Abstract
With the advent of deep learning algorithms, fully automated radiological image analysis is within reach. In spine imaging, several atlas- and shape-based as well as deep learning segmentation algorithms have been proposed, allowing for subsequent automated analysis of morphology and pathology. The first “Large Scale Vertebrae Segmentation Challenge” (VerSe 2019) showed that these perform well on normal anatomy, but fail in variants not frequently present in the training dataset. Building on that experience, we report on the largely increased VerSe 2020 dataset and results from the second iteration of the VerSe challenge (MICCAI 2020, Lima, Peru). VerSe 2020 comprises annotated spine computed tomography (CT) images from 300 subjects with 4142 fully visualized and annotated vertebrae, collected across multiple centres from four different scanner manufacturers, enriched with cases that exhibit anatomical variants such as enumeration abnormalities (n = 77) and transitional vertebrae (n = 161). Metadata includes vertebral labelling information, voxel-level segmentation masks obtained with a human-machine hybrid algorithm and anatomical ratings, to enable the development and benchmarking of robust and accurate segmentation algorithms.
Measurement(s) | vertebra |
Technology Type(s) | computed tomography |
Factor Type(s) | imaging centre • scanner manufacturer |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.14716968
Similar content being viewed by others
Background & Summary
Numerous applications of computer-aided diagnostics (CADx) are currently being developed beginning to gradually reshape the future of radiological clinical practice and research1,2,3,4,5,6,7. In spinal imaging, different deep learning approaches have been used for vertebral labelling and segmentation tasks in the form of convolutional neural networks (CNN), graph convolutional networks (GCN) or point clouds (PC) to analyse bone structures8,9,10,11,12,13. Various applications of computer algorithms have shown great potential to detect vertebral fractures and to measure bone mineral density (BMD)13,14,15,16,17. However, most of these approaches are largely data dependent, as the algorithms require extensive datasets with corresponding metadata for the development, training and validation to enable efficient models18. Aiming at the task of improving automated quantification of spinal morphology and pathology by vertebral labelling and segmentation, the first iteration of the “Large Scale Vertebrae Segmentation Challenge” (VerSe 2019)17,19 was held at the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2019, Shenzhen, China). The segmentation challenge received considerable participation from the scientific community with more than 250 registrations and 20 participating teams19. As part of the VerSe 2019 challenge a large dataset was provided addressing the previous severe shortage of publicly-available, large, accurately annotated CT spine data in the community by releasing 160 CT image series and their voxel-level annotations comprised of a large variety in fields of view and spatial resolutions as well as spinal and vertebral pathologies20.
Building on the data, experience, and learning from the VerSe 2019 challenge, we proposed to organise a second iteration of the vertebrae segmentation challenge at the MICCAI 2020 in Lima, Peru. For the VerSe 2020 challenge, we aimed to substantially increase the existing dataset to 300 subjects. While retaining the richness of the VerSe 2019 dataset, the VerSe 2020 dataset was complemented with images from different institutions and four major scanner manufacturers. In addition, we aimed to include rare anatomical variants such as numeric aberrations and cervicothoracic or lumbosacral transitional vertebrae. As such variants have a low prevalence in the population, they are under-represented, if present at all, in unselected training datasets like that presented at the VerSe 2019 challenge. Consequently, deep learning-based algorithms fail in such cases. Thus, focus was given to enrich the VerSe 2020 dataset with rare anatomical variants to improve derived model performance as previously described21. We envisioned the creation of an extended annotated CT dataset to provide consistent and reliable ground truth data for algorithm training and benchmarking.
The proposed Verse 2020 dataset was released as part of the second iteration of the “Large Scale Vertebrae Segmentation Challenge” hosted at the MICCAI conference held in Lima, Peru (https://verse2020.grand-challenge.org/)19. The dataset was split into a training dataset, a public test dataset, and a private test dataset building on the preexisting VerSe 2019 dataset20 published for the MICCAI conference in 2019, with an overlap of 105 CT image series now comprising 319 image series of 300 subjects. To date, this dataset represents the largest publicly available CT imaging dataset of the spine with corresponding metadata including labelling information, voxel-level segmentations of all fully visualized vertebrae and definition of enumeration abnormalities and transitional vertebrae.
In summary, the successful segmentation challenges held at the MICCAI conferences in 2019 and 2020 based on these public datasets confirm, that reliable, fully-automated deep learning algorithms for segmentation of the spine can be trained and that algorithm performance benefits from large and diverse datasets.
Therefore, we regard the VerSe 2020 dataset as an important step towards clinical translation of CADx algorithms for spine imaging, which may soon supplement the radiologist’s work in daily routine. We are convinced that in the near future, patients will greatly benefit from CADx extracting even more relevant information from medical imaging than currently possible.
Methods
Subject selection
This retrospective evaluation of imaging data was approved by the local institutional review board and written informed consent was waived (Proposal 27/19 S-SR).
Inclusion criteria for the dataset: Subjects older than 18 years were included, who had received CT imaging of the spine showing a minimum of 7 fully visualized vertebrae without counting sacral vertebrae or transitional vertebrae. Exam dates were limited to the time between February 5th, 2016 and March 1st, 2020. The minimum required spatial resolution was defined as 1.5 mm in the craniocaudal direction, 1 mm in the anterior-posterior direction and 3 mm in the left-right direction to allow for a sufficient delineation of vertebral deformities22. As in VerSe 2019, traumatic fractures and bony metastases were excluded. Other osseous changes such as Schmorl nodes, hemangioma, degenerative changes, or the presence of foreign materials for kyphoplasty or spondylodesis intentionally remained part of the dataset to reflect the widest possible spectrum of spine morphology. Aiming at a > 100% increase compared to the 141 subjects from Verse 2019 and providing a dataset with 50% multivendor data and 50% anatomical variants, we composed a dataset consisting of 300 subjects, including 86 subjects from VerSe 2019 and 214 new subjects. In order to select new subjects, we searched the institutional picture archive and communication system (PACS) regarding two aspects: 1) CT studies imported from other institutions, that were acquired on scanner hardware different from that installed in our institution; 2) CT reports documenting the presence of spinal anatomical variants including enumeration abnormalities, transitional vertebrae or cervical ribs. In both queries, we aimed for a balanced composition of cases: From the first query, we selected 20 subjects from external Toshiba scanners, 20 subjects from external GE scanners and 30 subjects from external Siemens scanners. We added to 30 subjects from Ben Glocker’s23 dataset and 50 subjects from VerSe 2019 (10 from Siemens scanners and 40 from Philips scanners), to form the multivendor dataset with 150 subjects. The query for anatomical variants of spinal anatomy such as thoracolumbar and lumbosacral transitional vertebrae, cervical ribs, thoracic and lumbar short ribs as well as enumeration variants revealed 308 subjects. Adding to 36 VerSe 2019 subjects with anatomical variants we selected another 114 subjects to form the 150 subjects of the dataset with anatomical variants. The final dataset comprised 300 subjects with a total of 319 image series, as some subjects comprised two separate image series (e.g. thoracic spine and lumbar spine). Subject characteristics and data subset stratification are listed in Table 1. All selected imaging series were categorized regarding their primary attribute (e.g. Toshiba scanner, Castellvi grade 4 transitional vertebra, or numeric aberration with 4 lumbar vertebrae) and each subgroup was randomly split to the training, public validation and private test subsets as demonstrated in Fig. 1.
Composition of the VerSe 2020 dataset: original data derived from the preexisting VerSe 2019 dataset, the Glocker23 dataset and newly added subjects based on a pacs search for subjects with anatomical variants of the spine as well as images from different scanner vendors and imaging centers.
Despite the wide inclusion criteria, very rare variants might still be missing and osseous changes other than the included anatomical variants or cases with new foreign material may still limit its generalizability. Also, the sacrum including lumbar transitional vertebrae, i.e. vertebrae partially fused with the sacrum, have not been segmented in this dataset. Future works, potentially based on the presented data, will need to address complete segmentation of the spine including the sacrum. Another future focus may be the inclusion of more pathological changes, e.g. only two cases with half-vertebrae are included in VerSe 2020, metastases and dislocated traumatic fractures were excluded. In this regard, it first remains to be discussed how these pathologies should be labelled and segmented.
Vertebral segmentation
The labelling of the vertebrae and the segmentation process are essential steps in processing spine data. All subsequent analyses such as the detection and grading of fractures, calculation of bone mineral density but also analysis of spinal shape, curvature, and deformity such as scoliosis rely on these initial tasks.
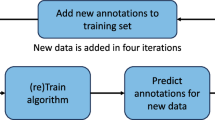
For the proposed dataset, we used a semi-automated in-house developed algorithm to generate segmentation masks of the vertebrae step-by-step as illustrated in Fig. 2: first, the CT input data was anonymized by conversion to Neuroimaging Informatics Technology Initiative (NIfTI) format (https://nifti.nimh.nih.gov/nifti-1). To ensure full anonymity, defacing was achieved by deleting the raw data in a manually segmented mask of the face. In case of the 86 re-used VerSe 2019 subject datasets, the spatial resolution has previously been reduced to 1 mm isotropic or in sagittal 2 mm/ 3 mm series to 1 mm in-plane resolution. In case of the 214 newly added patient datasets, the original resolution was kept. Second, a deep-learning framework (publicly accessible under: https://anduin.bonescreen.de) was used to label and segment individual vertebrae. In brief, at first a low-spatial resolution CNN is used for the detection of all osseous spinous structures, resulting in a low-resolution heatmap. This was used to automatically generate a spine bounding box containing the spine. Second, the Btrfly Net was used to label the vertebrae, with the option to manually correct the centroids if needed. Third, a U-Net was used in order to segment each vertebra defined by each single vertebral label in separate patches, defined by bounding boxes around the vertebral labels11,24,25,26. These vertebral masks were created at 1 mm3 isotropic resolution and subsequently merged into one multi-label segmentation mask with individually labelled vertebrae. Originally, both Btrfly Net and U-Net were trained with the VerSe 2019 data and have continuously been improved using the labels and segmentation masks derived from this dataset for repetitive training. Third, the segmentation output of the algorithm was manually corrected in a laborious process by two specifically trained medical students using the open-source software ITK-SNAP27. This manual correction was performed in the original imaging space, but was limited to an accuracy of 1 mm, similar to the output of the U-net; i.e. segmentation errors in smaller structures than 1mm were not corrected. Finally, corrections were reviewed and corrected or approved by a neuroradiologist to achieve the highest possible consistency of the presented segmentation masks. Despite the good performance of the baseline algorithm, the correction of on average 15 objects of interest with approximately 105 voxels took considerable effort.
Schematic overview on the image processing by the in-house developed algorithm publicly accessible under https://anduin.bonescreen.de (green boxes indicate fully automated algorithms) and the steps of manual interaction (blue boxes).
Anatomical variants
As anatomical variants of the spine can be frequently observed, both numeric and morphologic changes have been intentionally included in the VerSe 2020 dataset. Tins and Balain have reported numeric anatomical variations of the spine to be more frequent (7.7%) than transitional vertebrae (3.3%) with a tendency of male subjects to show more additional vertebrae and females to show more missing vertebrae28. Numeric aberrations of the cervical spine are rare, but additional cervical ribs can be observed in a clinical setting with a prevalence of 0.05%–6.1%29. According to the literature, variations of the thoracolumbar region are generally rarely reported and often overlooked, despite potential clinical implications e.g. for surgical planning. Wigh et al. differentiated the presence of accessory ossification centres and stump ribs as compared to the last pair of ribs at a thoracic vertebra based on length and morphology. Based on their data, any vertebra presenting with ribs shorter than 38 mm on both sides was defined as a transitional thoracolumbar vertebra30. While there are different approaches to classify transitional vertebrae, computer algorithms need a clear definition to which part of the spine a vertebra belongs. In our dataset, we used this threshold and vertebrae with at least one rib measuring more than 38 mm from the costovertebral joint to the osseous tip and showing a typical diagonal downward alignment were classified as thoracic vertebrae. In cases with both ribs smaller than 38 mm, e.g., when only small ossification centres were present with a horizontal alignment, the vertebra was considered lumbar. Of note, after primary application of rib length and rib angulation, the classification was consistent with the presumed vertebral body shape e.g., thoracic or lumbar shape of processes and facet joint orientation in all cases. In detail, joint facets at the thoracic spine usually show an angulation in medial-lateral direction, whereas at the lumbar spine an almost orthogonal orientation in anterior-posterior direction can be observed31,32. Using this rib-centred nomenclature, a consistent enumeration was achieved in almost all cases, while only 2 cases (581 and 606) remained ambiguous, decreasing the regular number of thoracic vertebrae and simultaneously increasing the number of lumbar vertebrae (7/11/6/5).
Variants of the lumbosacral region are frequently observed and according to Thawait et al.29 can be found in 4%–30% of examined cases. Regarding the lumbosacral region, we used the well-established Castellvi classification to lumbosacral transitional vertebrae mainly based on the morphology of the transverse process of the last lumbar vertebra and whether it is fused with the sacrum or not33. Owing to the Castellvi classification we did not segment vertebrae that showed partial fusion with the sacrum (Castellvi grades III and IV) and did not include them for further analysis. Of note, in our database search, no “lumbalized” sacral vertebra with four sacral vertebrae remaining fused could be identified.
If present in the scan, vertebrae were labelled starting at the first cervical or thoracic vertebra. If T1 was not visible within the scan’s field-of-view, the thoracic spine was considered to have 12 vertebrae, as enumeration errors in the thoracic spine are much less frequent compared to the lumbar spine28.
Data Records
Data repositories and storage
The Verse 2020 dataset comprises 319 CT image series from 300 subjects and 4142 vertebrae encompassing 581 cervical, 2255 thoracic, and 1306 lumbar vertebrae as listed in Table 1. The stratification of anatomical variants in the VerSe dataset along with the corresponding ratings is listed in Table 2. The Dataset with its division into test, training, and private data subsets has been made publicly available under the creative commons license CC BY-SA 4.0 hosted at the open science framework https://doi.org/10.17605/OSF.IO/T98FZ34. More information regarding the segmentation challenge algorithms submitted by the participants of the MICCAI VerSe challenges in 2019 and its second iteration in 2020 can be found at https://verse2019.grand-challenge.org/ and https://verse2020.grand-challenge.org/ as well as in the publication by Sekuboyina et al.19. Worth of note, there is an overlap of 86 subjects and 105 imaging series between the VerSe 2020 dataset and the previously published VerSe 2019 imaging dataset, which is separately available for public use under the creative commons license CC BY-SA 4.0 at https://doi.org/10.17605/OSF.IO/NQJYW. In a previous publication Loeffler et al. used the VerSe 2019 dataset to automatically detect and grade vertebral fractures and to calculate bone mineral density from a subset of the provided scans20.
Data structure and file formats
All medical imaging files were converted into Neuroimaging Informatics Technology Initiative (NIfTI) format (https://nifti.nimh.nih.gov/nifti-1). Segmentation masks are also saved in NIfTI format and labels of all 4142 segmented vertebrae are provided in JSON format.
For organizational reasons of the segmentation challenge, all CT data (NIfTI format) in the Verse dataset was separated into a training dataset (100 subjects), a public test dataset (100 subjects), and a private test dataset (100 subjects) as previously described and demonstrated in Fig. 1. Corresponding metadata is provided in the additional documents with the datasets.
CT imaging and scan provenience
CT scans included were intentionally chosen to be heterogeneous to ensure the best possible training and generalization of the algorithms. Therefore, data from the four major scanner vendors including Philips, Siemens, Toshiba, and GE from a variety of different multidetector CT scanner types of each vendor was included. The majority of images (45.8%) was acquired by Philips, 32.3% by Siemens, 6.3% by Toshiba, and 6.3% by GE scanners as shown in Table 1 and, on a patient-level in Supplemental Table 1. There is no information regarding the scanner vendor provenience of the Glocker dataset23, therefore they are listed as scans of “unknown” origin. Part of these examinations was carried out with additional administration of oral and/or intravenous contrast medium of various manufacturers. All included imaging series were based on edge-enhancing reconstructions, as this is the clinical standard for bone CT-imaging. Because the dataset comprises multi-centre imaging data from different scanner vendors, isotropic data was not available in all cases, replicating a typical clinical scenario.
Anatomical variants
From the overall 300 subjects with 319 scans, 90 scans (28.2%) demonstrated stump ribs. 135 scans visualized at least the lower part of the cervical spine and of these, 18 scans (13.3%) showed cervical ribs. From a total of 145 scans showing the complete thoracic spine, 131 scans presented with 12 thoracic vertebrae (90.3%), 8 scans showed 11 thoracic vertebrae (5.5%), and 6 scans demonstrated with 13 thoracic vertebrae (4.1%). From the 252 scans depicting the lumbosacral region, 165 showed 5 lumbar vertebrae (65.5%), 3 scans presented with 4 lumbar vertebrae (1.2%), and 85 scans showed 6 lumbar vertebrae (33.7%). Lumbosacral transitional vertebrae were graded according to the Castellvi classification, detailed in Table 2. In accordance with other authors and due to the difficulty of classifying Castellvi type I as well as the lack of clinical relevance, only Castellvi II–IV were classified35,36,37. Anatomical variants for each subject and scan are listed in Supplemental Table 1.
Technical Validation
The presented medical imaging data was derived from the institutional picture archiving system and therefore fully complies with the legal standards and quality controls for the acquisition of medical imaging in Germany and the European Union, as well as the industrial standards of the scanner vendors. Segmentation masks were prepared and annotated at voxel-level by a human-machine hybrid algorithm with manual checks and corrections by specifically trained medical students. Afterwards, the masks were reviewed, corrected, and finally approved by a neuroradiologist. The anatomical ratings were carried out by two neuroradiologists in a consensus reading. The resulting NIFTI datasets have successfully been processed by all algorithms of the 13 participants of the VerSe 2020 challenge. Of these, the best performing algorithm achieved a mean vertebral identification rate of 95.6% with a mean localisation error of less than 2 mm. Concerning segmentation, the best mean Dice score of 91.7%.
Usage Notes
Additional metadata e.g., additional anatomical ratings, fracture grading, bone mineral density measurements will be continuously updated and published at the open science framework website https://doi.org/10.17605/OSF.IO/T98FZ. Due to the medical nature of the data, it is published under the creative commons license CC BY-SA 4.0; however, upon reasonable request, respecting our patients, special arrangements for use and re-distribution can be made directly with the authors.
Code availability
A compiled, executable version of the in-house developed segmentation algorithm which was used to annotate the VerSe dataset is publicly accessible through a website (https://anduin.bonescreen.de). This final version has been trained with the entire VerSe dataset. The entire dataset and its helper code (including the data reading, writing, and evaluation scripts) can be accessed here: https://github.com/anjany/verse.
For an overview of the various algorithms submitted as part of the segmentation challenges held at the MICCAI conferences in 2019 and 2020 based on the public VerSe datasets, kindly refer to Sekuboyina et al.19. These nicely demonstrate the feasibility and potential of automated segmentation tasks and processing of spine CT imaging.
References
Kickingereder, P. et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. The Lancet Oncology 20, 728–740 (2019).
Sim, Y. et al. Deep Convolutional Neural Network-based Software Improves Radiologist Detection of Malignant Lung Nodules on Chest Radiographs. Radiology 294, 199–209 (2020).
Jungo, A., Balsiger, F. & Reyes, M. Analyzing the Quality and Challenges of Uncertainty Estimations for Brain Tumor Segmentation. Front Neurosci 14, 282 (2020).
Menze, B. H. et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE transactions on medical imaging 34, 1993–2024 (2015).
Rebsamen, M. et al. Divide and Conquer: Stratifying Training Data by Tumor Grade Improves Deep Learning-Based Brain Tumor Segmentation. Front Neurosci 13, 1182 (2019).
Rajpurkar, P. et al. CheXaid: deep learning assistance for physician diagnosis of tuberculosis using chest x-rays in patients with HIV. NPJ Digit Med 3, 115 (2020).
Kiani, A. et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med 3, 23 (2020).
Janssens, R., Zeng, G. & Zheng, G. Fully automatic segmentation of lumbar vertebrae from CT images using cascaded 3D fully convolutional networks. IEEE Int Symp Biomed Imaging 2018, 893–897 (2018).
Lessmann, N., van Ginneken, B., de Jong, P. A. & Isgum, I. Iterative fully convolutional neural networks for automatic vertebra segmentation and identification. Med Image Anal 53, 142–155 (2019).
Sekuboyina, A., Kukačka, J., Kirschke, J.S., Menze, B.H. & Valentinitsch, A. Attention-Driven Deep Learning for Pathological Spine Segmentation. 108-119 (Springer International Publishing, Cham, 2018).
Sekuboyina, A., Valentinitsch, A., Kirschke, J. & Menze, B. A Localisation-Segmentation Approach for Multi-label Annotation of Lumbar Vertebrae using Deep Nets. Preprint at https://arxiv.org/abs/1703.04347 (2017).
Ruiz-Espana, S., Diaz-Parra, A., Arana, E. & Moratal, D. A fully automated level-set based segmentation method of thoracic and lumbar vertebral bodies in Computed Tomography images. Annu Int Conf IEEE Eng Med Biol Soc 2015, 3049–3052 (2015).
Burns, J. E., Yao, J. & Summers, R. M. Vertebral Body Compression Fractures and Bone Density: Automated Detection and Classification on CT Images. Radiology 284, 788–797 (2017).
Burns, J. E., Yao, J., Munoz, H. & Summers, R. M. Automated Detection, Localization, and Classification of Traumatic Vertebral Body Fractures in the Thoracic and Lumbar Spine at CT. Radiology 278, 64–73 (2016).
Pickhardt, P. J. et al. Population-based opportunistic osteoporosis screening: Validation of a fully automated CT tool for assessing longitudinal BMD changes. Br J Radiol 92, 20180726 (2019).
Valentinitsch, A. et al. Regional analysis of age-related local bone loss in the spine of a healthy population using 3D voxel-based modeling. Bone 103, 233–240 (2017).
Loffler, M. T. et al. Improved prediction of incident vertebral fractures using opportunistic QCT compared to DXA. Eur Radiol 29, 4980–4989 (2019).
Hosny, A., Parmar, C., Quackenbush, J., Schwartz, L. H. & Aerts, H. Artificial intelligence in radiology. Nat Rev Cancer 18, 500–510 (2018).
Sekuboyina, A. et al. VerSe: a vertebrae labelling and segmentation benchmark for multi-detector CT images. Med Image Anal 73, 102166 (2021).
Löffler, M.T., et al. A Vertebral Segmentation Dataset with Fracture Grading. Radiology: Artificial Intelligence 2 (2020).
Baum, T. et al. Automatic detection of osteoporotic vertebral fractures in routine thoracic and abdominal MDCT. Eur Radiol 24, 872–880 (2014).
Bauer, J. S. et al. Detection of osteoporotic vertebral fractures using multidetector CT. Osteoporos Int 17, 608–615 (2006).
Glocker, B., Zikic, D., Konukoglu, E., Haynor, D. R. & Criminisi, A. Vertebrae localization in pathological spine CT via dense classification from sparse annotations. Med Image Comput Comput Assist Interv 16, 262–270 (2013).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 234-241 (Springer International Publishing, Cham, 2015).
Sekuboyina, A., et al. Btrfly Net: Vertebrae Labelling with Energy-Based Adversarial Learning of Local Spine Prior. 649-657 (Springer International Publishing, Cham, 2018).
Sekuboyina, A., Rempfler, M., Valentinitsch, A., Menze, B.H. & Kirschke, J.S. Labeling Vertebrae with Two-dimensional Reformations of Multidetector CT Images: An Adversarial Approach for Incorporating Prior Knowledge of Spine Anatomy. Radiology: Artificial Intelligence 2 (2020).
Yushkevich, P. A. et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128 (2006).
Tins, B. J. & Balain, B. Incidence of numerical variants and transitional lumbosacral vertebrae on whole-spine MRI. Insights Imaging 7, 199–203 (2016).
Thawait, G., Thawait, S., Chhabra, A., Fayad, L. & Carrino, J. An epidemiological imaging study of spinal segment variants in a nonsymptomatic population. Radiological Society of North America Scientific Assembly and Annual Meeting 2011 (2011).
Wigh, R. E. The thoracolumbar and lumbosacral transitional junctions. Spine (Phila Pa 1976) 5, 215–222 (1980).
Du Plessis, A. M., Greyling, L. M. & Page, B. J. Differentiation and classification of thoracolumbar transitional vertebrae. Journal of Anatomy 232, 850–856 (2018).
Rawls, A. & Fisher, R.E. Developmental and Functional Anatomy of the Spine. in The Genetics and Development of Scoliosis (eds. Kusumi, K. & Dunwoodie, S.L.) 1-29 (Springer International Publishing, Cham, 2018).
Castellvi, A. E., Goldstein, L. A. & Chan, D. P. Lumbosacral transitional vertebrae and their relationship with lumbar extradural defects. Spine (Phila Pa 1976) 9, 493–495 (1984).
Kirschke, J. S., Sekuboyina, A. & Löffler, M. T. VerSe20. Open Science Framework https://doi.org/10.17605/OSF.IO/T98FZ (2021).
Hou, L. et al. Lumbar plain radiograph is not reliable to identify lumbosacral transitional vertebra types according to Castellvi classification principle. BMC Musculoskelet Disord 21, 333 (2020).
Hsieh, C. Y., Vanderford, J. D., Moreau, S. R. & Prong, T. Lumbosacral transitional segments: classification, prevalence, and effect on disk height. J Manipulative Physiol Ther 23, 483–489 (2000).
Paik, N. C., Lim, C. S. & Jang, H. S. Numeric and morphological verification of lumbosacral segments in 8280 consecutive patients. Spine (Phila Pa 1976) 38, E573–578 (2013).
Acknowledgements
This work was supported by the European Research Council (ERC) with Starting Grant No. 637164 “iBack” to Jan S. Kirschke and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) with Project No. 432290010 to Thomas Baum.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
H.L. manuscript preparation, data review/review, segmentation corrections, image rating. D.S. manuscript preparation, data review. A.S. code development, project design, data review, manuscript preparation. L.M. data preparation/review, manuscript preparation, segmentation. M.L. code development, project design, manuscript preparation, data review/preparation. A.B. code development, manuscript preparation, data review/preparation. M.H. code development, manuscript preparation, data review/preparation. G.T. code development, project design, manuscript preparation, data review/preparation. K.G. data preparation/review, manuscript preparation, segmentation. E.N. data preparation/review, manuscript preparation, segmentation. T.B. manuscript preparation, data review, algorithm/code development. B.W. manuscript preparation, data review. B.M. manuscript preparation, data review. R.B. manuscript preparation, data review. C.Z. data review, manuscript preparation, study design. J.K. manuscript preparation, data review, segmentation corrections, image rating, algorithm/code development, study design
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Liebl, H., Schinz, D., Sekuboyina, A. et al. A computed tomography vertebral segmentation dataset with anatomical variations and multi-vendor scanner data. Sci Data 8, 284 (2021). https://doi.org/10.1038/s41597-021-01060-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-01060-0
This article is cited by
-
An Open Dataset of Annotated Metaphase Cell Images for Chromosome Identification
Scientific Data (2023)
-
A Review of CT-Based Fracture Risk Assessment with Finite Element Modeling and Machine Learning
Current Osteoporosis Reports (2022)
-
Automated detection of the contrast phase in MDCT by an artificial neural network improves the accuracy of opportunistic bone mineral density measurements
European Radiology (2022)