Abstract
We describe a collection of T1-, diffusion- and functional T2*-weighted magnetic resonance imaging data from human individuals with albinism and achiasma. This repository can be used as a test-bed to develop and validate tractography methods like diffusion-signal modeling and fiber tracking as well as to investigate the properties of the human visual system in individuals with congenital abnormalities. The MRI data is provided together with tools and files allowing for its preprocessing and analysis, along with the data derivatives such as manually curated masks and regions of interest for performing tractography.
Measurement(s) | brain white matter |
Technology Type(s) | magnetic resonance imaging |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.16726246
Similar content being viewed by others
Background & Summary
We present CHIASM, the human brain albinism, and achiasma dataset, a unique collection of magnetic resonance imaging (MRI) data of brains with congenital abnormalities in the visual system. The unique feature of these subjects is the varied amount of crossing found in a specific structure –the optic chiasm– across participants with albinism. More specifically, it is well established1 that the number of crossing fibers crossing at the human optic chiasma to reach the contralateral brain hemisphere (right and left respectively) varies between certain groups. The percentage of fiber crossing at the chiasm has been reported for normal-sighted (control) participants to be about 53%2. In brains affected by albinism instead, the number of crossing fibers at the optic chiasm grows above 53%3. Crossing fibers within the optic chiasma in individuals affected by chiasm hypoplasia is lower than 53%4. Finally, data from individuals with achiasma have been shown to completely lack neuronal fiber crossing at the optic chiasm1,5,6.
The data covers four participant groups: the controls (n = 8, Fig. 1a), albinism (n = 9, Fig. 1b), chiasma hypoplasia (n = 1; Fig. 1c) and achiasma (n = 1; Fig. 1d), and comprises three different MRI modalities: (A) T1-weighted (T1w; Fig. 1, top row) images, (B) diffusion-weighted images (DWI, Fig. 1, middle row) and (c) T2*-weighted functional MRI (fMRI; Fig. 1, bottom row) images. More specifically: (A) T1w images are provided together with further derivatives (masks and labels obtained through segmentation, white matter mask manually curated in optic chiasm region), (B) DWI data was acquired using high angular7,8 and spatial9,10,11 resolution and is provided with further derivatives, such as tractography results, (C) fMRI data is provided for the subset of participants from control and albinism groups (n = 4 and n = 6, respectively) and is accompanied with meta-files describing stimulus and acquisition. All the data, both in the raw and preprocessed form are available on the cloud computing platform https://brainlife.io11 and Github repository https://github.com/rjpuzniak/CHIASM.
Overview of structural abnormalities of the optic chiasm and provided MRI data. (a) Exemplary control participant (CON1). (b) Exemplary participant with albinism (ALB1). (c) Participant with chiasma hypoplasia (CHP1). (d) Participant with achiasma (ACH1). The fMRI data is not provided due to severe nystagmus and motion compromising the quality of data. Top, middle and bottom rows display respectively T1w, DW, and fMRI data. Images show pseudo-axial views of a T1w image cropped to the brain mask.
The data we present here can be of value to the scientific community for multiple reasons. First, it can serve as a reference dataset to support basic research for clarifying the neuroscientific underpinnings of the different conditions. Currently, there are no reference datasets available covering similar conditions measured with high-resolution DWI data. Second, this dataset can be used by investigators to validate independent results and advance studies on the disease and neuroplasticity mechanisms. This is possible as chiasmal malformations induce abnormal representations within the visual pathways, which are expected to trigger neuroplastic mechanisms e.g. to resolve potential sensory conflicts1,12,13,14,15,16,17,18,19. Finally, the dataset presented here can be used to advance tractography methods development. The field of brain tractography has faced a long-lasting challenge commonly referred to as the “crossing fibers problem” or simply CFP18,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35. CFP can lead to poor estimates of the number of crossing fibers through brain regions containing multiple fiber populations36,37. It has been established that up to 90% of total brain white matter volume might have crossing fibers38. Advancing methods for accurate tracking in regions with crossing fibers is fundamental in clarifying the role of white matter in human health and disease18,39,40. As of today, several important approaches to tractography evaluation and validation have been proposed. These approaches can be classified into four primary categories: synthetic phantoms41,42, physical phantoms43, biological phantoms44,45, and statistical9,32,46,47,48,49. Most of these approaches have helped advance tractography methods, but major challenges remain30,31,42. The data made available here opens the possibility to assess crossing strength at the optic chiasm by first using anatomical data (T1w, DWI) to model the crossing at the optic chiasm and cross-validating the proposed findings with functional estimates (fMRI) of misrouting based on the BOLD signal4,50,51 (see Suppl. Table 1). This provides a unique opportunity for testing novel tractography methods that assess crossing strength by providing an independent modality for their evaluation.
Methods
MRI Data sources
The described MRI data was analyzed in previously published studies4,50,51, where acquisition protocols and data properties are detailed.
Participants
A single participant with achiasma, a single participant with chiasm hypoplasia, 9 participants with diagnosed albinism, and 8 control participants [no neurological or ophthalmological history; normal visual acuity (≥1.0 with Freiburg Visual Acuity Test52) and normal stereo vision53,54] were recruited for the MRI measurements. Each participant was instructed about the purpose of the study and the methods involved and gave written informed study participation and data sharing consent. The study was approved by the Ethics Committee of the Otto-von-Guericke University Magdeburg, Magdeburg, Germany. The patients and control participants underwent ophthalmological examination (Suppl. Table 1), which incorporated methods described in55,56.
MRI Data acquisition
MRI data was acquired with a Siemens MAGNETOM Prisma 3 Tesla scanner with the Syngo MR D13D software and a 64-channel head coil. The acquisition protocol for T1w and DW data was initiated by a localizer scan, followed by a whole-brain T1w 3D-MPRAGE scan and two DW scans - respectively with anterior-posterior (A-P) and posterior-anterior (P-A) phase-encoding direction. T1w and DW images were collected during a single continuous scanning session, fMRI data was acquired in separate sessions (patients data was acquired on two consecutive days). T1w images were obtained in sagittal orientation using a 3D-MPRAGE sequence (TE/TR = 4.46/2600 ms, TI = 1100 ms, flip angle = 7°, resolution = 0.9 × 0.9 × 0.9 mm3, FoV: 230 × 230 mm²; image matrix: 256 × 256 × 176, acquisition time = 11 min:06 s57) and corrected simultaneously during acquisition for gradient nonlinearity distortions. Each individual’s T1w data was screened by a radiologist for unexpected abnormalities present in the data. Apart from the abnormalities given in Methods (Participants), no clinically relevant abnormalities were detected.
DWI were acquired with Echo-Planar Imaging (EPI) sequence (TE/TR = 64.0/9400 ms, b-value 1600 s/mm², resolution 1.5 × 1.5 × 1.5 mm³, FoV 220 × 220 mm², anterior to posterior (A-P) phase-encoding direction, acquisition time = 22 min:24 s, no multi-band). The b-value was chosen with regard to reported optimal values for resolving two-way crossing58 (1500–2500 s/mm2). Scans were performed with 128 gradient directions, so the obtained DWI data can be described as High Angular Resolution Diffusion Imaging7 (HARDI) data. The redundantly high number of gradient directions for the maximal angular contrast provided by a b-value of 1600 s/mm2 supported residual bootstrapping. This enhanced the effective signal-to-noise ratio (SNR), which is an important feature considering the reduced SNR of the DWI of the optic chiasm. The gradient scheme, initially generated using E. Caruyer’s tool for q-space sampling59 for 3 shells acquisition, was narrowed to the single shell in order to address the acquisition time constraints. DW volumes were evenly intersected by 10 non-diffusion weighted (b-value = 0, hereafter referred to as b0) volumes for the purpose of motion correction. The second DW series were acquired with reversed phase-encoding direction in comparison to the previous scan, specifically posterior to anterior (P-A). Apart from that, all scan parameters were identical to ones corresponding to the preceding acquisition. Acquisition of two DW series with opposite phase-encoding directions enhanced the correction of geometrically induced distortions60. Furthermore, the additional scans improve the signal-to-noise ratio (SNR) of the total DWI data.
fMRI data was acquired from 4 controls and 6 participants with albinism (see Suppl. Tables 1 and 2) with T2*-weighted EPI sequence (TE/TR = 30.0/1500 ms, flip angle = 70°, resolution 2.5 × 2.5 × 2.5 mm³, FoV 210 × 210 mm2, acquired with multi-band and in-plane acceleration factor = 2) during visual stimulation. Visual stimulation was performed in either the left, right or both visual hemifields in separate runs. A single repetition comprised 168 volumes acquired within 252 seconds. Each of these three stimulation conditions was repeated three times, resulting in a total of nine functional runs acquired within a single session. The visual stimulation is detailed in51. Briefly, it employed a moving high-contrast checkerboard pattern61 presented within the aperture of a drifting bar (width: 2.5°) within a circular aperture (radius: 10°). The bar aperture was moving in four directions (upwards, downwards, left, and right) across the stimulus window in 20 evenly spaced steps within 30 s. The sequence of the visual stimulation runs was interspersed by equally long (30 s) mean luminance blocks with zero contrast. The stimuli, generated with Psychtoolbox62,63 in MATLAB (Mathworks, Natick, MA, USA), were projected onto a screen (resolution 1140 × 780 pixels) placed at the magnet bore. The participants viewed the stimuli monocularly with their dominant eye (see Suppl. Tables 1 and 2) via an angled mirror at a distance of 35 centimeters, and were instructed to fixate on a central dot and respond with a button press to dot color changes.
Data preprocessing
Data preprocessing was mainly performed online, using web services available on the brainlife.io platform (https://brainlife.io), with a few exceptional steps done offline. The source code for the Apps used for online preprocessing is to be found at https://github.com/brainlife. The offline preprocessing involved conversion of DICOM data to NIfTI format, data anonymization, and, in the case of DW data, correction of gradient nonlinearity distortions and alignment to T1w image. The scripts for all of the offline preprocessing steps are available on https://github.com/rjpuzniak/CHIASM. Data preprocessing was meant to provide minimally processed data and standardized T1w, DWI, and fMRI data files.
The following software packages were used for data preprocessing: MRtrix28,64, FMRIB’s FSL65,66,67, ANTs68,69, FreeSurfer70, dcm2niix71, MIPAV72, VISTASOFT package (including mrVista and mrDiffusion tools; https://github.com/vistalab/vistasoft), AFNI73, fMRIPrep74, Mindboogle75, Nipype76, Nilearn77 and Human Connectome Project gradunwarp package (https://github.com/Washington-University/gradunwarp). The computing environment of the brainlife.io uses Docker (https://docker.com) as well as Singularity containers (https://sylabs.io/singularity/ and https://singularity.lbl.gov).
Preprocessing of the T1w data
In the offline preprocessing steps, T1w images were converted into the NIfTI format using dcm2niix71 and subsequently anonymized through the removal of facial features using mri_deface algorithm78 from FreeSurfer 6.0.0. Anonymized T1w images were aligned to the Anterior Commissure - Posterior Commissure (ACPC) plane using the mrAnatAverageAcPcNifti.m command from mrDiffusion package (https://github.com/vistalab/vistasoft/wiki/ACPC-alignment). The outcome T1w images were used as the reference image for the coregistration of DWI. Further, T1w images were automatically segmented into five-tissue-type (cerebrospinal fluid, white, grey, and subcortical grey matter, and eventual pathological tissue; 5TT) segmented images79 through the use of commands from FSL 6.0.365,80,81,82. Finally, the T1w data was uploaded to brainlife.io, where it was segmented once again using FreeSurfer 7.1.1 (Fig. 2a, top row). Detailed information about the preprocessing code is provided in the Code Availability section (Table 1).
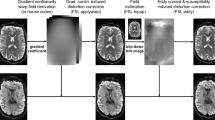
Qualitative overview of preprocessing for a representative participant (CON1). (a) Axial view of surfaces of white and pial matter (blue and red color, respectively) overlaid on T1w (top row), non-diffusion weighted (b0; middle row) and fMRI (T2*; bottom row) images. (b) Axial (top row), sagittal (middle row) and coronal (bottom row) views of fMRI images. The red contour marks brain mask estimated from BOLD signal, magenta contour marks combined CSD and WM masks, where voxels with partial GM volume were removed, blue contour marks the top 2% most variable voxels within brain mask.
Preprocessing of the DWI data
The DICOM DWI data preprocessing followed the well-established outline proposed by the Human Connectome Project (HCP) consortium83. Initially, DW files were converted offline into NIfTI format using dcm2niix and uploaded to brainlife.io. Next, the DWI data was corrected online for the Rician noise using dwidenoise84,85 and for Gibbs ringing using mrdegibbs86 commands from MRtrix 3.0. The following step of preprocessing involved estimation of the susceptibility-induced off-resonance field in the DW data with FSL’s topup command60,65 using two DW series with opposite phase encoding directions. The output of topup was subsequently fed to eddy command87 in order to correct for susceptibility- and eddy current-induced off-resonance field, as well as the motion correction. The topup and eddy command were implemented through dwifslpreproc command from MRtrix 3.0, which final output was a single file containing the corrected DW series. In the final online preprocessing step, the data was corrected for the field biases using dwibiascorrect from MRtrix 3.0, which, in turn, used the N4 algorithm from ANTS88 in order to estimate the MR field inhomogeneity. At this stage, the DWI data were downloaded and corrected in an offline mode for the gradient nonlinearities distortions. This step involved using the gradunwarp package and information about Legendre coefficients in spherical harmonics for the scanner’s gradient coil, provided by the vendor (stored in the https://github.com/rjpuzniak/CHIASM repository). As for the final step of preprocessing, the DWI data was coregistered to T1w data using the Boundary-Based Registration (Fig. 2a, middle row). At first, the transformation matrix from DWI to T1w image space was estimated with the epi_reg command from FLIRT89,90,91, a part of FSL 6.0.3 package. The transformation matrix was subsequently applied to DWI data by the flirt command from the same package, and to the corresponding b-vectors by shell script from HCP repository (https://github.com/Washington-University/HCPpipelines/blob/master/global/scripts/Rotate_bvecs.sh). The resulting data, in NIfTI format, have been uploaded to brainlife.io and were published as a preprocessed DW data set. Detailed information about the preprocessing code is provided in the Code Availability section (Table 2).
Preprocessing of the fMRI data
The fMRI data was converted into NIfTI format using dcm2niix. Subsequent preprocessing was performed online using two Apps wrapping the fMRIPrep tool74: fMRIPrep - Surface output92 (which output data as the surface vertices) and fMRIPrep - Volume output93 (which output data in volumetric format). The preprocessing, in both cases, involved correction for susceptibility distortions using antsRegistration from ANTs 2.3.3, registration to T1w image using bbregister command from FreeSurfer 6.0.1 (Fig. 2a, bottom row), slice-time correction using 3dTshift from AFNI and correction for head-motion. Additionally, the BOLD (blood-oxygen-level-dependent) data was subject to Component-Based Noise Correction (CompCor)94, which uses information principal components from noise-driven regions (defined as top 2% variable voxels in BOLD image; Fig. 2b, blue contour) in order to reduce the standard deviation of resting-state BOLD data. The noise-driven regions selection was limited only to voxels not affected by gray matter partial volume (Fig. 2b, pink contour). The output files created during preprocessing with fMRIPrep Apps are described in detail in section Data Records. Detailed information about the preprocessing pipeline is provided in the HTML report files generated by fMRIPrep application in the Data Records, while the code is provided in the Code Availability section (Table 3).
Drawing of the optic chiasm mask
Due to the limited accuracy of the automatically generated optic chiasm mask (Fig. 3a), manual segmentation was necessary to ensure the proper anatomical definition of the structures in each participant. The procedure comprised the following steps:
-
1)
Initial segmentation of voxels unambiguously belonging to the optic chiasm (i.e. outer voxels affected by partial volume effects were excluded). This segmentation was performed only in an axial view and was done in multiple slices covering optic nerves, optic chiasm,and optic tract.
-
2)
Second step where voxels affected by partial volume effects, previously omitted, were included. The two main criteria for the inclusion of candidate voxels were (a) relative intensity (compared to neighboring voxels identified in the previous step) and the coherence/continuity of the optic chiasm structure (already defined by voxels selected in the previous step).
-
3)
A third and final step involved corrections performed in axial, coronal, and sagittal views at the same time. The main criterion here was to assure the continuous borders.
Masks and regions of interest used to define the location of the optic chiasm, tract, and nerve. Left-hand column, the pseudo-axial view of the anatomical image of exemplary control participant (CON6) and overlaid optic-chiasm white matter mask before (a) and after (b) manual correction. Middle column, four representative ROIs covering the optic nerves and optic tracts in pseudo-axial (c) and sagittal (d) view. Right-hand column, four representative ROIs covering a cross-section of optic nerves (e) and optic tracts (f) in pseudo-coronal view.
The outcome masks covered posterior optic nerves, whole optic chiasm, and anterior optic tracts (Fig. 3b) and were used for correction of white matter definition in previously generated 5TT masks. The corrected white matter masks were extracted from 5TT images using mrconvert command, transformed to the space of T1w image (in order to ensure matching of QForm and SForm transformation matrices) with flirt command from FSL and mrconvert commands from MRtrix, and uploaded to the brainlife.io. Detailed information about the code for preprocessing can be found in the Code Availability section (Table 4).
Drawing regions of interest in the optic chiasm
Four ROIs were manually drawn and curated in each participant (Fig. 3c) on the T1w images. These ROIs identified the anterior and posterior aspects of the optic chiasm in each individual. The two anterior ROIs identified the location of the left and right optic nerve (Fig. 3c,e,yellow, and magenta). The posterior ROIs identified the left and right optic tract (Fig. 3c,f, cyan, and red). Once created, ROIs were transformed to the space of T1w image with mrconvert and mrtransform and thresholded with mrthreshold commands from MRtrix (in order to remove interpolation artifacts) and uploaded to the brainlife.io. Detailed information about the code for preprocessing can be found in the Code Availability section (Table 4).
Diffusion signal reconstruction, tractography, and statistical evaluation
The whole-brain tractography was performed using MRtrix 0.2.1264. The tractography was based on diffusion tensor (DT) and constrained spherical deconvolution (CSD) models and was performed using both deterministic and probabilistic methods25,28,95,96,97,98,99. The DT model was used for deterministic tracking, in the case of CSD both deterministic and probabilistic tracking was applied. The tractography utilized Anatomically-Constrained Tractography79, where the tracking was restricted to gray matter-white matter boundary from the newly created FreeSurfer 7.1.1 segmentation of the provided T1w image to ensure the agreement of the obtained tracks with the underlying anatomical structure. Additionally, an Ensemble Tractography100 framework was used, which negates bias of different parameter options in tractography by merging results of several tracking approaches, with each generating tracks with different properties18,41,101,102,103,104. Merging all tractograms mitigates the bias introduced from the variability of individual tractography parameters. Adhering to this, the final tractogram consisted of streamlines generated using: (A) deterministic tracking97 based on Diffusion Tensor95,105, (B) deterministic tracking25 based on Constrained Spherical Deconvolution model106 (CSD), and (C) probabilistic CSD-based tracking using iFOD2 algorithm28. The tractography was performed for several harmonic orders L = 2, 4, 6, 8, 10, 12, which were estimated using dwi2reponse tournier107 and dwi2fod msmt_csd108 commands from MRtrix 3.0. The full set of parameters guiding tractography is as follows: step size 0.15 mm for (A) and (B), 0.75 mm for (C); minimal length 7.5 mm, maximal length 200 mm, maximum angle between consecutive steps 5, 10, 20, 40, 80°, 15,000 fibers per parameters combination. The tracking has been performed using Ensemble Tracking (dwi)109 brainlife.io application.
The whole-brain tractogram was evaluated and optimized using the Linear Fasiscle Evaluation method9,49 (LiFE). Over the course of the evaluation process, every streamline is assigned a weight indicating its unique contribution in explaining the measured diffusion signal based on a tensor fit of the preprocessed diffusion data. Streamlines with non-zero weights are deemed as significant, while others are being discarded. The brainlife.io application implementing LiFE evaluation9 can be found at110.
Data Records
The data includes T1w, DW, and (if available) fMRI images of: single participant with achiasma (ACH1), single participant with chiasma hypoplasia (CHP1), 9 participants with albinism (ALB1 - ALB9) and 8 control participants (CON1 - CON8). The data from control participants are provided under an open license. To assure anonymity of the participants with clinical conditions, their data are made available upon direct request (as regulated by the Data Use Agreement, Suppl. Box 1).
The data is publicly accessible via brainlife.io platform11 at https://doi.org/10.25663/brainlife.pub.9111. When downloaded, the files are organized as defined by brainlife.io DataTypes (https://brainlife.io/docs/user/datatypes/ and https://brainlife.io/datatypes), and, if applicable, as the most updated version of the Brain Imaging Data Structure specification112 (BIDS). Due to the developmental nature of the BIDS format, at the present time, it does not support all the data derivative types presented here; the data records detailed below are presented according to brainlife.io Data Types. The data files stored for each subject on brainlife.io can be divided into three general categories: (A) source data, which consist of anonymized and aligned to the anterior commissure - posterior commissure (AC-PC) space T1w image, raw DW and fMRI data in NIfTI format, (B) preprocessed data, which consist of preprocessed DW and fMRI data, as described in Data Preprocessing section and (C) data derivatives, as described in Data Derivatives section. Additionally, the fMRI NIfTI data stored on brainlife.io are provided together with (D) MrVista.mat files (further referred to as “fMRI meta-files”), which are necessary for the analysis of the former. Those files are stored in a separate Open Science Framework (OSF) repository: https://doi.org/10.17605/osf.io/XZ29Q113 and are described in detail in the ‘fMRI meta-files’ section.
Source data
Source data (raw) files consist of two DWI datasets, one T1w set per participant, and, in the case of 6 participants from the albinism group and 4 controls, fMRI T2*-weighted images.
DW source data
Source DWI data covers two DW series acquired with opposite phase encoding directions (PEDs) - Anterior-Posterior (AP, Box 1a) and Posterior-Anterior (PA, Box 1b), as indicated by the tags.
fMRI source data
The source fMRI data is available for 6 participants with albinism (ALB1, ALB5, ALB6, ALB7, ALB8 and ALB9) and 4 controls (CON1, CON2, CON3 and CON8) and incorporates BOLD series acquired in 6 runs (3 runs corresponding to monocular stimulation of right visual hemifield, and 3 runs for left), except for ALB5 (3 runs for right and 2 for left hemifield) and CON1 (2 runs for right and 3 for left hemifield). Importantly, the fMRI files for the participant are stored in separate sessions (e.g. CON1/run4, Box 2).
T1w source data
Source T1w images (already anonymized and aligned to ACPC plane) were uploaded to participant’s main folder (e.g. CON1/, Box 3a), and, if applicable, all fMRI sessions (e.g. CON1/run4, CON1/run5 etc., Box 3b).
Preprocessed data
Preprocessed files are divided into 2 main categories: DW and fMRI files. The former are stored in each participant’s main folder, whereas fMRI files, if provided, are stored in folders corresponding to separate sessions.
DW preprocessed data
Preprocessed DW data consists of two files, tagged as “preprocessed” and “clean”. The ‘preprocessed’ tag marks the data (Box 4), which has been processed online, but lacks correction for gradient nonlinearity distortions and was not aligned to T1w image (those last two steps were performed offline) - the details are described in the “Data preprocessing” section.
The “clean” tag marks the data which has been completely preprocessed and aligned to T1w image (Box 5). Consequently, the files tagged as “clean” are recommended for further analyses.
fMRI preprocessed data
fMRI data processing was performed both for surface and volume representations of the data, and in both cases several output files were created.
In case of surface output92, the output files consist of surface vertices (3D mesh), for pial and white matter, as well as inflated representation, defined for both hemispheres (Box 6a), surface data in NIfTI format containing measures at each vertices (Box 6b), surface time series data in CIFTI format (Box 6c), HTML preprocessing report (Box 6d), volumetric mask of brain (Box 6e) and confounds (nuisance regressors) representing fluctuations with a potential non-neuronal origin, identified using CompCor (Box 6f).
Volumetric preprocessing output93 shares a majority of files with surface preprocessing (Box 6c–f), except for files containing data in surface representation (Box 6a,b). Furthermore, two additional files are included in the volumetric input: brain mask based on BOLD image (Box 7a) and volumetric BOLD image (Box 7b).
Data derivatives
Provided data derivatives consist of manually curated and automatically generated white matter masks, custom ROIs, T1w image segmentation, tractograms, and filtered tractograms.
Manually curated and automatically generated masks
White matter masks manually curated in the optic chiasm region (Fig. 3b; creation described in “Data preprocessing” section) sampled to match the original T1w image resolution (Box 8):
Additional white matter mask (created from FreeSurfer segmentation of white matter), generated by brainlife.io App performing tractography109 (Box 9):
Custom ROIs
We provide a set of four masks covering the left and right optic nerve and left and right optic tract (Fig. 3e). ROIs (Box 10) are provided as individual NIfTI files containing the left and right optic tract (OT) and the left and right optic nerve (ON). Data in the files contain a ‘1’ for each voxel within the ROIs, 0 otherwise. These ROIs can be used for tracking start-end.
T1w image segmentation
A FreeSurfer (v 7.1.1.) segmentation of T1w image, which was generated as a part of data preprocessing (see section “Data preprocessing”) and was used in tractography (Box 11a) and fMRI data preprocessing (Box 11b) is provided exclusively in brainlife.io Data Types format.
Tractograms and filtering results
The results of tractography performed for the purpose of technical validation of the DW data (Box 12a) and results of its filtering with LiFE (Box 12b) are provided as part of the repository:
fMRI meta-files
fMRI meta-files (for a subset of 6 albinism and 4 control participants, for which fMRI source data are provided) are available on the Open Science Framework (OSF) platform: https://doi.org/10.17605/osf.io/XZ29Q113. The files are in MATLAB format.mat and provide all the necessary information for performing the retinotopy data analysis using the MrVista package (https://github.com/vistalab/vistasoft). For each participant there is a total of 6 files: description of visual stimulus presented during left and right visual hemifield stimulation (Box 13a,b, respectively), full information about the acquisition parameters, participant’s response and stimulus for left and right visual hemifield stimulation (Box 13c,d, respectively; this also includes contents of visual stimulus corresponding to given hemifield stimulation as in Box 13a,b, respectively), file containing all parameters necessary for initialization of session in MrVista (such as paths to files required in analysis; Box 13e) and mrSession file storing all information about the analysis (Box 13f):
Technical Validation
This section provides a quality assessment of the published DW and fMRI data and is based on a previously published approach11 comprising qualitative and quantitative measures.
Qualitative assessment
The qualitative assessment involves (A) demonstration of the quality of alignment between anatomical, DWI, and fMRI images, and (B) demonstration of reconstruction of diffusion signal and tractography in the optic chiasm.
Registration of anatomical, DW, and fMRI data
A critical step in data preprocessing is to obtain the precise alignment between images of various modalities (T1w, DWI, and fMRI images; for a detailed description see Methods). The quality of registration is demonstrated by overlaying FreeSurfer’s 7.1.1 segmentation contours of white and pial matter (Fig. 2a; blue and red colors, respectively) on top of T1w image (from which they were derived; Fig. 2a, top row), DWI (Fig. 2a, middle row) and BOLD image (Fig. 2a, bottom row) for a representative participant (CON1).
Diffusion signal reconstruction and tractography in the optic chiasm
Considering the role of optic chiasm malformations as a major factor driving group differences, the quality assessment included diffusion signal modeling and tractography in this structure. Figure 4a displays representative optic chiasms in the T1w images, next to aligned dMRI b0 images (Fig. 4b). The DWI signal in each voxel was modeled using a CSD106,114 model in a process where an estimated single fiber response (SFR; Lmax = 6) was used as a deconvolution kernel in the process of calculating the fiber orientation distribution function20 (FOD; Lmax = 12) from acquired DWI. Figure 4c demonstrates the fit of calculated FOD in an optic chiasm region. Figure 4d demonstrates tracking results in the region of the optic chiasm. Presented results are limited only to probabilistic CSD-based tractography (iFOD2 algorithm, step size = 0.75 mm, FOD cutoff amplitude = 0.06, maximum angle between successive steps = 45°) based on already calculated ODFs (Lmax = 12; Fig. 4c), which was done between pairs of ROIs and within manually corrected white matter mask defined in the Data derivatives paragraph of the Methods section. For the purpose of clarity, only 0.25% of the total number of generated streamlines is displayed.
Diffusion signal reconstruction and tractography. The rows correspond to exemplary participants representing, respectively from top to bottom: control (CON1), albinism (ALB1), chiasma hypoplasia (CHP1) and achiasma (ACH1). (a) Pseudo-axial view of T1w image of the optic chiasm. (b) Pseudo-axial view of b0 image of the optic chiasm. (c) Pseudo-axial view of the anatomical image with overlaid estimated fiber orientation distribution function (FOD; Lmax = 12). (d) Pseudo-axial view of an anatomical image with overlaid results of tractography performed between pairs of ROIs defined in Methods.
Quantitative assessment
The quantitative validation includes (A) assessment of participants’ motion during DW data acquisition, (B) SNR in raw and preprocessed DW images, (C) modeling of DW data, (D) tractography, (E) temporal SNR of fMRI data, and (F) pRF-mapping.
Participant’s motion during DW data acquisition
The participants’ motion in DWI has been calculated for concatenated AP and PA series (acquired subsequently during a single scanning session, see Methods) by calculating the RMS of each voxels’ displacement using the Eddy command from FSL. The displacement calculation used the first acquired voxels as a reference for all volumes and included only voxels within the brain mask. The slow, yet steadily increasing drift visible for all participants (Fig. 5a) can be well tracked with b0 images intersecting DW series, which benefited motion correction. While, the lowest RMS was observed for the control group (which can be justified by the inclusion of trained control participants, well accustomed to MRI scanning), no proof for inequality of mean displacement RMS between participants with albinism and controls was found (Student’s t-test p-value = 0.11). It should be noted that the increased motion shown by achiasmatic participant ACH1 matches the observations from the fMRI scanning session, where the data had to be discarded due to extensive motion.
Quantitative assessment of motion, SNR, and modeling of DW data, as well as the quality of derived tractograms. (a) Left plot displays motion in the DW volumes (expressed as Root Mean Square of voxel displacements within brain mask, calculated with respect to the first volume) where 138 volumes with AP PED were followed by 138 volumes with opposite PED. Markers indicate the values calculated for the b0 volumes. The right-hand plot displays the mean motion RMS calculated for each participant. The color code and markers shape correspond to groups: orange circle - control, magenta square - albinism, violet up-pointing triangle - achiasma, blue down-pointing triangle - chiasma hypoplasia. (b) SNR in b0 images for each participant calculated from callosal voxels (selected from raw AP PED DW series, raw PA PED DW series, and corrected DWI) and optic chiasm (only for corrected DWI). (c) Values of zero phase (m = 0) coefficients of SFR calculated for each participant. (d) Results of evaluation of tractogram using LiFE method. Y-axis displays the number of fascicles remaining after filtering with LiFE, x-axis displays voxel-wise error between original signal and the one predicted from optimized tractograms. The bright markers correspond to the published datasets, while the dimmed display results for representative datasets from other publicly available DW repositories (such as 3T and 7T HCP datasets).
The output data of eddy describing motion for each participant is available online on the Github repository (https://github.com/rjpuzniak/CHIASM/tree/main/Plots/Fig.5_Motion), where it is provided together with the MATLAB code of Fig. 5a.
SNR in DW data
In order to evaluate the quality of the DWI data, the signal-to-noise ratio (SNR) of raw and preprocessed images was measured. The computations were performed for b0 (Fig. 5b) and diffusion-weighted (along with X-, Y- and Z-axis; Suppl. Fig. 1) for corpus callosum and optic chiasm voxels. Specifically, the SNR was defined as the mean ratio of the signal in voxels (from the selected structure) to standard deviation of noise (measured in voxels outside the brain), as described in37,115. In the case of the corpus callosum, the SNR was calculated separately for b0 images of two raw DW series (one with AP PED and one with opposite PA PED) and for the fully preprocessed DW image (“corrected”). As expected, the comparable SNR of corrected images is increased while the preprocessing (unwarping) is performed (Fig. 5b).
Comparable analysis of SNR in optic chiasm was obstructed by the severe geometry-induced distortions present in this region, which introduce spatial warping of the chiasm. Although theoretically this problem can be addressed by using new sets of optic chiasm masks (created separately for images warped in AP and PA directions), drawing new masks on top of DW images in heavily distorted regions is practically extremely challenging and is highly likely to introduce inaccuracies. Instead, the SNR was calculated for the fully preprocessed (corrected) DWI images, where the optic chiasm mask was cropped from a manually curated white matter mask (Fig. 5b). The observed higher SNR in optic chiasm (compared to corpus callosum) could be due to multiple reasons that were not tested by the authors. We speculate that it might be the result of using a 64-channel Radio Frequency coil, which measures stronger signals from peripheral brain regions in comparison to deeper regions such as the corpus callosum. The brainlife.io application implementing the SNR computation in the corpus callosum can be found at116 which follows the outlined SNR calculation strategy presented in37,115.
Coefficients of the single fiber response (SFR) function
The quality of modeling of DWI was assessed by plotting the zero phase coefficient of the SFR function (calculated for Lmax = 6 with dwi2response command from MRtrix, which used a iterative algorithm for SFR voxels selection and response function estimation;107 Fig. 5c). The observed plot is in agreement with theoretical expectations, where successive non-zero (even) terms are of opposite signs and of decreasing absolute value.
Quality of tractography
Finally, the quality of the created tractogram has been assessed with the LiFE algorithm (see Diffusion signal reconstruction, tractography and statistical evaluation in Methods). Figure 5d demonstrates the correlation between the number of fascicles of non-zero weights (y-axis) and Connectome Root Mean Square Error (RMSE;9,103). The data points for all CHIASM participants demonstrate a high number of weighted fascicles (positively contributing to measured signal, y-axis on Fig. 5d) combined with a low connectome’s RMSE (measuring the discrepancy between signal predicted from weighted fascicles and measured signal, y-axis). Those measures replicate previous findings of high-quality diffusion scans (HCP/O3D11,117).
tSNR of fMRI data
The quality of fMRI images was assessed using a temporal SNR (tSNR) measure (Fig. 6). Specifically, the tSNR in the BOLD images was calculated for two areas - whole brain volume (derived from BOLD images with fMRIPrep - Volume Output93 App) and primary visual cortex (V1) mask derived from T1w image using Benson’s atlas118,119,120. The tSNR has been calculated using the code provided in121 (https://github.com/psychoinformatics-de/studyforrest-data-aligned). The mean values of tSNR calculated for participants with albinism and controls (51.0 and 57.7, respectively) correspond well to the tSNR previously reported for voxel volumes of 16.625 mm3 122.
BOLD image temporal SNR in V1 and whole-brain volume. (a) Histograms of tSNR calculated in whole brain volume (orange) and V1 region (defined according to Benson’s atlas; violet). (b) Whole brain (orange) and V1 (violet) masks overlaid on maps of tSNR calculated for representative participants (top - control participant CON1, bottom - participant with albinism ALB1).
Population Receptive Fields (pRF) Mapping
The pRF-mapping data derivatives and methods were described in a previous publication51. Briefly: The pRF sizes and positions can be estimated from the fMRI data and visual stimulus position time course. The BOLD response of each voxel can be predicted using a circular 2D-Gaussian model of the neuronal populations receptive field defined by three stimulus-referred parameters i.e. x0, y0, σ where x0 and y0 are the coordinates of the receptive field center and σ is it’s spread61. The predicted BOLD signal can be calculated by convolution of the stimulus sequence for the respective pRF-model and its three parameters with the canonical hemodynamic response function123. Based on this, the optimal pRF parameters can be found by minimizing the residual sum of squared errors (RSS) between the predicted and observed BOLD time-course. Only voxels will be retained whose explained variance exceeded a threshold of 15%.
Usage Notes
This data is organized according to BIDS standard112, whenever applicable (additionally, in all cases the data is organized according to brainlife.io Data Types), and are stored in documented standard NIfTI format. The data is to be accessed at the brainlife.io computing platform either by (A) the web interface of brainlife.io and/or (B) a command-line interface (https://github.com/brainlife/cli). CLI offers means to query and download partial or full data. This utility is further expanded when using a web interface, which in addition to selection and download of data allows for online processing with provided brainlife.io Apps Table 5.
Code availability
The processing was performed mainly using brainlife.io services (https://brainlife.io/apps), which together with the code are available online. The offline preprocessing was performed with freely accessible neuroimaging tools. The preprocessing steps, together with the references to the Software/Apps are provided separately for the T1w, DW, fMRI data, and hand-curated ROIs and mask (Tables 1–4, respectively). The web links to source code are provided separately in the Table 5.
References
Hoffmann, M. B. & Dumoulin, S. O. Congenital visual pathway abnormalities: a window onto cortical stability and plasticity. Trends Neurosci. 38, 55–65 (2015).
Kupfer, C., Chumbley, L. & Downer, J. C. Quantitative histology of optic nerve, optic tract and lateral geniculate nucleus of man. J. Anat. 101, 393–401 (1967).
Hoffmann, M. B., Lorenz, B., Morland, A. B. & Schmidtborn, L. C. Misrouting of the Optic Nerves in Albinism: Estimation of the Extent with Visual Evoked Potentials. Invest. Ophthalmol. Vis. Sci. 46, 3892–3898 (2005).
Ahmadi, K. et al. Triple visual hemifield maps in a case of optic chiasm hypoplasia. NeuroImage 215, 116822 (2020).
Hoffmann, M. B. et al. Plasticity and stability of the visual system in human achiasma. Neuron 75, 393–401 (2012).
Davies-Thompson, J., Scheel, M., Lanyon, L. J. & Barton, J. J. S. Functional organisation of visual pathways in a patient with no optic chiasm. Neuropsychologia 51, 1260–1272 (2013).
Tuch, D. S. et al. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn. Reson. Med. 48, 577–582 (2002).
Frank, L. R. Characterization of anisotropy in high angular resolution diffusion-weighted MRI. Magn. Reson. Med. 47, 1083–1099 (2002).
Caiafa, C. F. & Pestilli, F. Multidimensional encoding of brain connectomes. Sci. Rep. 7, 11491 (2017).
Wu, X. et al. High-resolution whole-brain diffusion MRI at 7T using radiofrequency parallel transmission. Magn. Reson. Med. 80, 1857–1870 (2018).
Avesani, P. et al. The open diffusion data derivatives, brain data upcycling via integrated publishing of derivatives and reproducible open cloud services. Sci. Data 6, 69 (2019).
Guillery, R. W. Neural abnormalities of albinos. Trends Neurosci. 9, 364–367 (1986).
Morland, A. B., Baseler, H. A., Hoffmann, M. B., Sharpe, L. T. & Wandell, B. A. Abnormal retinotopic representations in human visual cortex revealed by fMRI. Acta Psychol. (Amst.) 107, 229–247 (2001).
Hagen, E. A. H. von dem, Houston, G. C., Hoffmann, M. B., Jeffery, G. & Morland, A. B. Retinal abnormalities in human albinism translate into a reduction of grey matter in the occipital cortex. Eur. J. Neurosci. 22, 2475–2480 (2005).
Bridge, H. et al. Changes in brain morphology in albinism reflect reduced visual acuity. Cortex 56, 64–72 (2014).
Ogawa, S. et al. White matter consequences of retinal receptor and ganglion cell damage. Invest. Ophthalmol. Vis. Sci. 55, 6976–6986 (2014).
Allen, B., Spiegel, D. P., Thompson, B., Pestilli, F. & Rokers, B. Altered white matter in early visual pathways of humans with amblyopia. Vision Res. 114, 48–55 (2015).
Rokem, A. et al. The visual white matter: The application of diffusion MRI and fiber tractography to vision science. J. Vis. 17, 4 (2017).
Yoshimine, S. et al. Age-related macular degeneration affects the optic radiation white matter projecting to locations of retinal damage. Brain Struct. Funct. 223, 3889–3900 (2018).
Tournier, J.-D., Calamante, F., Gadian, D. G. & Connelly, A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage 23, 1176–1185 (2004).
Anderson, A. W. Measurement of fiber orientation distributions using high angular resolution diffusion imaging. Magn. Reson. Med. 54, 1194–1206 (2005).
Peled, S., Friman, O., Jolesz, F. & Westin, C.-F. Geometrically Constrained Two-Tensor Model for Crossing Tracts in DWI. Magn. Reson. Imaging 24, 1263–1270 (2006).
Staempfli, P. et al. Resolving fiber crossing using advanced fast marching tractography based on diffusion tensor imaging. NeuroImage 30, 110–120 (2006).
Descoteaux, M., Angelino, E., Fitzgibbons, S. & Deriche, R. Regularized, fast, and robust analytical Q-ball imaging. Magn. Reson. Med. 58, 497–510 (2007).
Descoteaux, M., Deriche, R., Knosche, T. R. & Anwander, A. Deterministic and Probabilistic Tractography Based on Complex Fibre Orientation Distributions. IEEE Trans. Med. Imaging 28, 269–286 (2009).
Wedeen, V. J. et al. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. NeuroImage 41, 1267–1277 (2008).
Dell’Acqua, F. et al. A modified damped Richardson–Lucy algorithm to reduce isotropic background effects in spherical deconvolution. NeuroImage 49, 1446–1458 (2010).
Tournier, J.-D., Calamante, F. & Connelly, A. MRtrix: Diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol. 22, 53–66 (2012).
Cheng, J., Deriche, R., Jiang, T., Shen, D. & Yap, P.-T. Non-Negative Spherical Deconvolution (NNSD) for estimation of fiber Orientation Distribution Function in single-/multi-shell diffusion MRI. NeuroImage 101, 750–764 (2014).
Thomas, C. et al. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc. Natl. Acad. Sci. 111, 16574–16579 (2014).
Reveley, C. et al. Superficial white matter fiber systems impede detection of long-range cortical connections in diffusion MR tractography. Proc. Natl. Acad. Sci. 112, E2820–E2828 (2015).
Rokem, A. et al. Evaluating the Accuracy of Diffusion MRI Models in White Matter. PLOS ONE 10, e0123272 (2015).
Shi, Y. & Toga, A. W. Connectome imaging for mapping human brain pathways. Mol. Psychiatry 22, 1230–1240 (2017).
Aydogan, D. B. et al. When tractography meets tracer injections: a systematic study of trends and variation sources of diffusion-based connectivity. Brain Struct. Funct. 223, 2841–2858 (2018).
Schilling, K. G. et al. Limits to anatomical accuracy of diffusion tractography using modern approaches. NeuroImage 185, 1–11 (2019).
Oouchi, H. et al. Diffusion Anisotropy Measurement of Brain White Matter Is Affected by Voxel Size: Underestimation Occurs in Areas with Crossing Fibers. Am. J. Neuroradiol. 28, 1102–1106 (2007).
Jones, D. K., Knösche, T. R. & Turner, R. White matter integrity, fiber count, and other fallacies: The do’s and don’ts of diffusion MRI. NeuroImage 73, 239–254 (2013).
Jeurissen, B., Leemans, A., Tournier, J., Jones, D. K. & Sijbers, J. Investigating the prevalence of complex fiber configurations in white matter tissue with diffusion magnetic resonance imaging. Hum. Brain Mapp. 34, 2747–2766 (2012).
Wandell, B. A. Clarifying Human White Matter. Annu. Rev. Neurosci. 39, 103–128 (2016).
Pestilli, F. Human white matter and knowledge representation. PLoS Biol. 16 (2018).
Fillard, P. et al. Quantitative evaluation of 10 tractography algorithms on a realistic diffusion MR phantom. NeuroImage 56, 220–234 (2011).
Maier-Hein, K. H. et al. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 8, 1349 (2017).
Perrin, M. et al. Validation of q-ball imaging with a diffusion fibre-crossing phantom on a clinical scanner. Philos. Trans. R. Soc. B Biol. Sci. 360, 881–891 (2005).
Roebroeck, A. et al. High-resolution diffusion tensor imaging and tractography of the human optic chiasm at 9.4 T. NeuroImage 39, 157–168 (2008).
Spees, W. M. et al. MRI-based assessment of function and dysfunction in myelinated axons. Proc. Natl. Acad. Sci. 115, E10225–E10234 (2018).
Daducci, A., Palù, A. D., Lemkaddem, A. & Thiran, J. A convex optimization framework for global tractography. in 2013 IEEE 10th International Symposium on Biomedical Imaging 524–527, https://doi.org/10.1109/ISBI.2013.6556527 (2013).
Daducci, A., Dal Palù, A., Lemkaddem, A. & Thiran, J.-P. COMMIT: Convex optimization modeling for microstructure informed tractography. IEEE Trans. Med. Imaging 34, 246–257 (2015).
Smith, R. E., Tournier, J.-D., Calamante, F. & Connelly, A. SIFT: Spherical-deconvolution informed filtering of tractograms. NeuroImage 67, 298–312 (2013).
Pestilli, F. et al. LiFE: Linear Fascicle Evaluation a new technology to study visual connectomes. J. Vis. 14, 1122–1122 (2014).
Puzniak, R. J. et al. Quantifying nerve decussation abnormalities in the optic chiasm. NeuroImage Clin. 24, 102055 (2019).
Ahmadi, K. et al. Population receptive field and connectivity properties of the early visual cortex in human albinism. NeuroImage 202, 116105 (2019).
Bach, M. The Freiburg Visual Acuity test–automatic measurement of visual acuity. Optom. Vis. Sci. Off. Publ. Am. Acad. Optom. 73, 49–53 (1996).
Donzis, P. B., Rappazzo, J. A., Bürde, R. M. & Gordon, M. Effect of Binocular Variations of Snellen’s Visual Acuity on Titmus Stereoacuity. Arch. Ophthalmol. 101, 930–932 (1983).
Lang, J. I. & Lang, T. J. Eye Screening with the Lang Stereotest. Am. Orthopt. J. 38, 48–50 (1988).
Schmitz, B. et al. Configuration of the Optic Chiasm in Humans with Albinism as Revealed by Magnetic Resonance Imaging. Invest. Ophthalmol. Vis. Sci. 44, 16–21 (2003).
Hoffmann, M. B. et al. Visual Pathways in Humans With Ephrin-B1 Deficiency Associated With the Cranio-Fronto-Nasal Syndrome. Invest. Ophthalmol. Vis. Sci. 56, 7427–7437 (2015).
Mugler, J. P. & Brookeman, J. R. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE). Magn. Reson. Med. 15, 152–157 (1990).
Sotiropoulos, S. N. et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. NeuroImage 80, 125–143 (2013).
Caruyer, E., Lenglet, C., Sapiro, G. & Deriche, R. Design of Multishell Sampling Schemes with Uniform Coverage in Diffusion MRI. Magn. Reson. Med. 69, 1534–1540 (2013).
Andersson, J. L. R., Skare, S. & Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage 20, 870–888 (2003).
Dumoulin, S. O. & Wandell, B. A. Population receptive field estimates in human visual cortex. NeuroImage 39, 647–660 (2008).
Brainard, D. H. The Psychophysics Toolbox. Spat. Vis. 10, 433–436 (1997).
Pelli, D. G. The VideoToolbox software for visual psychophysics: transforming numbers into movies. Spat. Vis. 10, 437–442 (1997).
Tournier, J.-D. et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage 202, 116137 (2019).
Smith, S. M. et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 23, S208–S219 (2004).
Woolrich, M. W. et al. Bayesian analysis of neuroimaging data in FSL. NeuroImage 45, S173–S186 (2009).
Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W. & Smith, S. M. FSL. NeuroImage 62, 782–790 (2012).
Avants, B. B. et al. The optimal template effect in hippocampus studies of diseased populations. NeuroImage 49, 2457–2466 (2010).
Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54, 2033–2044 (2011).
Fischl, B. FreeSurfer. NeuroImage 62, 774–781 (2012).
Rorden, C. & Brett, M. Stereotaxic display of brain lesions. Behav. Neurol. 12, 191–200 (2000).
McAuliffe, M. J. et al. Medical Image Processing, Analysis and Visualization in clinical research. in Proceedings 14th IEEE Symposium on Computer-Based Medical Systems. CBMS 2001 381–386, https://doi.org/10.1109/CBMS.2001.941749 (2001).
Cox, R. W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. Int. J. 29, 162–173 (1996).
Esteban, O. et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116 (2019).
Klein, A. et al. Mindboggling morphometry of human brains. PLOS Comput. Biol. 13, e1005350 (2017).
Gorgolewski, K. et al. Nipype: A Flexible, Lightweight and Extensible Neuroimaging Data Processing Framework in Python. Front. Neuroinformatics 5 (2011).
Abraham, A. et al. Machine learning for neuroimaging with scikit-learn. Front. Neuroinformatics 8 (2014).
Bischoff‐Grethe, A. et al. A technique for the deidentification of structural brain MR images. Hum. Brain Mapp. 28, 892–903 (2007).
Smith, R. E., Tournier, J.-D., Calamante, F. & Connelly, A. Anatomically-constrained tractography: Improved diffusion MRI streamlines tractography through effective use of anatomical information. NeuroImage 62, 1924–1938 (2012).
Zhang, Y., Brady, M. & Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001).
Smith, S. M. Fast robust automated brain extraction. Hum. Brain Mapp. 17, 143–155 (2002).
Patenaude, B., Smith, S. M., Kennedy, D. N. & Jenkinson, M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage 56, 907–922 (2011).
Glasser, M. F. et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 80, 105–124 (2013).
Veraart, J. et al. Denoising of diffusion MRI using random matrix theory. NeuroImage 142, 394–406 (2016).
Veraart, J., Fieremans, E. & Novikov, D. S. Diffusion MRI noise mapping using random matrix theory. Magn. Reson. Med. 76, 1582–1593 (2016).
Kellner, E., Dhital, B., Kiselev, V. G. & Reisert, M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn. Reson. Med. 76, 1574–1581 (2016).
Andersson, J. L. R. & Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage 125, 1063–1078 (2016).
Tustison, N. J. et al. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010).
Jenkinson, M. & Smith, S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 5, 143–156 (2001).
Jenkinson, M., Bannister, P., Brady, M. & Smith, S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage 17, 825–841 (2002).
Greve, D. N. & Fischl, B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage 48, 63–72 (2009).
Levitas, D., Hunt, D., Faskowitz, J. & Hayashi, S. fMRIPrep - Surface Output. brainlife.io https://doi.org/10.25663/brainlife.app.267 (2019).
Levitas, D., Hunt, D., Faskowitz, J. & Hayashi, S. fMRIPrep - Volume Output. brainlife.io https://doi.org/10.25663/brainlife.app.160 (2019).
Behzadi, Y., Restom, K., Liau, J. & Liu, T. T. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage 37, 90–101 (2007).
Basser, P. J., Pajevic, S., Pierpaoli, C., Duda, J. & Aldroubi, A. In vivo fiber tractography using DT-MRI data. Magn. Reson. Med. 44, 625–632 (2000).
Tuch, D. S., Belliveau, J. W. & Wedeen, V. J. A Path Integral Approach to White Matter Tractography. in Proceedings of the 8th Annual Meeting of ISMRM 791 (2000).
Lazar, M. et al. White matter tractography using diffusion tensor deflection. Hum. Brain Mapp. 18, 306–321 (2003).
Behrens, T. E. J., Berg, H. J., Jbabdi, S., Rushworth, M. F. S. & Woolrich, M. W. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? NeuroImage 34, 144–155 (2007).
Sherbondy, A. J., Dougherty, R. F., Napel, S. & Wandell, B. A. Identifying the human optic radiation using diffusion imaging and fiber tractography. J. Vis. 8, 12.1–1211 (2008).
Takemura, H., Caiafa, C. F., Wandell, B. A. & Pestilli, F. Ensemble Tractography. PLOS Comput. Biol. 12, e1004692 (2016).
Bastiani, M., Shah, N. J., Goebel, R. & Roebroeck, A. Human cortical connectome reconstruction from diffusion weighted MRI: The effect of tractography algorithm. NeuroImage 62, 1732–1749 (2012).
Côté, M.-A. et al. Tractometer: Towards validation of tractography pipelines. Med. Image Anal. 17, 844–857 (2013).
Pestilli, F., Yeatman, J. D., Rokem, A., Kay, K. N. & Wandell, B. A. Evaluation and statistical inference for living connectomes. Nat. Methods 11, 1058–1063 (2014).
Neher, P. F., Descoteaux, M., Houde, J.-C., Stieltjes, B. & Maier-Hein, K. H. Strengths and weaknesses of state of the art fiber tractography pipelines – A comprehensive in-vivo and phantom evaluation study using Tractometer. Med. Image Anal. 26, 287–305 (2015).
Basser, P. J. & Pierpaoli, C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J. Magn. Reson. B 111, 209–219 (1996).
Tournier, J.-D., Calamante, F. & Connelly, A. Robust determination of the fibre orientation distribution in diffusion MRI: Non-negativity constrained super-resolved spherical deconvolution. NeuroImage 35, 1459–1472 (2007).
Tournier, J.-D., Calamante, F. & Connelly, A. Determination of the appropriate b value and number of gradient directions for high-angular-resolution diffusion-weighted imaging. NMR Biomed. 26, 1775–1786 (2013).
Jeurissen, B., Tournier, J.-D., Dhollander, T., Connelly, A. & Sijbers, J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. NeuroImage 103, 411–426 (2014).
Hayashi, S., Kitchell, L., McPherson, B. & Caron, B. Ensemble Tracking (dwi). brainlife.io https://doi.org/10.25663/bl.app.103 (2018).
Caron, B., Pestilli, F., Berto, G. & Hayashi, S. LiFE (dwi). brainlife.io https://doi.org/10.25663/bl.app.104 (2018).
Puzniak, R., McPherson, B. & Pestilli, F. The human brain albinism and achiasma dataset: A biological testbed for the crossing-fibers problem with ground truth. brainlife.io https://doi.org/10.25663/brainlife.pub.9 (2019).
Gorgolewski, K. J. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044 (2016).
Puzniak, R. & Pestilli, F. CHIASM, the human brain albinism and achiasma MRI dataset. Open Science Framework https://doi.org/10.17605/osf.io/XZ29Q (2021).
Descoteaux, M., Angelino, E., Fitzgibbons, S. & Deriche, R. Apparent diffusion coefficients from high angular resolution diffusion imaging: estimation and applications. Magn. Reson. Med. 56, 395–410 (2006).
Descoteaux, M., Deriche, R., Le Bihan, D., Mangin, J.-F. & Poupon, C. Multiple q-shell diffusion propagator imaging. Med. Image Anal. 15, 603–621 (2011).
Hunt, D. Compute SNR on Corpus Callosum. brainlife.io https://doi.org/10.25663/brainlife.app.120 (2018).
Van Essen, D. C. et al. The WU-Minn Human Connectome Project: an overview. NeuroImage 80, 62–79 (2013).
Benson, N. C. et al. The Retinotopic Organization of Striate Cortex Is Well Predicted by Surface Topology. Curr. Biol. 22, 2081–2085 (2012).
Benson, N. C., Butt, O. H., Brainard, D. H. & Aguirre, G. K. Correction of Distortion in Flattened Representations of the Cortical Surface Allows Prediction of V1-V3 Functional Organization from Anatomy. PLOS Comput. Biol. 10, e1003538 (2014).
Benson, N. C. & Winawer, J. Bayesian analysis of retinotopic maps. eLife 7, e40224 (2018).
Sengupta, A. et al. A studyforrest extension, retinotopic mapping and localization of higher visual areas. Sci. Data 3, 160093 (2016).
Triantafyllou, C. et al. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. NeuroImage 26, 243–250 (2005).
Friston, K. J. et al. Event-Related fMRI: Characterizing Differential Responses. NeuroImage 7, 30–40 (1998).
Hayashi, S. Freesurfer 7.1.1. brainlife.io https://doi.org/10.25663/brainlife.app.462 (2020).
McPherson, B. mrtrix3 preprocess. brainlife.io https://doi.org/10.25663/bl.app.68 (2018).
Acknowledgements
R.J.P., K.A., and M.B.H. were supported by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 641805 and by the German research foundation (DFG; HO2002/10-3). B.M. was partially supported by NIH 5T32MH103213-05 to William Hetrick. NSF IIS 1636893, NSF IIS 1912270, NIH NIBIB 1R01EB029272, NSF BCS 1734853 and a Microsoft Faculty Fellowship granted to F.P. We thank all participants in the project for their time and effort. We thank Soichi Hayashi for support with uploading the data on brainlife.io, and Denise Scheermann and Martin Kanowski for their contribution in the data acquisition.
Author information
Authors and Affiliations
Contributions
J.K., K.A., R.J.P., A.H., M.B.H., A.G., A.B.M., and I.G. acquired the data. T.L. performed screening of the data. R.J.P., B.M., F.P. and M.B.H. processed the data. R.J.P., J.K., B.M., M.B.H., and F.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Puzniak, R.J., McPherson, B., Ahmadi, K. et al. CHIASM, the human brain albinism and achiasma MRI dataset. Sci Data 8, 308 (2021). https://doi.org/10.1038/s41597-021-01080-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-01080-w