Genet 9, e1003907 (2013).
Qi, Y. et al. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development 128, 2723–2733 (2001).
Dai, Z. M. et al. Stage-specific regulation of oligodendrocyte development by Wnt/beta-catenin signaling. J Neurosci 34, 8467–8473 (2014).
Zhao, C. et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nature communications 7, 10883 (2016).
Zhu, Y. et al. Control of oligodendrocyte generation and proliferation by Shp2 protein tyrosine phosphatase. Glia 58, 1407–1414 (2010).
Hall, A., Giese, N. A. & Richardson, W. D. Spinal cord oligodendrocytes develop from ventrally derived progenitor cells that express PDGF alpha-receptors. Development 122, 4085–4094 (1996).
Pringle, N. P., Mudhar, H. S., Collarini, E. J. & Richardson, W. D. PDGF receptors in the rat CNS: during late neurogenesis, PDGF alpha-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development 115, 535–551 (1992).
Stolt, C. C. et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16, 165–170 (2002).
Fyffe-Maricich, S. L., Karlo, J. C., Landreth, G. E. & Miller, R. H. The ERK2 mitogen-activated protein kinase regulates the timing of oligodendrocyte differentiation. J Neurosci 31, 843–850 (2011).
Ishii, A., Furusho, M., Dupree, J. L. & Bansal, R. Role of ERK1/2 MAPK signaling in the maintenance of myelin and axonal integrity in the adult CNS. J Neurosci 34, 16031–16045 (2014).
Ishii, A., Furusho, M., Dupree, J. L. & Bansal, R. Strength of ERK1/2 MAPK Activation Determines Its Effect on Myelin and Axonal Integrity in the Adult CNS. J Neurosci 36, 6471–6487 (2016).
Ishii, A., Furusho, M. & Bansal, R. Sustained activation of ERK1/2 MAPK in oligodendrocytes and schwann cells enhances myelin growth and stimulates oligodendrocyte progenitor expansion. J Neurosci 33, 175–186 (2013).
Zhu, Y. et al. Necl-4/SynCAM-4 is expressed in myelinating oligodendrocytes but not required for axonal myelination. PLoS One 8, e64264 (2013).
Mastracci, T. L., Lin, C. S. & Sussel, L. Generation of mice encoding a conditional allele of Nkx2.2. Transgenic research 22, 965–972 (2013).
Lu, Q. R. et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86 (2002).
Lappe-Siefke, C. et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet 33, 366–374 (2003).
Acknowledgements
This work was supported by National Key Basic Research Program of China (2013CB531300), the National Natural Science Foundation of China (Grant No. 31101642, 31372150, 31471955). We are grateful to Fan Wang (Department of Cell Biology, Duke University Medical Center) for providing Tmeff2-KO mice.
Author information
Authors and Affiliations
Contributions
H.H., X.Z. and M.Q. conceived and designed the experiments; H.H., T.P., R.M., A.Y. and X.Z. performed the experiments and statistically analyzed the data, H.H. and M.Q. wrote the manuscript and Z.Z. revised it critically.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Huang, H., Teng, P., Mei, R. et al. Tmeff2 is expressed in differentiating oligodendrocytes but dispensable for their differentiation in vivo . Sci Rep 7, 337 (2017). https://doi.org/10.1038/s41598-017-00407-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-00407-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.