Abstract
Data comprise intracranial EEG (iEEG) brain activity represented by stereo EEG (sEEG) signals, recorded from over 100 electrode channels implanted in any one patient across various brain regions. The iEEG signals were recorded in epilepsy patients (N = 10) undergoing invasive monitoring and localization of seizures when they were performing a battery of four memory tasks lasting approx. 1 hour in total. Gaze tracking on the task computer screen with estimating the pupil size was also recorded together with behavioral performance. Each dataset comes from one patient with anatomical localization of each electrode contact. Metadata contains labels for the recording channels with behavioral events marked from all tasks, including timing of correct and incorrect vocalization of the remembered stimuli. The iEEG and the pupillometric signals are saved in BIDS data structure to facilitate efficient data sharing and analysis.
Measurement(s) | intracranial electroencephalogram measurement • gaze • pupil size trait |
Technology Type(s) | intracranial electroencephalography (iEEG) • gaze tracking • Pupillometry |
Factor Type(s) | patient • task |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.16847566
Similar content being viewed by others
Background & Summary
Intracranial EEG (iEEG) monitoring is a standard diagnostic procedure used to localize and treat sources of epileptogenic activities in patients suffering from pharmacoresistant focal epilepsy1. The iEEG signals are acquired directly from brain structures using various types of implanted electrode contacts, ranging from standard clinical macro-contacts to special micro-contacts. Apart from their clinical value, the acquired signals present a unique opportunity to investigate physiological processes in the human brain, including those related to memory and cognition2,3,4, at a large scale of neural activities5. Most of the studies employing these signals in cognitive tasks are limited by clinical and research recording systems, multiple formats for acquiring and storing data files, and separate data structures for electrophysiological, behavioral, pupillometric, and brain imaging data.
The iEEG recording systems used in the clinical setting are typically limited to low sampling rates and inefficient compression formats6. Systems used for research recordings offer excellent sampling rates of up to 40 kHz and efficient compression formats to store continuously sampled signals from hundreds of channels over multiple days of the seizure localization procedure6,7. This allows large-scale recordings of electrophysiological processes sampled both at the macro- and at the micro-level of brain organization2,5,8. However, these large-scale recordings, which require high temporal (sampling rate) and spatial (size and number of channels) resolution, grow rapidly in size. The large-scale iEEG recordings present a challenge for efficient storing, organization, and processing6,7. In addition, these rare iEEG datasets need to be synchronized with data from behavioral tasks and with brain imaging data for electrode localization. All these various sources of data could ideally be stored in one environment with a structure universal for various types of brain signals and tasks. Recently, the widely-recognized Brain Imaging Data Structure (BIDS)9 was proposed for a universal organization of iEEG data10 that would accommodate the large scale of electrophysiological signals sampled in any behavioral task.
Well organized datasets are critical for rapid sharing and exploration of the iEEG data and open the way for large datasets with recordings collected from national and international clinical centers10. The largest database of iEEG recordings during memory tasks was collected in a BRAIN initiative project called Restoring Active Memory11 with recordings collected from over 300 patients at multiple clinical centers. Although the project yielded a whole series of seminal papers about human memory enhancement12,13,14 and the neural mechanisms of memory processing15,16,17,18, the dataset revealed multiple challenges for use of this and similar datasets by researchers outside of such projects. One challenge is related to combining anatomical data for electrode localization19. Others are related to new structures and formats used in such datasets that require various custom-made toolboxes for processing the data. Marking events and metadata in the data signals presents yet another challenge that is addressed by unified database structures10. Adding complementing behavioral data, e.g. from pupillometry 20, introduce another challenge for organizing the datasets20 in a unified structure.
Therefore, there is a growing demand for new open-access and universal datasets with the human iEEG task recordings. There are several recent reports using large-scale iEEG recordings of ECoG, local field potential, and single unit spiking activities during memory tasks21,22,23 that could be cross-validated or reproduced in one combined large database from these various tasks. In addition, the same findings that were observed for a given frequency range (70–150 Hz) could be reproduced e.g. in the higher ripple and fast ripple frequency bands or in the single and multi-unit activity ranges (>1000 Hz), where other memory-associated activities were reported24,25,26,27. Currently, these datasets are limited to only a selected range of these activities instead of a broad scale of wide-bandwidth signals.
The purpose of sharing this dataset is to provide researchers with high quality and accessible recordings of iEEG signals during tasks stored in a universal structure together with pupillometry signals. This will help to advance the understanding of the multiple brain activities and cognitive processes reflected in the eye movements that are engaged during memory performance. Here, we aim to combine measures from large-scale electrophysiology and cognitive psychology in one universal structure for efficient analysis and sharing to accelerate novel discoveries and deepen the knowledge about human memory and the associated brain functions.
Methods
Ethics declaration
Data were collected from patients at Mayo Clinic in accordance with the research protocol approved by the local IRB (15-006530). Informed consent was obtained from each participant and all procedures were performed in accordance with the relevant guidelines and regulations approved by the Mayo Clinic IRB.
Data collection
We used intracranial recordings made during free recall memory tasks in patients undergoing iEEG monitoring as part of clinical treatment for drug-resistant epilepsy. Electrophysiological signals were collected from electrodes implanted into the brain parenchyma with 32 kHz sampling frequency. The signals were collected from standard clinical penetrating depth electrodes (AdTech Inc., PMT Inc.) implanted into the brain parenchyma. The depth electrode contacts were separated by 5–10 mm spacing. The patients were implanted with the depth electrodes using the stereo EEG (sEEG) procedure, which involved stereotactic planning of implantation trajectories in 3D anatomical space of the brain. Note that the ECoG data are not published within the here presented version of this dataset but will be added in the future versions. For each patient, the number and placement of the electrodes were determined by a clinical team with the goal of localizing epileptogenic brain regions as part of the patient’s evaluation for epilepsy surgery. The postimplantation anatomical location of electrode contacts was determined by expert reviewers. The anatomical terminology followed the ‘Atlas of the Human Brain’28
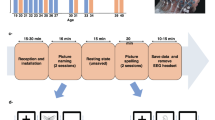
Following electrode implantation, each patient participated in a battery of four cognitive tasks, probing verbal memory and eye movements with tracking of pupil size and gaze position on the task display screen. All tasks were performed by the implanted patients resting comfortably in a hospital bed within one experimental task session a day. Task events were annotated automatically in the iEEG recordings with TTL pulses generated in the task computer and transmitted to a parallel input port for the iEEG signal amplifier (Neuralynx Inc., Bozeman MT). Behavioral responses (vocalizations of remembered word stimuli and eye movements on the screen) were processed after the experiment using microphone recordings and the pupillometric signals acquired on the task computer (i4tracking system, Medicton Group Ltd., Prague, Czech Republic). Task calibration, progression through the trials, and switching between the tasks were operated by the experimenter on the task computer without any need for patients to press the keyboard or make any motor movement except for the eye movements and the vocalizations. Patients were instructed to lay still in the bed, refrain from moving, and focus their gaze on the center of the screen. Only the task monitor was presented in front of the patients. Position of the monitor for optimal pupil detection was determined by visually checking real-time pupil detection before starting the task battery. In addition, an automatic eye tracking calibration check was conducted between the tasks to ensure high quality of the pupillometry data. The general flow of one task session is depicted in Fig. 1.
The battery started with a Smooth Pursuit (SP) task where patients follow a horizontally moving dot in the middle of the screen with constant slow acceleration (Fig. 1). SP controls not only for any oculomotor and/or sensory deficits but also for sensorimotor (non-verbal) working memory with known neurophysiological substrates impaired in specific brain disorders29.
The SP task was followed by the main verbal memory task for Free Recall (FR) of word lists30, in which patients were asked to remember 12 words presented one at a time on a computer screen, and then to recall them in any order after a short distractor task with simple algebraic equations, as we described before in Kucewicz et al. (2017)17 (Fig. 1). In each complete FR session there were 15 such word lists (180 different word epochs in total). Patients were free to stop the task at any point. Interictal epileptiform activities31,32,33 and seizures, including micro-seizures8, were previously reported in these recordings and could affect performance in the tasks. In contrast to the previous studies with the FR task, we repeated the same word lists on subsequent FR sessions on the following days of the hospital stay at regular intervals (24 and 72 hours after the first FR session). Thus, the same word lists were used for a patient and presented in the same order to enable study of reinstatement of specific electrophysiological patterns during the task and their reactivation during delayed recall in the task34,35,36,37.
After the FR task, another eye movement task was administered to reduce interference of the word stimuli before the final task using the same pool of common English nouns. This task is a classic paradigm for saccadic and anti-saccadic (AS) eye movements38 directed toward a target colored dot shifting position either to the right or to the left side of the screen center (Fig. 1). AS was used to probe non-verbal sensorimotor working memory for holding information about the color of the presented dot, which determines either a pro-saccade (green), i.e. follow the coming shift to either side, or an anti-saccade (red), i.e. do not follow the shift and saccade to the opposite side. There were 120 trials of each type that were pseudo-randomly distributed between two blocks of 60 trials with equal number of the pro- and anti-saccade trials. Like the SP task, AS was described with a well-defined network of the underlying neural substrates and mechanisms, which are impaired in a range of brain disorders39.
The battery was concluded with another version of the verbal memory task that used 6 pairs of words displayed one after another for subsequent cued recall (in contrast to the list of 12 words presented for non-cued free recall in FR), following the same distractor delay as in FR, to probe paired-associate learning (PAL)30. In PAL, the 15 lists of word pairs are initially presented in the same order across the sessions, but the order of the paired associate words (one word from each pair) during the cued recall phase is randomized. In this recall stage, one of the words from each pair (randomly selected) is presented as the cue for recall (Fig. 1). This design was employed to control for any effects of the predicted sequence of words, as in the previous studies22,37. The same pool of common English nouns was used for both FR and PAL tasks (randomly assigned for each list without replacement) to enable investigation of the iEEG activities in response to the same words in the context of two different tasks. Overall, within any session each of the 180 words was presented at least 2 times. There was at least one arithmetic equation presented per each list for control comparison of the word-induced iEEG responses.
In summary, each task session comprised a battery of the four tasks administered in the following order: SP, FR, AS, PAL. The aim was to record all tasks during one task session but this was not always possible. The recordings were collected from a minimum of one daily task session with at least one task completed (Table 1).
Tracking of eye movements and pupil dilation
Recording of gaze position and pupil size was performed using a custom-made application developed from the i4tracking system (Medicton group Inc., Prague, Czech Republic) designed for clinical use40. The recording was performed on a laptop computer connected to a 24-inch monitor screen with resolution of 1680 × 1050 px, where the gaze position was tracked by high-resolution (2048 × 1088 px) and high-speed (up to 150 Hz sampling rate) external camera device placed right under the screen in front of the patient. Stimuli were displayed on the screen using font size of 100 pt and were viewed from a distance of approx. 60 cm. Pupil position and size were detected by the camera device, corresponding to approx. 0.1 mm per pixel in the eye image. The camera device was positioned to capture the face area from forehead to the mouth. Two sources of infrared light were emitted from the camera to capture the reflected light for pupil detection. Other sources of infrared light in the room were eliminated. Raw images from the camera were sampled at the rate of 150 Hz and were saved for extracting pupil size and position using a detection algorithm, which worked by fitting a general ellipse equation over the estimated pupil image. The pupil size in pixels was also converted to millimeters using measured interpupillary distance (IPD) and the IPD in the camera images. The reported pupil area was computed using the corresponding vertical and horizontal diameters in the ellipse area equation. Gaze position was determined by projecting the movement of the estimated center of the pupil onto the monitor screen area with the use of corneal reflection. Gazes outside of the screen area as well as the eye-blinks were treated as missing-samples. Vocal responses of the patients during the recall phase of the task were recorded using an external USB microphone and manually annotated after the experiments to mark the time and identity of the uttered words in custom audio editing software.
Before presentation of the task word lists, the eye tracker was calibrated for each recruited patient. In the calibration procedure patients were asked to focus their gaze on nine points presented consecutively at specific positions across the diagonals and centers of the side edges of the display screen. Calibration was repeated before starting any new task in the session to ensure accurate estimate of the pupil size. Moreover, patients were instructed not to move their heads and focus gaze on the screen throughout all phases of the task trials. Despite these efforts, the quality of the eye-tracking was lower toward the end of each task due to slight shifts in the focus with changes in the head position. All stimuli were presented on the screen in a light gray color on light gray background to minimize pupil responses to changing lighting and contrast. The testing was conducted in a room with low-light conditions that remained constant throughout the testing procedure.
Format conversion
The time series data (Nuralynx iEEG, i4tracking pupillometry and audio) were processed by a software pipeline (Fig. 2), aligned in time based on TTL pulses that indicated specific annotated events in the tasks (Fig. 3), stored in Multiscale Electrophysiology Format (MEF) version 36, and organized in BIDS data structure for further viewing and automated processing. All data were de-identified during the conversion process by omitting any information that could lead to identification of the patient, such as name and date of birth.
Data Records
Currently the data comprises recordings from 10 patients with 65 tasks performed. Summary of the tasks performed with recordings from a specified number of implanted contacts is presented in Table 1.
The raw time series data (iEEG, audio, pupillometry) were stored in the MEF3 format6 due to its capability for handling data compression, variable sampling frequency across signals, and any signal discontinuities (Fig. 3). The MEF C library is publicly available at https://github.com/msel-source/meflib. The high level API readers exist for python (https://github.com/msel-source/pymef) and for Matlab (https://github.com/MaxvandenBoom/matmef). Overview of the processing structure is summarized in Fig. 2.
The data and metadata are organized in BIDS data structure for iEEG10, which is supported by the International Neuroinformatics Coordinating Facility (https://www.incf.org/). The structure contains human readable metadata and is widely used by the neuroscience community, enabling automatic processing by a wide range of available tools. The metadata are stored in .tsv and .json files and contain information about anatomical location of implanted electrodes (anatomical structure and coordinates in scanner coordinate space), channel sampling frequencies, channel quality, and behavioral events (e.g. word presentations, recalled words, saccades). The structure of the data follows the BIDS standard version 1.2.1 with the folder levels arranged into a subject-session-iEEG-task-run order. The BIDS runs are individual performances of a particular task by one patient separated by 24–72 hours.
The root dataset folder contains readme and dataset description files with general information about the dataset and participants.tsv describing the metadata of individual patients. The “code” folder contains an ipython notebook with scripts to guide the users and provide easy access to the data. The BIDS subject and session levels do not contain data files, but have structural purpose. The BIDS session level was implemented in case the same patients are reimplanted in the future. The ieeg level contains multiple files and folders directly related to individual recordings. Each run represents one performance of a particular task. The .mefd folders are MEF session folders containing raw data. The file _electrodes.tsv provides information about electrode and contact positions in coordinates and individual anatomical structures. The _channels.tsv files provide information about individual channel type, sampling frequency, filters and data quality. The _events.tsv files provide events that happened during the recording. The events without the trial_type information are the original ttl events as sent by the stimulation laptop. In FR and PAL the events with trial_type information provide detailed description with specific words during encoding and recall along with distractor phase events. In SP the events represent the change in moving dot direction. In AP the events show information about fixation and the following anti/pro-saccade event. The .json files provide general descriptions of the recordings and descriptions of the columns in .tsv files that are not specified in BIDS standard.
The dataset is available under the terms of EBRAINS data usage agreement for human data (Supplement 1) via the EBRAINS Knowledge Graph41.
Technical Validation
The iEEG data were first visually checked for corrupt signals coming from contacts that were either damaged or implanted outside of the brain tissue. The information about the signal quality was stored in the BIDS _channels.tsv files for each subject-task-run. The patient verbal responses were manually marked in the signals using vocal recordings to indicate the start of word vocalization (Fig. 3).
To validate the iEEG signals we first investigated the proper alignment of word presentation with the signals, we averaged the iEEG responses across all trials to confirm clear event-related potentials evoked on a subset of the electrode channels following the word presentation (Fig. 4). To confirm our previous results (Kucewicz et al. 2017, 2019)17,18 and further confirm the technical validity of iEEG data, we analyzed the changes in the spectral power density in forgotten and recalled words across 44 contacts within 5 patients (Fig. 5). Only patients with electrodes implanted in the middle temporal gyrus were selected. We observed significant differences of this so-called Subsequent Memory Effect in the theta frequency band during presentation of recalled and forgotten words using Kruskal-Wallis, alpha = 0.05.
To validate the synchronization of pupillometry signals and their scientific utility we performed analysis of pupil dilation during a FR task. Pupil dilatation was dynamically modulated during different stages of the task (Fig. 6a). We further evaluated the Subsequent Memory Effect as the difference in pupil dilation during encoding of recalled and forgotten words (Fig. 6b) as reported previously20. Significantly larger pupil dilation was observed during presentation of recalled words around 0.5 s after stimulus presentation. Both of these results are congruent with the previous report from a different group of patients (Kucewicz et al. 2018)20.
Example of the eye tracking data during FR, sub-004. (a) Pupil dilation is modulated by different phases of the free recall verbal memory task. The pupil size was averaged over all trials of one patient in the FR task. Shaded areas mark epochs of word presentation on the screen and their recall with blank screen. (b) Recalled and forgotten words show different pupil responses during memory encoding. The pupil responses to word presentation were averaged with respect to subsequently recalled (red) and forgotten (blue) words. Shaded areas mark word presented on the screen.
Usage Notes
For easy manipulation with the dataset we recommend usage of BIDS compatible libraries such as pybids42 or bids-matlab (https://github.com/bids-standard/bids-matlab) along with the libraries for MEF3 reading mentioned in the data records section. The folder “code” in the dataset root directory contains a jupyter notebook with example code for reading the data in python programming language. The signals are not preprocessed intentionally to provide the data in a raw form and allow the users to perform their own preprocessing. The iEEG signals exhibiting pathological activity are not excluded but can be detected on demand using the provided code (see Code availability). The pupillometry signals contain blinking artifacts and artifacts caused by clinical settings such as movement due to discomfort or pain. The pupillometry signal artifacts can be eliminated by using published methods43. The data stored in MEF can be viewed using PySigView44, a custom made python viewer, or the SignalPlant program45. The iEEG recordings were limited to 128 channels even though the number of implanted contacts was higher. We will provide support to the users if they encounter any problems using the above mentioned methods.
Limitations
The dataset comprises a relatively small number of recordings. We intend to add recordings over time in new versions of this dataset which will also include parallel recorded electrocorticography (ECoG) data for some cases. Currently the contact coordinate information stored in _electrodes.tsv files are provided in MRI scanner space and is therefore not applicable across patients. Our priority was to provide the anatomical localization data in the raw form to allow conversion of the patient-specific coordinates into a preferred coordinate and anatomical labeling frame, if needed. We will provide support to users if they wish to convert the coordinates to any specific coordinate system. The data set currently does not include the brain images for the sake of protecting patient privacy46. This issue will be addressed in the future versions of the dataset. In the new datasets, we are also planning to include micro-contact signal recordings, which present additional challenges for interpolating their coordinates relative to the neighboring macro-contacts.
Code availability
To provide an easy way to detect and analyze electrophysiological activity in iEEG signals we make our codes available at GitLab (https://gitlab.com/brainandmindlab/memory_encoding). Currently, the repository contains a python script to automatically process the individual BIDS layers of the dataset (subject, task, run, channel) using the EPYCOM library47. The library is focused on iEEG processing and contains a set of algorithms for automated detection of high frequency oscillations24,48,49, interictal epileptiform discharges50 and for computation of univariate and bivariate feature calculation51,52. The repository will be gradually updated with scripts for statistical analyses and result visualizations.
References
Rosenow, F. & Lüders, H. Presurgical evaluation of epilepsy. Brain 124, 1683–1700 (2001).
Lhatoo, S. D., Kahane, P. & Luders, H. O. Invasive Studies of the Human Epileptic Brain: Principles and Practice. (Oxford University Press, USA, 2019).
Fried, I., Rutishauser, U., Cerf, M. & Kreiman, G. Single Neuron Studies of the Human Brain: Probing Cognition. (MIT Press, 2014).
Engel, A. K., Moll, C. K. E., Fried, I. & Ojemann, G. A. Invasive recordings from the human brain: clinical insights and beyond. Nature Reviews Neuroscience 6, 35–47 (2005).
Kucewicz, M. T. et al. Combined Single Neuron Unit Activity and Local Field Potential Oscillations in a Human Visual Recognition Memory Task. IEEE Trans. Biomed. Eng. 63, 67–75 (2016).
Stead, M. & Halford, J. J. Proposal for a Standard Format for Neurophysiology Data Recording and Exchange. J. Clin. Neurophysiol. 33, 403–413 (2016).
Brinkmann, B. H., Bower, M. R., Stengel, K. A., Worrell, G. A. & Stead, M. Large-scale electrophysiology: acquisition, compression, encryption, and storage of big data. J. Neurosci. Methods 180, 185–192 (2009).
Stead, M. et al. Microseizures and the spatiotemporal scales of human partial epilepsy. Brain 133, 2789–2797 (2010).
Gorgolewski, K. J. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data 3, 160044 (2016).
Holdgraf, C. et al. iEEG-BIDS, extending the Brain Imaging Data Structure specification to human intracranial electrophysiology. Sci Data 6, 102 (2019).
Underwood, E. Researchers aim for an electrical memory prosthesis. Science 345, 250–250 (2014).
Kucewicz, M. T. et al. Evidence for verbal memory enhancement with electrical brain stimulation in the lateral temporal cortex. Brain 141, 971–978 (2018).
Ezzyat, Y. et al. Closed-loop stimulation of temporal cortex rescues functional networks and improves memory. Nat. Commun. 9, 365 (2018).
Kucewicz, M. T. et al. Electrical Stimulation Modulates High γ Activity and Human Memory Performance. eNeuro 5 (2018).
Solomon, E. A. et al. Widespread theta synchrony and high-frequency desynchronization underlies enhanced cognition. Nat. Commun. 8, 1704 (2017).
Ezzyat, Y. et al. Direct Brain Stimulation Modulates Encoding States and Memory Performance in Humans. Curr. Biol. 27, 1251–1258 (2017).
Kucewicz, M. T. et al. Dissecting gamma frequency activity during human memory processing. Brain 140, 1337–1350 (2017).
Kucewicz, M. T. et al. Human Verbal Memory Encoding Is Hierarchically Distributed in a Continuous Processing Stream. eNeuro 6 (2019).
Stolk, A. et al. Integrated analysis of anatomical and electrophysiological human intracranial data. Nat. Protoc. 13, 1699–1723 (2018).
Kucewicz, M. T. et al. Pupil size reflects successful encoding and recall of memory in humans. Sci. Rep. 8, 4949 (2018).
Norman, Y. et al. Hippocampal sharp-wave ripples linked to visual episodic recollection in humans. Science 365 (2019).
Vaz, A. P., Wittig, J. H. Jr, Inati, S. K. & Zaghloul, K. A. Replay of cortical spiking sequences during human memory retrieval. Science 367, 1131–1134 (2020).
Vaz, A. P., Inati, S. K., Brunel, N. & Zaghloul, K. A. Coupled ripple oscillations between the medial temporal lobe and neocortex retrieve human memory. Science 363, 975–978 (2019).
Kucewicz, M. T. et al. High frequency oscillations are associated with cognitive processing in human recognition memory. Brain 137, 2231–2244 (2014).
Minxha, J., Adolphs, R., Fusi, S., Mamelak, A. N. & Rutishauser, U. Flexible recruitment of memory-based choice representations by the human medial frontal cortex. Science 368 (2020).
Urgolites, Z. J. et al. Spiking activity in the human hippocampus prior to encoding predicts subsequent memory. Proc. Natl. Acad. Sci. USA 117, 13767–13770 (2020).
Kamiński, J. et al. Persistently active neurons in human medial frontal and medial temporal lobe support working memory. Nat. Neurosci. 20, 590–601 (2017).
Mai, J. K., Majtanik, M. & Paxinos, G. Atlas of the Human Brain. (Academic Press, 2015).
Fukushima, K., Fukushima, J., Warabi, T. & Barnes, G. R. Cognitive processes involved in smooth pursuit eye movements: behavioral evidence, neural substrate and clinical correlation. Front. Syst. Neurosci. 7, 4 (2013).
Kahana, M. J. Foundations of Human Memory. (Oxford University Press, 2014).
Matsumoto, J. Y. et al. Network oscillations modulate interictal epileptiform spike rate during human memory. Brain 136, 2444–2456 (2013).
Horak, P. C. et al. Interictal epileptiform discharges impair word recall in multiple brain areas. Epilepsia 58, 373–380 (2017).
Kleen, J. K. et al. Hippocampal interictal epileptiform activity disrupts cognition in humans. Neurology 81, 18–24 (2013).
Staresina, B. P., Alink, A., Kriegeskorte, N. & Henson, R. N. Awake reactivation predicts memory in humans. Proc. Natl. Acad. Sci. USA 110, 21159–21164 (2013).
Staresina, B. P., Henson, R. N. A., Kriegeskorte, N. & Alink, A. Episodic reinstatement in the medial temporal lobe. J. Neurosci. 32, 18150–18156 (2012).
Johnson, J. D., McDuff, S. G. R., Rugg, M. D. & Norman, K. A. Recollection, familiarity, and cortical reinstatement: a multivoxel pattern analysis. Neuron 63, 697–708 (2009).
Yaffe, R. B. et al. Reinstatement of distributed cortical oscillations occurs with precise spatiotemporal dynamics during successful memory retrieval. Proc. Natl. Acad. Sci. USA 111, 18727–18732 (2014).
Munoz, D. P. & Everling, S. Look away: the anti-saccade task and the voluntary control of eye movement. Nat. Rev. Neurosci. 5, 218–228 (2004).
Coe, B. C. & Munoz, D. P. Mechanisms of saccade suppression revealed in the anti-saccade task. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372 (2017).
Doležal, J. & Fabian, V. 41. Application of eye tracking in neuroscience. Clinical Neurophysiology 126, e44 (2015).
Cimbalnik, J. et al. Intracranial electrophysiological recordings from the human brain during memory tasks with pupillometry. EBRAINS https://doi.org/10.25493/GKNT-T3X (2021).
Yarkoni, T. et al. PyBIDS: Python tools for BIDS datasets. Journal of Open Source Software 4, 1294 (2019).
Mathôt, S., Fabius, J., Van Heusden, E. & Van der Stigchel, S. Safe and sensible preprocessing and baseline correction of pupil-size data. Behav. Res. Methods 50, 94–106 (2018).
Cimbalnik, J. ICRC-BME/PySigView: First release for zenodo. Zenodo https://doi.org/10.5281/zenodo.2590814 (2019).
Plesinger, F., Jurco, J., Halamek, J. & Jurak, P. SignalPlant: an open signal processing software platform. Physiol. Meas. 37, N38–48 (2016).
Schwarz, C. G. et al. Identification of Anonymous MRI Research Participants with Face-Recognition Software. N. Engl. J. Med. 381, 1684–1686 (2019).
Cimbalnik, J., Klimes, P. & Travnicek, V. ICRC-BME/epycom: EPYCOM-beta. Zenodo https://doi.org/10.5281/zenodo.4030570 (2020).
Cimbálník, J., Hewitt, A., Worrell, G. & Stead, M. The CS algorithm: A novel method for high frequency oscillation detection in EEG. J. Neurosci. Methods 293, 6–16 (2018).
Gardner, A. B., Worrell, G. A., Marsh, E., Dlugos, D. & Litt, B. Human and automated detection of high-frequency oscillations in clinical intracranial EEG recordings. Clin. Neurophysiol. 118, 1134–1143 (2007).
Barkmeier, D. T. et al. High inter-reviewer variability of spike detection on intracranial EEG addressed by an automated multi-channel algorithm. Clinical Neurophysiology 123, 1088–1095 (2012).
Cimbalnik, J. et al. Multi-feature localization of epileptic foci from interictal, intracranial EEG. Clin. Neurophysiol. 130, 1945–1953 (2019).
Klimes, P. et al. NREM sleep is the state of vigilance that best identifies the epileptogenic zone in the interictal electroencephalogram. Epilepsia 60, 2404–2415 (2019).
Acknowledgements
Cindy Nelson, Aiswarya Lakshmi Nandakumar, M.D., and Tal Pal Attia provided technical, clinical and administrative assistance in patient testing and data collection at Mayo Clinic. This dataset would not have been collected without collaborations with local departments of neurosurgery, radiology, and neurology, nurses, and EEG technicians, and the dedicated effort and participation of patients and their families. Efforts of M.T.K., M.L., C.T. and the data collection process were supported from the First Team grant no. POIR.04.04.00-00-4379/17 of the Foundation for Polish Science, co-financed by the European Union under the European Regional Development Fund. Work of J.C. was supported by grant no. PPN/ULM/2019/1/00240 of the Polish National Agency for Academic Exchange under the ULAM program. This research was supported by the National Institutes of Health R01-NS92882, project no. LQ1605 from the National Program of Sustainability II (MEYS CR), Ministry of Youth and Sports of the Czech Republic project no. LH15047 (KONTAKT II). V.M. is a member of the Biomedical Engineering and Physiology Graduate Program and is supported by Mayo Clinic Graduate School of Biomedical Sciences.
Author information
Authors and Affiliations
Contributions
M.K. and J.D. designed the battery of cognitive tasks. J.C. and M.K. designed and created the database structure. J.C. wrote and developed toolboxes and used them to pre-process that data. J.D. and M.D. wrote and developed the code for the tasks with eye-tracking. C.T., M.L., V.M. and B.J. performed the experiments and collected the data. J.C. created and uploaded the database to EBRAINS portal. J.V.G. and G.W. implanted the electrodes and provided access to the patient recordings at Mayo Clinic. J.C. and M.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
JD and MD are employed in Medicton Group Ltd. (Prague, Czech Republic) as engineers working on the I4 tracking system. M.D. is the chief executive officer of the Medicton Group Ltd., which developed the I4 tracking system.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Cimbalnik, J., Dolezal, J., Topçu, Ç. et al. Intracranial electrophysiological recordings from the human brain during memory tasks with pupillometry. Sci Data 9, 6 (2022). https://doi.org/10.1038/s41597-021-01099-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-01099-z