Abstract
In contrast to genotypic sex determination (GSD), temperature-dependent sex determination (TSD) in amniotic vertebrates eludes intuitive connections to Fisherian sex-ratio theory. Attempts to draw such connections have driven over 50 years of research on the evolution of sex-determining mechanisms (SDM), perhaps most prominently among species in the order Testudines. Despite regular advancements in our understanding of this topic, no efforts have been published compiling the entirety of data on the relationships between incubation temperature and offspring sex in any taxonomic group. Here, we present the Reptilian Offspring Sex and Incubation Environment (ROSIE) database, a comprehensive set of over 7,000 individual measurements of offspring sex ratios in the order Testudines as well as SDM classifications for 149 species. As the name suggests, we plan to expand the taxonomic coverage of ROSIE to include all non-avian reptiles and will regularly release updates to maintain its comprehensive nature. This resource will enable crucial future research probing the ecology and evolution of SDM, including the presumed sensitivity of TSD to rapid environmental change.
Measurement(s) | sex-determining mechanism • offspring sex ratio • sex determination • incubation environment |
Technology Type(s) | systematic review study design |
Sample Characteristic - Organism | Testudines • Reptile |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.16415784
Similar content being viewed by others
Background & Summary
Assuming the cost of producing males and females is equal, theory predicts that gonochoristic populations should reach an equilibrium sex ratio of 1:1, a value easily produced at fertilization by the meiotic processes of most forms of genotypic sex determination (GSD)1,2. That species therefore would deviate from this sex-determining mechanism (SDM) is remarkable, but deviate they do3,4. Perhaps best known among the alternative mechanisms is temperature-dependent sex determination (TSD), where sex is irreversibly determined after fertilization by temperatures experienced in the nest4,5,6, a mechanism that commonly produces single-sex clutches7,8,9,10,11. Despite its evident potential to produce non-Fisherian sex ratios, TSD is remarkably widespread among vertebrates; first described in the lizard Agama agama by Madeleine Charnier in 196612, it was subsequently confirmed in turtles13, fishes14, crocodilians15, and tuatara16. Within these latter two orders, all species display TSD, whereas both GSD and TSD occur in squamates, turtles, and fishes3,4. This curious SDM diversity and phylogenetic distribution has spurred a bounty of research over the past 50 + years on the ecology and evolution of SDM, focusing primarily on the mechanism and potential adaptive value of TSD (reviewed by6,17,18,19,20,21).
Among taxa in which it is present, the order Testudines has arguably contributed most to our understanding of TSD. Though first described in a lizard, it was the works of Claude Pieau13 and Chester Yntema22 in Emys orbicularis and Testudo graeca, and Chelydra serpentina, respectively, that brought TSD to the attention of the broader research community. When the community responded by attributing the phenomenon to differential mortality, James Bull and Richard Vogt23 confirmed that it was incubation temperature that directly influenced offspring sex in a community of North American turtles; they were also the first to use the term “temperature-dependent sex determination”. Since those pioneering years, chelonian studies have dominated the published literature on TSD, accounting for approximately 50% of all offspring sex-ratio studies in non-avian reptiles despite only comprising ~3% of the species in this group24.
Investigations of aspects of the ecology and evolution of TSD in chelonians are published routinely, and the state of our understanding of SDM in reptiles more broadly is regularly summarized every few years6,17,18,19,20,21. However, despite the metronomic publication of knowledgeable reviews, limited effort has been made to compile and publish the data from which this knowledge is drawn. In particular, only two efforts have attempted to organize chelonian offspring sex-ratio data, each with their own shortcomings. Paukstis & Janzen25 represents the first effort, which includes offspring sex-ratio data spanning the diversity of non-avian reptiles, but only includes results from constant-temperature incubation experiments and can no longer be considered up to date. The more recent compilation24 likewise spans non-avian reptiles while also including measurements of additional phenotypes beyond sex that are influenced by incubation temperatures. However, this database excludes studies on natural incubation and exogenous hormone application, two topics often investigated in the ecological/evolutionary literature18,26,27,28. In addition, the authors’ methods largely excluded data outside the scope of Web of Science (e.g., unpublished theses/dissertations, select journals).
Such offspring sex-ratio data are necessary to characterize TSD, but a complete understanding of SDM evolution is impossible without comprehensive data on the taxonomic distribution of both TSD and its counterpart GSD as well. Several sources present these data for turtles but suffer from shortcomings much like those described above. For example, the Tree of Sex Database4 has, to the best of our knowledge, not been directly updated since its initial release in 2014. In addition, both it and subsequent publications that indirectly expand its taxonomic coverage (e.g.29,) have not taken advantage of SDM classifications presented in gray literature, such as publications from conservation breeding programs or unpublished theses/dissertations.
Here, we present the Reptilian Offspring Sex and Incubation Environment (ROSIE) database, a comprehensive compilation of offspring sex ratios and SDM in chelonians, with future plans to include data from all non-avian reptiles. Our database is easily updatable, and can be used to address a variety of key questions, including:
-
1)
What is the ancestral SDM in chelonians, and how often have transitions between mechanisms occurred?
-
2)
How does the relationship between incubation temperature and offspring sex (i.e., the sex-ratio reaction norm) evolve within and among species?
-
3)
To what extent does TSD vary geographically? Temporally? Among species? Within species? Among clutches? Across generations?
Methods
We obtained hatchling sex-ratio data in turtles using Web of Science (v.5.35) to search for research published since the discovery of TSD (196612) until 31 December 2020. On two separate occasions (17 June 2020 and 7 January 2021), we searched all databases for topics including the following terms: sex AND determin* AND incubat*, along with either turtle* or a wildcard version of each species’ taxonomic name to account for suffix variation (e.g., apalon* mutic* for both Apalone muticus and Apalone mutica). Taxonomy followed the 356 chelonian species identified in Turtle Taxonomy Working Group30. We reviewed additional publications and gray literature known to contain hatchling sex-ratio data, as well as research referenced within sources from the systematic search. Altogether, these methods returned 910 sources for evaluation.
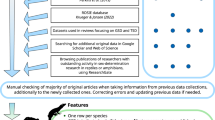
We evaluated sources obtained in the literature search based on the full text, and exclusions fell into the following categories: (1) inaccessible (n = 14), (2) study species was not a turtle (n = 116), (3) hatchling sex ratios were not reported (n = 269), (4) hatchling sex ratios were estimated based on incubation temperatures or durations (n = 48), and (5) hatchling sex-ratio data were previously reported elsewhere (n = 63). After exclusion, 400 sources remained for data extraction (Fig. 1).
Workflow of literature search and source screening process. Web of Science searches on 17 June 2020 and 7 January 2021 returned 798 unique sources for screening, supplemented by 112 sources known to contain offspring sex-ratio data identified outside the Web of Science results. After screening, we extracted data from 400 sources for inclusion in the database.
From each source, we extracted data on incubation conditions and offspring sex measurements, including additional variables such as hatching success, incubation duration, and sexing methodology (Online-only Table 1)31. When variable values (mean incubation duration, sex ratio, etc.) were not provided in the text, tables, or figure legends, we extracted values from figures using WebPlotDigitizer (v4.432). For a number of sources (n = 42), we contacted the corresponding authors to request relevant materials to clarify sample sizes or other questions about the data. We examined all data and exclusions twice to ensure accuracy and, to avoid data replication, we examined data and manuscripts from lab/author groups to determine whether multiple sources analyzed the same information. When sources shared data, we excluded measurements from the more recent source(s) unless (1) additional samples were included, or (2) data were presented in a different format (e.g., sex ratio per shelf in each incubator vs. sex ratio per whole incubator).
We gathered chelonian SDM information in a stepwise manner. First, we compiled classifications from existing databases4,29, which were next evaluated for accuracy and supplemented with SDM classifications based on the offspring sex-ratio data collected as described above. Finally, we performed extensive online searches to identify sources supplying SDM classifications for additional species. Where possible for each species, we include relevant citations, SDM classifications, and classification confidence based on a combination of available data, data in closely related species, and author expertise.
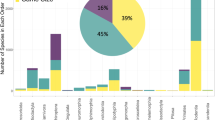
Our offspring sex-ratio compilation contains data on 32.9% (117/356) of recognized chelonian species (Fig. 2), though the taxonomic distribution is highly skewed; just 7 species from 3 families (Chelydridae: Chelydra serpentina; Cheloniidae: Caretta caretta, Chelonia mydas, Lepidochelys olivacea; Emydidae: Trachemys scripta, Chrysemys picta, Emys orbicularis) were the focus of nearly half of all studies (45.2%; 241/533; note that some sources contain data on multiple species, hence the difference between total sources [n = 400] and total studies [n = 533]). The geographic distribution is likewise biased with most studies on wild populations of North American species, starkly contrasting the sampling of African populations (Fig. 3; but note that several African species are represented in captive colonies located outside the continent). Study design is likewise biased, with most (269/400) sources focusing solely or partly on the results of constant-temperature incubation, whereas 85 employ other forms of controlled or semi-controlled incubation conditions (e.g., fluctuating temperatures, temperature switch experiments, room temperature incubation), 124 contain results from natural regimes, and 16 do not define incubation conditions. In addition, 71 studies investigate the influence of chemical applications on offspring sex ratios. In all, the database contains over 7,000 individual measurements of offspring sex ratios, ranging from data on individual eggs to a whole population’s nesting season and representing the sexing of nearly 200,000 turtle hatchlings and embryos.
Taxonomic coverage of the database. We downloaded the phylogeny of the order Testudines from a recently published paper33, and it covers 279 of 356 turtle species spanning all 14 recognized turtle families (labeled)30. The tree includes 111 species found in the offspring sex-ratio database but excludes 6 species that are not represented in the phylogeny (Batagur affinis, Homopus femoralis, Lepidochelys kempii, Natator depressus, Kinosternon alamosae, and Podocnemis lewyana). Additionally, it includes 124 species found in the SDM database but excludes 25 species (Acanthochelys radiolata, Batagur affinis, Chelodina oblonga, Chelonoidis becki, Chelonoidis darwini, Chelonoidis donfaustoi, Chelonoidis duncanens, Chelonoidis guntheri, Chelonoidis microphyes, Chelonoidis phantasticus, Chelonoidis vandenburghi, Chelonoidis vicina, Eretmochelys imbricata, Homopus areolatus, Homopus femoralis, Kinosternon alamosae, Kinosternon stejnegeri, Lepidochelys kempii, Lissemys ceylonensis, Mesoclemmys gibba, Natator depressus, Nilssonia nigricans, Podocnemis lewyana, Pseudemydura umbrina, and Rhinemys rufipes).
Our SDM database contains confident SDM classifications for 149 chelonian species (Fig. 2) and unsupported or unlikely classifications for an additional 13 species. Of those with confidently assigned SDM, 24% (36/149) exhibit GSD, 19 of which are also represented in the sex-ratio database (Fig. 2). Besides one species with an unconfident SDM classification (Chitra chitra), the remaining species in the sex-ratio database (n = 97) comprise a subset of the 113 species confidently known to exhibit TSD. Overall, our collection of chelonian SDM represents a near 50% increase in taxonomic coverage relative to recently published summaries (149 vs 101 species29).
As indicated by the name, we plan to expand ROSIE to encompass all non-avian reptile species. In our next update, we will incorporate data from the remaining reptilian orders (Crocodylia, Rhynchocephalia, and Squamata) following the methods described herein, including all data published through the end of 2020. Once ROSIE has reached this final taxonomic scope, we will push updates every other year to include newly available data and maintain the up-to-date nature of this resource.
Data Records
This database is hosted by GitHub (https://github.com/calebkrueger/ROSIE), and the raw data can be accessed via a unique, stable DOI through Zenodo31. The database consists of csv files of (1) extracted offspring sex ratio and incubation environment data with complete references, (2) SDM classifications, (3) excluded sources with complete references and exclusion criteria, and (4) metadata.
Technical Validation
The data have been thoroughly checked for accuracy by C.J.K. prior to release. The authors urge users to report errors or submit additional data and updates by emailing the corresponding author. Any errors identified can readily be corrected in future updates, which will occur biannually.
Code availability
No custom code was used in the creation of this database.
References
Fisher, R. A. The Genetical Theory of Natural Selection (Oxford University Press, 1930).
Bull, J. J. Evolution of Sex Determining Mechanisms (Benjamin Cummings, 1983).
Bachtrog, D. et al. Sex determination: Why so many ways of doing it? PLoS Biol. 12, e1001899 (2014).
The Tree of Sex Consortium. Tree of Sex: A database of sexual systems. Sci. Data 1, 140015 (2014).
Ewert, M. A., Jackson, D. R. & Nelson, C. E. Patterns of temperature‐dependent sex determination in turtles. J. Exp. Zool. 270, 3–15 (1994).
Valenzuela, N. & Lance, V. A. (eds.) Temperature-Dependent Sex Determination in Vertebrates (Smithsonian Books, 2004).
Mrosovsky, N. & Provancha, J. Sex ratio of hatchling loggerhead sea turtles: Data and estimates from a 5-year study. Can. J. Zool. 70, 530–538 (1992).
Janzen, F. J. Vegetational cover predicts the sex ratio of hatchling turtles in natural nests. Ecology 75, 1593–1599 (1994).
Doody, J. S. et al. Nest site choice compensates for climate effects on sex ratios in a lizard with environmental sex determination. Evol. Ecol. 20, 307–330 (2006).
Wapstra, E. et al. Climate effects on offspring sex ratio in a viviparous lizard. J. Anim. Ecol. 78, 84–90 (2009).
Refsnider, J. M., Milne-Zelman, C., Warner, D. A. & Janzen, F. J. Population sex ratios under differing local climates in a reptile with environmental sex determination. Evol. Ecol. 28, 977–989 (2014).
Charnier, M. Action de la température sur la sex-ratio chez l’embryon d’Agama agama (Agamidae, Lacertilien). C. R. Séances Soc. Biol. Fil. 160, 620–622 (1966).
Pieau, C. Sur la proportion sexuelle chez les embryons de deux Chelonians (Testudo graeca et Emys orbicularis L.) issues d’oeufs incubes artificiallement. C. R. Acad. Sci. D 272, 3071–3074 (1971).
Conover, D. O. & Kynard, B. E. Environmental sex determination: Interaction of temperature and genotype in a fish. Science 213, 577–579 (1981).
Ferguson, M. W. J. & Joanen, T. Temperature of egg incubation determines sex in Alligator mississippiensis. Nature 296, 850–853 (1982).
Cree, A., Thompson, M. B. & Daugherty, C. H. Tuatara sex determination. Nature 375, 543 (1995).
Bull, J. J. Sex determination in reptiles. Q. Rev. Biol. 55, 3–21 (1980).
Janzen, F. J. & Paukstis, G. L. Environmental sex determination in reptiles: Ecology, evolution, and experimental design. Q. Rev. Biol. 66, 149–179 (1991).
Ciofi, C. & Swingland, I. R. Environmental sex determination in reptiles. Appl. Anim. Behav. Sci. 51, 251–265 (1997).
Warner, D. A. Sex determination in reptiles. in Hormones and Reproduction of Vertebrates, Volume 3: Reptiles (eds. Norris, D. O. & Lopez, K. H.) 1-38 (Academic Press, 2011).
Miyagawa, S., Yatsu, R. & Iguchi, T. Environmental control of sex determination and differentiation in reptiles. in Reproductive and Developmental Strategies: The Continuity of Life (eds. Kobayashi, K., Kitano, T., Iwao, Y. & Kondo, M.) 367-390 (Springer Japan KK, 2018).
Yntema, C. L. Effects of incubation temperatures on sexual differentiation in the turtle, Chelydra serpentina. J. Morphol. 150, 453–461 (1976).
Bull, J. J. & Vogt, R. C. Temperature-dependent sex determination in turtles. Science 206, 1186–1188 (1979).
Noble, D. W. A. et al. A comprehensive database of thermal developmental plasticity in reptiles. Sci. Data 5, 180138 (2018).
Paukstis, G. L. & Janzen, F. J. Sex determination in reptiles: Summary of effects of constant temperatures of incubation on sex ratios of offspring. Smithson. Herpetol. Inf. Serv. 83, 1–28 (1990).
Paitz, R. T., Gould, A. C., Holgersson, M. C. N. & Bowden, R. M. Temperature, phenotype, and the evolution of temperature-dependent sex determination: How do natural incubations compare to laboratory incubations? J. Exp. Zool. B 314, 86–93 (2010).
Crews, D. Temperature-dependent sex determination: The interplay of steroid hormones and temperature. Zool. Sci. 13, 1–13 (1996).
Warner, D. A., Addis, E., Du, W.-G., Wibbels, T. & Janzen, F. J. Exogenous application of estradiol to eggs unexpectedly induces male development in two turtle species with temperature-dependent sex determination. Gen. Comp. Endocrinol. 206, 16–23 (2014).
Bista, B. & Valenzuela, N. Turtle insights into the evolution of the reptilian karyotype and the genomic architecture of sex determination. Genes 11, 416 (2020).
Turtle Taxonomy Working Group. Turtles of the world: Annotated checklist and atlas of taxonomy, synonymy, distribution, and conservation status (8th Ed.). Chelonian Res. Monogr. 7, 1–292 (2017).
Krueger, C. J. & Janzen, F. J. The Reptilian Offspring Sex and Incubation Environment (ROSIE) Database. Zenodo https://doi.org/10.5281/zenodo.4670573 (2021).
Rohatgi, A. WebPlotDigitizer. https://automeris.io/WebPlotDigitizer/index.html (2020).
Thomson, R. C., Spinks, P. Q. & Shaffer, H. B. A global phylogeny of turtles reveals a burst of climate-associated diversification on continental margins. Proc. Natl. Acad. Sci. USA 118, e2012215118 (2021).
Acknowledgements
We thank those colleagues who provided clarification of results, copies of manuscripts, or raw data, including P. Andrade, K. Bieser, B. Bock, A. Carter, D. Crews, W.-G. Du, S. Freedberg, A. Georges, A. Grosse, J. Gutiérrez, D. Holliday, W. Hopkins, F. Ihlow, G. Kuchling, D. Ligon, A. Lolavar, A. Malvasio, M. Massey, S. McGaugh, J. McGlashan, V. Mislin, L. Neuman-Lee, R. Paitz, J. Phillips, S. Podreka, P. Praschag, H. Prokop, J. Refsnider, T. Rhen, D. Shaver, M. Thompson, N. Valenzuela, J. Van Dyke, R. Vogt, D. Warner, T. Wibbels, and J. Wyneken. Our ongoing empirical work on this topic has been funded primarily by the U.S. National Science Foundation. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. 1744592. This is Kellogg Biological Station contribution number 2301.
Author information
Authors and Affiliations
Contributions
C.J.K. conducted literature searches, record filtering, and data extraction. C.J.K. and F.J.J. wrote and edited the data descriptor.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Table
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Krueger, C.J., Janzen, F.J. ROSIE, a database of reptilian offspring sex ratios and sex-determining mechanisms, beginning with Testudines. Sci Data 9, 22 (2022). https://doi.org/10.1038/s41597-021-01108-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-01108-1
This article is cited by
-
HerpSexDet: the herpetological database of sex determination and sex reversal
Scientific Data (2023)