Abstract
Computed Tomography Coronary Angiography (CTCA) is a non-invasive method to evaluate coronary artery anatomy and disease. CTCA is ideal for geometry reconstruction to create virtual models of coronary arteries. To our knowledge there is no public dataset that includes centrelines and segmentation of the full coronary tree. We provide anonymized CTCA images, voxel-wise annotations and associated data in the form of centrelines, calcification scores and meshes of the coronary lumen in 20 normal and 20 diseased cases. Images were obtained along with patient information with informed, written consent as part of the Coronary Atlas. Cases were classified as normal (zero calcium score with no signs of stenosis) or diseased (confirmed coronary artery disease). Manual voxel-wise segmentations by three experts were combined using majority voting to generate the final annotations. Provided data can be used for a variety of research purposes, such as 3D printing patient-specific models, development and validation of segmentation algorithms, education and training of medical personnel and in-silico analyses such as testing of medical devices.
Similar content being viewed by others
Background & Summary
Coronary artery disease is a leading cause of death worldwide1, causing a large body of research to focus on the understanding of coronary anatomy and blood flow, disease progression and treatment options2,3,4. With rapid advancements in computation, additive manufacturing and other technologies capable of taking advantage of virtual organ models, computational models of coronary arteries have been increasingly used in research, including the designing and testing of medical devices, as well as for education and training purposes5.
While different modalities can be used to image coronary arteries, only Computed Tomography Coronary Angiography (CTCA) is non-invasive and has sufficient sub-millimetre resolution to allow reconstruction of the small coronary arteries. Therefore it is commonly used and as a result ideal as underlying modality for subsequent image segmentation and virtual coronary artery reconstruction. This commonly required manual refinement after initial automatic threshold due to the small scale, lack of clear contrast with the surrounding tissue and common image artefacts, especially for calcified lesions. Segmentation of the full coronary tree is particularly difficult as even with highest resolution CTCA machines today, distal vessels are only captured via a few image pixels. As a result, despite a wealth of CTCA data available to date, there are extremely few virtual coronary models publicly available and the use of reconstruction workflows on a large-scale patient-specific basis is cost and time intensive.
Traditional segmentation methods are extremely time consuming6, generally requiring semi-automated segmentation closely supervised by a human expert to guide the algorithm and correct errors. Additionally, the segmentations produced are highly sensitive to the individual expert and hence consistent segmentation between different experts is difficult. This has led to no public datasets currently available for use in applications that require accurate patient-specific coronary models. Related datasets are limited, including the ‘Visible Heart Project’, which, however focuses on educational images and videos using magnetic resonance imaging. Although access may be provided to limited CTCA images, these are without annotations or reconstructed models7. Also available is the ‘The Rotterdam dataset’8,9, which is primary public dataset, but focused on stenosis detection and stenosis evaluation with sub-voxel accuracy. This dataset may only be used for its stated purpose of stenosis detection and lumen segmentation, and is also no longer publicly available from the challenge website (https://coronary.bigr.nl/).
To overcome the problems with these traditional segmentation methods, we created high quality segmentations of the coronary arteries, to serve as both a benchmark dataset for newly developed segmentation methods and pre-existing segmentation for further processing, for example investigating differences in helicity between stented idealized and patient-specific vessels10. This was part of the ‘Automated Segmentation of Coronary Arteries’ (ASOCA) Challenge11,12 we facilitated during the Medical Image Computing and Computer Assisted Intervention (MICCAI) 2020 conference to focus on the development of automated segmentation algorithms using this data, providing a convenient system for submission of results and automated evaluation and ranking.
The coronary artery CTCA images were available to us through the Coronary Atlas13, an ongoing collection of CTCA images and associated clinical and demographics data used to investigate differences in coronary anatomy14 and haemodynamic behaviour between patients15,16,17. A set of 40 patient-specific coronary artery tree data is provided here, including anonymized CTCA images in .nrdd format, combined high-quality manual voxel annotations derived from 3 experts, and other associated data such as centrelines, smoothed meshes in .stl format and calcification scores. These served as the training dataset for the ASOCA challenge. Our dataset is the only public dataset of annotations and associated data of the full coronary tree in 20 normal and 20 disease cases. Additionally, a separate set of 20 CTCA images (the test set images for the ASOCA challenge) is provided primarily to facilitate participating in the challenge. In order to not compromise the integrity of the challenge, no other information is provided with these images. Researchers can participate on the challenge website (asoca.grand-challenge.org), using the training data to develop segmentation algorithms and submit results to the challenge website for automatic evaluation and scoring.
In summary, the current dataset has several advantages over previously available coronary artery datasets. While our dataset is based solely on CTCA and can not provide sub-voxel segmentation and stenosis identification as accurate as the Rotterdam dataset, we do however provide high quality segmentation of all coronary vessels visible in CTCA. In contrast our dataset is available to all researchers including commercial projects. Further, our inclusion of all arteries larger than 1 mm rather than selected vessel segments allows for expanded applications such as more complex simulations, and more comprehensive training and educational applications. The balanced set of normal and diseased patients ensures effects of disease can be independently studied, as well as ensuring that newly developed segmentation algorithms can robustly handle cases with disease. The dataset is sufficiently large and balanced for training machine learning models. Device manufacturers and researchers with an interest in cardiovascular modelling, prediction and treatment of coronary artery disease can analyse this data directly or combine it with other available datasets. The smooth surface meshes and centrelines can be directly used for computational modelling16, directly 3D printed for experiments18,19,20,21, assist in developing and testing medical devices such as stents22,23,24, and can be used for Virtual Reality applications for education and training25,26,27. Moreover, our dataset allows for the development and benchmarking of new segmentation algorithms aiming to efficiently annotate the coronary arteries automatically as per ASOCA challenge28.
Methods
Patient cohort
Forty patients were randomly selected from a retrospective dataset based on the calcification, stenosis and image quality reported by the cardiologist. Images must have acceptable quality as described by the cardiologist. The dataset was divided into twenty normal patients with no evidence of stenosis and non-obstructive disease, and twenty patients with evidence of calcium scores higher than 0 and obstructive disease. The calcification score in the diseased group ranged between 1 and 986 with a mean of 254. Obstructions in the diseased group ranged from 30% to 70% stenosis. Patients were included during routine procedures after written and informed consent and approval from University of New South Wales Human Research Ethics Committee (Ref. 022961).
Imaging
The CTCA imaging was undertaken using a multi-detector CT scanner (GE Lightspeed 64 multi-slice scanner, USA) using retrospective ECG gating. A contrast medium (Omnipaque 350) was used for imaging and the patient heart rate was controlled around 60bpm by administration of beta blockers. The end diastolic time step was saved for analysis and the images exported as DICOM files. Images were converted to Nearly Raw Raster Data (NRRD) format during the anonymization process and the intensity rescaled to Hounsfield units based on the appropriate DICOM tags.
Annotation
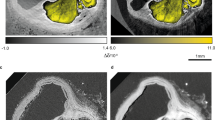
The open-source software 3D Slicer (version 4.3)29 was used to manually annotate the coronary arteries images. The annotation process was performed independently by three annotators, who were instructed to segment the left and right coronary trees starting at the aortic root. Thresholding at a cut-off chosen by the expert was used to generate an initial rough segmentation of the vessel, followed by manually correcting the vessel contours in each slice. All coronary vessels with a diameter larger 1 mm, representing 1–2 voxels, were included in the segmentation. In segments showing significant imaging artefacts affecting the vessel that would make further segmentation unfeasible, the rest of the vessel was ignored. A sample of the annotated CTCA images and the resulting 3D reconstructions are shown in Fig. 1. Figure 2 shows a diseased case with calcified plaque and stenosis.
Post processing
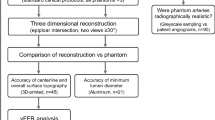
The annotations are combined to produce a final segmentation of the arteries by majority vote among the three annotations, i.e. including regions where at least two of the annotators agreed. Small vessels (<1 mm, i.e. 1–2 voxels) were removed if they were mistakenly included. The segmentations are available as voxel-wise annotations, as well as smoothed surface meshes. Surface meshes were produced from the annotations using the Flying Edges algorithm30. It should be noted that with voxel-wise labelling as used in this dataset rather than a tubular parametrization, further smoothing would be necessary to recover a smooth vessel shape. The annotations were smoothed using Taubin’s algorithm31, implemented in the open-source Vascular Modelling Tool Kit (VMTK, https://www.vmtk.org)32,33, with a passband of 0.03 and 30 iterations before being exported as an STL file. Taubin’s smoothing method is commonly used when processing vessel segmentations34 and is expected to preserve topology and volume of the vessels35. These settings correspond to the smoothing used in the Coronary Atlas to calculate shape parameters. The raw annotations provided can be used to produce surface meshes with different smoothing settings if needed. Vessel centrelines were extracted manually by marking the inlet and outlet points on the mesh for automated centreline calculation in VMTK, as shown in Fig. 3.
ASOCA test data set
An additional 10 normal and 10 diseased CTCA cases, separate to the 20 normal and 20 diseased used for the training data, were selected based on the same criteria to serve as the test set for the ASOCA challenge. These cases will be distributed alongside the annotated dataset to facilitate further participation in the challenge. Ground truth annotation and other associated data for these cases is not publicly available.
Data Records
The dataset is available on the UK Data Service (https://reshare.ukdataservice.ac.uk/855916/)36. Patients are labelled sequentially from 1 to 20, with normal and diseased patients labelled separately (i.e. Normal_1…Normal_20 represent the normal patients and Diseased_1…Diseased_20 represent diseased patients). CTCA scans are provided as Nearly Raw Raster Data (NRRD) file labelled sequentially based on patient name (Normal_1.nrrd, Normal_2.nrrd…). This naming convention is used for the rest of the data folders. The annotations folder contains the final annotation for each patient. This represents the voxel-wise annotations, with the background voxels assigned a value of 0 and the foreground (vessel lumen) assigned a value of 1. Both the CTCA images and annotations have anisotropic resolution, a common characteristic of most CT machines, with the z-axis resolution of 0.625 mm and the in-plane resolution ranging from 0.3 mm to 0.4 mm depending on the patient. The SurfaceMeshes directory contains smooth surface meshes generated from the voxel annotations. These meshes are provided in STL format, with an average of 37,000 vertices to capture the arterial geometry.
The centrelines folder contains centrelines of the coronary arteries for each patient, provided in VTK Poly Data (VTP) format that allows for efficient storage of centreline data. Figure 3 shows a sample of the extracted centreline and underlying surface mesh. The spreadsheet DiseaseReports.xlsx reports calcium score and stenoses levels for each patient.
Technical Validation
Dice Similarity Coefficient (DSC)37 is frequently used to measure the degree of overlap between annotations. DSC is defined as in Eq. 1 for two sets of voxels A and B. Similarly, Hausdorff Distance (HD) as shown in Eq. 2 measures the distance of corresponding points between annotations. In practice commonly the 95th percentile HD is used rather than the maximum in order to reduce sensitivity to outliers38.
We used DSC (Table 1) and 95th percentile HD (Table 2) to compare variability between annotators compared to the final ground truth generated for each case. The average Dice Score among the three annotators was 85.6% ± 7.7% (mean ± standard deviation) and an average HD of 5.92 ± 7.3 mm (mean ± standard deviation). The concordance between annotators was higher for normal cases compared to diseased (87.4% vs 83.9%, p = 0.01 using Welch’s t test39), due presence of stenosis and calcified plaques complicating the annotation of diseased images. Hausdorff Distance showed similar results (4.45 mm in normal cases vs 7.38 mm in diseased, p = 0.028). A Dice Score of 1 (indicating perfect agreement) is difficult to achieve, as this dataset attempts to segment the full coronary artery tree including small arteries near the limit of CTCA imaging resolution. This Dice Score and Hausdorff Distance indicates high agreement between the annotators and is unlikely to adversely affect usage of this dataset. Table 3 shows the Hausdorff Distance between centre of the voxel labels and the smoothed mesh.
Usage Notes
These recommendations focus on free, open-source software, however as the dataset is provided commonly used formats commercially available software suites will can also be utilised. CTCA and ground-truth data is provided in NRRD format, compatible with all common medical imaging software such as 3D Slicer29 and ITK-SNAP40. 3D Slicer is the recommended software for working with this data, providing tools for common editing operations and various add-ons for specialised tasks. The centrelines are saved in VTK Poly Data (VTP) format, expected to be used with the Visualization Toolkit (VTK)41 and the Vascular Modelling Toolkit32,33. VMTK is also available as a 3D Slicer add-on. Surface meshes are provided in Standard Tessellation Language (STL), compatible with most mesh software. Both 3D Slicer and VMTK allow editing and processing STL meshes, including addition of flow extensions and generation of volume meshes for computational fluid dynamics simulations. Specific mesh editing software such as Meshlab42 can be used for more complex tasks. The dataset can be also be used to develop new segmentation algorithms and evaluate the performance on the standardised ASOCA challenge. Submission instructions are available on the challenge website (https://asoca.grand-challenge.org/SubmittingResults/).
The dataset can be used for research and commercial purposes. Researchers request access on the UK Data Service36 for access and provide evidence of ethics review and approval, or waiver regarding their project.
Code availability
The code for creation of this dataset, usage examples and evaluation code used in the challenge is available on GitHub (https://github.com/Ramtingh/ASOCADataDescription). Figures 1–3 were created with data included in the dataset. A copy of the raw data used is included in the repository under the corresponding folder to maker recreating these figures easier. 3D Slicer (version 4.3) was used in the preparation of the dataset and Figs. 1 and 2. Vascular Modelling Tool Kit (version 1.4) was used to calculate centerlines and generate Fig. 3.
References
World Health Organization. The atlas of heart disease and stroke. World Health Organization (2012).
Garca-Garca, H. M. et al. Computed tomography in total coronary occlusions (CTTO registry): radiation exposure and predictors of successful percutaneous intervention. EuroIntervention: journal of EuroPCR in collaboration with the Working Group on Interventional Cardiology of the European Society of Cardiology 4, 607–616 (2009).
Goodacre, S. et al. Systematic review, meta-analysis and economic modelling of diagnostic strategies for suspected acute coronary syndrome. Health Technol Assess 17, 1–188 (2013).
van den Boogert, T. et al. CTCA for detection of significant coronary artery disease in routine TAVI work-up. Netherlands Heart Journal 26, 591–599 (2018).
Li, Q. et al. An human-computer interactive augmented reality system for coronary artery diagnosis planning and training. J. Med. Syst. 41, 1–11 (2017).
Moccia, S., De Momi, E., El Hadji, S. & Mattos, L. S. Blood vessel segmentation algorithms–review of methods, datasets and evaluation metrics. Computer Methods and Programs in Biomedicine 158, 71–91 (2018).
Iaizzo, P. A. The visible heart project and free-access website ‘atlas of human cardiac anatomy’. EP Europace 18, iv163–iv172 (2016).
Schaap, M. et al. Standardized evaluation methodology and reference database for evaluating coronary artery centerline extraction algorithms. Med. Image Anal. 13, 701–714 (2009).
Kirişli, H. et al. Standardized evaluation framework for evaluating coronary artery stenosis detection, stenosis quantification and lumen segmentation algorithms in computed tomography angiography. Med. Image Anal. 17, 859–876 (2013).
Shen, C. et al. Secondary flow in bifurcations–important effects of curvature, bifurcation angle and stents. J. Biomech. 129, 110755 (2021).
Gharleghi, R. et al. Automated segmentation of normal and diseased coronary arteries - the ASOCA challenge. Computerized Medical Imaging and Graphics (2022).
Gharleghi, R., Samarasinghe, G., Sowmya, A. & Beier, S. Automated segmentation of coronary arteries https://doi.org/10.5281/zenodo.3819799 (2020).
Medrano-Gracia, P. et al. Construction of a coronary artery atlas from CT angiography. In Med. Image Comput. Comput. Assist. Interv., 513–520 (Springer, 2014).
Medrano-Gracia, P. et al. A study of coronary bifurcation shape in a normal population. J. Cardiovasc. Transl. Res. 10, 82–90 (2017).
Medrano-Gracia, P. et al. A computational atlas of normal coronary artery anatomy. EuroIntervention: journal of EuroPCR in collaboration with the Working Group on Interventional Cardiology of the European Society of Cardiology 12, 845–854 (2016).
Beier, S. et al. Impact of bifurcation angle and other anatomical characteristics on blood flow–a computational study of non-stented and stented coronary arteries. J. Biomech. 49, 1570–1582 (2016).
Beier, S. et al. Vascular hemodynamics with computational modeling and experimental studies. In Computing and Visualization for Intravascular Imaging and Computer-Assisted Stenting, 227–251 (Elsevier, 2017).
Gharleghi, R. et al. 3d printing for cardiovascular applications: From end-to-end processes to emerging developments. Ann. Biomed. Eng. 1–21 (2021).
Wang, K. et al. Dual-material 3d printed metamaterials with tunable mechanical properties for patient-specific tissue-mimicking phantoms. Addit. Manuf. 12, 31–37 (2016).
Beier, S. et al. Dynamically scaled phantom phase contrast MRI compared to true-scale computational modeling of coronary artery flow. J. Magn. Reson. Imaging 44, 983–992 (2016).
Yoo, S.-J., Spray, T., Austin, E. H. III, Yun, T.-J. & van Arsdell, G. S. Hands-on surgical training of congenital heart surgery using 3-dimensional print models. J. Thorac. Cardiovasc. 153, 1530–1540 (2017).
Antoine, E. E., Cornat, F. P. & Barakat, A. I. The stentable in vitro artery: an instrumented platform for endovascular device development and optimization. J. R. Soc. Interface 13, 20160834 (2016).
Zhong, L. et al. Application of patient-specific computational fluid dynamics in coronary and intra-cardiac flow simulations: Challenges and opportunities. Front. Physiol. 9, 742 (2018).
Sun, Z. & Jansen, S. Personalized 3D printed coronary models in coronary stenting. Quant. Imaging Med. Surg. 9, 1356 (2019).
Reinhard Friedl, M. Virtual reality and 3D visualizations in heart surgery education. In Heart Surg. Forum, 03054 (2001).
Dugas, C. M. & Schussler, J. M. Advanced technology in interventional cardiology: a roadmap for the future of precision coronary interventions. Trends Cardiovasc. Med. 26, 466–473 (2016).
Silva, J. N., Southworth, M., Raptis, C. & Silva, J. Emerging applications of virtual reality in cardiovascular medicine. JACC: Basic Transl. Sci. 3, 420–430 (2018).
Gharleghi, R. Ramtingh/ASOCA_MICCAI2020_Evaluation: MICCAI Evaluation. Zenodo https://doi.org/10.5281/zenodo.4460628 (2021).
Fedorov, A. et al. 3D slicer as an image computing platform for the quantitative imaging network. J. Magn. Reson. Imaging 30, 1323–1341 (2012).
Schroeder, W., Maynard, R. & Geveci, B. Flying edges: A high-performance scalable isocontouring algorithm. In IEEE 5th Symposium on Large Data Analysis and Visualization, 33–40 (IEEE, 2015).
Taubin, G., Zhang, T. & Golub, G. Optimal surface smoothing as filter design. In Comput. Vis. ECCV, 283–292 (Springer, 1996).
Antiga, L. & Steinman, D. A. Robust and objective decomposition and mapping of bifurcating vessels. IEEE Trans. Med. Imaging 23, 704–713 (2004).
Izzo, R., Steinman, D., Manini, S. & Antiga, L. The vascular modeling toolkit: a python library for the analysis of tubular structures in medical images. J. Open Source Softw. 3, 745 (2018).
Shum, J., Xu, A., Chatnuntawech, I. & Finol, E. A. A framework for the automatic generation of surface topologies for abdominal aortic aneurysm models. Ann. Biomed. Eng. 39, 249–259 (2011).
Antiga, L. et al. An image-based modeling framework for patient-specific computational hemodynamics. Med. Biol. Eng. Comput. 46, 1097–1112 (2008).
Gharleghi, R. et al. Computed tomography coronary angiogram images, annotations and associated data of normal and diseased arteries. Colchester, Essex: UK Data Service. https://doi.org/10.5255/UKDA-SN-855916 (2022).
Bertels, J. et al. Optimizing the dice score and jaccard index for medical image segmentation: Theory and practice. In Med. Image Comput. Comput. Assist. Interv., 92–100 (Springer, 2019).
Taha, A. A. & Hanbury, A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med. Imaging 15, 1–28 (2015).
Derrick, B., Toher, D. & White, P. Why welch’s test is type I error robust. Quant. Meth. Psych 12 (2016).
Yushkevich, P. A. et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128 (2006).
Schroeder, W., Martin, K. & Lorensen, B. The Visualization Toolkit–An Object-Oriented Approach To 3D Graphics, fourth edn (Kitware Inc., 2006).
Cignoni, P. et al. Meshlab: an open-source mesh processing tool. In Eurographics Italian chapter conference, vol. 2008, 129–136 (Salerno, Italy, 2008).
Acknowledgements
The authors would like to thank Jane Liggins and Miriam Hayward, Intra Imaging for assisting with the collection of this data. SB would like to acknowledge the Auckland Academic Health Alliance (AAHA) and the Auckland Medical Research Foundation (AMRF) their financial support and endorsement. This research was undertaken with the assistance of resources from the National Computational Infrastructure (NCI Australia), an NCRIS enabled capability supported by the Australian Government.
This research was conducted with approval from the University of New South Wales Human Research Ethics Committee (Ref. HC190145) and University of Auckland Human Participants Ethics Committee (Ref. 022961).
Author information
Authors and Affiliations
Contributions
R.G., D.A. and K.E. have contributed to annotation of the ground truth data and collation of the dataset in consultation with A.S. and S.O. R.G. has analysed the results. M.W., C.E. and S.B. have established the Coronary Atlas which provided the data for this study. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gharleghi, R., Adikari, D., Ellenberger, K. et al. Annotated computed tomography coronary angiogram images and associated data of normal and diseased arteries. Sci Data 10, 128 (2023). https://doi.org/10.1038/s41597-023-02016-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02016-2
This article is cited by
-
Mining multi-center heterogeneous medical data with distributed synthetic learning
Nature Communications (2023)