Abstract
Understanding how the living human brain functions requires sophisticated in vivo neuroimaging technologies to characterise the complexity of neuroanatomy, neural function, and brain metabolism. Fluorodeoxyglucose positron emission tomography (FDG-PET) studies of human brain function have historically been limited in their capacity to measure dynamic neural activity. Simultaneous [18 F]-FDG-PET and functional magnetic resonance imaging (fMRI) with FDG infusion protocols enable examination of dynamic changes in cerebral glucose metabolism simultaneously with dynamic changes in blood oxygenation. The Monash vis-fPET-fMRI dataset is a simultaneously acquired FDG-fPET/BOLD-fMRI dataset acquired from n = 10 healthy adults (18–49 yrs) whilst they viewed a flickering checkerboard task. The dataset contains both raw (unprocessed) images and source data organized according to the BIDS specification. The source data includes PET listmode, normalization, sinogram and physiology data. Here, the technical feasibility of using opensource frameworks to reconstruct the PET listmode data is demonstrated. The dataset has significant re-use value for the development of new processing pipelines, signal optimisation methods, and to formulate new hypotheses concerning the relationship between neuronal glucose uptake and cerebral haemodynamics.
Measurement(s) | brain activity |
Technology Type(s) | functional magnetic resonance imaging • FDG-Positron Emission Tomography |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.14977851
Similar content being viewed by others
Background & Summary
The enhanced abilities of the human brain to plan complex behaviour, make decisions, and process emotional and social contexts comes with heavy energy requirements. Although it accounts for only 2% of the body’s weight, the human brain accounts for 20% of its resting metabolism1,2. Oxygen and glucose are the two primary sources of energy in the human brain. The largest proportion of energy consumed by the brain, around 70–80%, occurs during neuronal computation and information processing3.
Understanding how the living human brain functions requires sophisticated in vivo neuroimaging technologies to characterise the complexity of neuroanatomy, neural function and brain metabolism. The ability to simultaneously image dynamic brain function and metabolic activity in living people was unimaginable just a decade ago. The technological developments that have led to simultaneous positron emission tomography (PET) – magnetic resonance imaging (MR)4 has now made it possible to simultaneously measure changes in blood oxygenation and glucose metabolism using simultaneous FDG-PET/BOLD-fMRI ([18 F]-fluorodeoxyglucose PET - blood oxygenation level dependent functional MRI). While BOLD-fMRI provides the capacity to measure changes in blood oxygenation in response to neuronal activity with high spatial resolution and moderate temporal resolution (~2 sec, sub-second with multiband acquisitions), the temporal resolution of traditional static FDG-PET studies is equal to the scan duration (i.e., a snapshot is taken across the scan period, 10–30 mins). As such, FDG-PET studies of human brain function have historically been limited in their capacity to measure the dynamic nature of neural activity.
Recent advances in FDG infusion protocols have made it possible to study dynamic changes in cerebral glucose metabolism simultaneously with dynamic changes in blood oxygenation. In a landmark study, Villien et al.5 adapted the slow infusion technique6 to show dynamic changes in glucose metabolism in response to checkerboard stimulation in the visual cortex with a temporal resolution of 1-min. Subsequent studies using this ‘functional PET’ (fPET) technique have extended these findings to demonstrate simultaneous BOLD and FDG activity in the visual and motor cortices7,8,9,10,11, in response to cognitive tasks12 and at rest13,14. The results of these studies indicate subtle commonalities and differences in patterns of blood oxygenation and glucose metabolism, which are not evident when measuring these responses separately.
Simultaneous FDG-fPET/BOLD-fMRI is a nascent technique which shows substantial promise for understanding the dynamic use of glucose and oxygen during neuronal activity. However, by comparison to BOLD-fMRI and other neuroimaging techniques, the processing pipelines are immature, and have not had the benefit of many years of work validating data preparation and signal detection optimisation. Furthermore, given the difficulty in acquiring these data – requiring access to a molecular MR scanner, access to radioisotopes and hot lab facilities, radiographer and nuclear medicine technologist specialists, as well as specialised MR-compatible equipment for dose delivery11 – only a handful of biomedical imaging facilities currently have the capability to acquire this multimodal data.
Here we describe the Monash vis-fPET-fMRI dataset, a simultaneous FDG-fPET/BOLD-fMRI dataset acquired from young healthy individuals. The dataset comprises unreconstructed fPET list-mode sinogram data, as well as PET image data reconstructed into 1-min time intervals. The dataset also includes simultaneously acquired reconstructed BOLD contrast MRI image data. The data was acquired using a flickering checkerboard embedded block design10, which allows the examination of a task-evoked signal with a known time course in a localised region-of-interest. The design is particularly suitable for development of novel processing and analysis pipelines. Note that this dataset uses low dose PET, with an average dose of 93MBq, range 39–105MBq (see Methods). One goal of the PET field has been to improve image quality while reducing the radiation exposure to the patient, two apparently contradictory aims15. One way to improve image quality without increasing radiation dose is to apply novel signal processing methods such as machine learning to low-dose PET images16. Recent advances in the development of machine learning algorithims for low dose PET have shown promise16,17,18, however additional studies using a diverse range of training sets is required to fully demonstrate the feasibility of low-dose acquisition protocols15. The dataset therefore shows promise for re-use in the development of methods to reduce the radiation exposure to patients without sacrificing image quality.
The release of unreconstructed and reconstructed PET data acquired during task-evoked visual cortex activation provides significant re-use value. This data has previously been used to test the effectiveness of the embedded block design to yield fMRI and fPET contrast10, to develop data fusion analyses based on independent component analysis (ICA)19, and to develop novel PET reconstruction methods based on Bowsher prior17. Examples of re-use may include disentangling the glucose metabolic and blood oxygenation level dependent responses to neuronal activity10,20, synergistic data reconstruction17,18,21 and fusion techniques19, novel multimodal attenuation correction procedures22, and refinement of fPET-fMRI data processing pipelines13,23.
Methods
All methods were reviewed by the Monash University Human Research Ethics Committee, following the Australian National Statement of Ethical Conduct in Human Research (2007). Administration of ionising radiation was approved by the Monash Health Principal Medical Physicist, following the Australian Radiation Protection and Nuclear Safety Agency Code of Practice (2005). For participants aged over 18-yrs, the annual radiation exposure limit of 5 mSv applies, and the effective dose in this study was 1.9 mSv. Informed consent was obtained from all human subjects in this dataset.
A video methods article demonstrating the data acquisition procedure was reported in Jamadar et al.11.
Participants
Participants (n = 10) were aged 18–49 years (mean 29 years), nine female, nine right handed. Data from an additional four participants’ were excluded for the following reasons: occluded cannula during infusion (2), scanner error (1), and one participant fell asleep during the protocol. Participants were screened for diabetes, personal or family history of neurological or neurodegenerative conditions, claustrophobia, MRI safety and nuclear medicine safety. Women were screened for current or possible pregnancy.
Prior to the scan, participants were directed to consume a high protein/low sugar diet for 24hrs, fast for six hours, and drink 2–6 glasses of water. Blood sugar level (BSL) was measured using an Accu-Chek Performa (model NC, Mannheim, Germany). Participants had BSL below 10mML (Table 1; note that BSL was measured but not recorded for one participant).
Demographic information, BSL and administered dose for each of the participants is provided in Table 1.
Stimuli and tasks
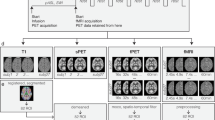
Figure 1 illustrates the workflow for the data acquisition.
Workflow & Paradigm. (a) Participants completed consent, safety screen, and demographic survey in the 30 mins prior to PET-MR scanning. (b) Next, participants were prepared for scanning. A cannula was placed in the forearm vein of each arm, then blood sugar level was measured. (c) Participants underwent a 90 min PET-MR scan while alternating between eyes closed rest and passive viewing of a visual checkerboard stimulus. i. Timing (minutes) of each component of the paradigm. Infusion and PET acquisition started at time 0. For the first 20-mins non-functional scans were taken while participants rested with their eyes closed. Then, participants completed 10 mins of flickering checkerboard stimulation. Embedded within this 10 min period was a fast on/off design (panel c.ii.), where the checkerboard was shown for 120 sec, and then for the remaining 8 mins, alternated off (16 sec, eye open fixated on cross) and on (32 sec). A 15 min eyes closed rest period then followed, then 5 min left hemifield stimulation, 5 min eyes closed rest, and 5 min right hemifield stimulation. During hemifield stimulation, participants viewed the stimulus with both eyes. Embedded within these 5-mins was a fast on/off design where the hemifield was shown for 150 sec, then alternated off (16 sec, eyes open fixated on cross), and on (32 sec). The protocol concluded with a 20 min eyes closed rest, and a final 10 min full checkerboard block, which had the same parameters as the first full checkerboard block. For participants collected with a 95 min scan, participants were instructed to lay with their eyes closed at the cessation of the final checkerboard block. (d) Representative activation results in the visual cortex. A joint independent component analysis (ICA) was conducted on the fMRI and fPET (1-min frames) timeseries. The first joint component is shown for fMRI and fPET. See Jamadar et al.10 for full report of methods and results of this analysis. Abbreviations: EPI, echo planar imaging; UTE, ultrashort echo time; GFM, gradient field map; ASL, arterial spin labelling, ICA, independent component analysis.
An ‘embedded’ block design was developed to produce signal contrast for both fast BOLD-fMRI and slow FDG-fPET measurements (Fig. 1c). The slow on/off task alternation periods (Fig. 1c.i.) were designed to provide FDG-fPET contrast (alternating task and rest over durations of minutes), and fast on/off alternation periods (Fig. 1c.ii.) were embedded in the stimulation periods designed to provide BOLD-fMRI contrast. We made the assumption that the fast on/off periods would alternate at a frequency too high to be detected by the fPET measurements.
Participants rested with eyes closed during the first 20 min block of MRI scans (localiser, T1, etc.). At 20 mins, a 10 min visual stimulation period was presented in an embedded 32/16 sec on/off design. The first 120 sec of the block was a sustained ‘on’ period; we predicted that this period would allow FDG-fPET signal to rise from resting levels. During the ‘on’ periods (i.e., 120 s & 32 s periods), the visual stimulus was a circular checkerboard (size 39 cm, visual angle 9°), presented on a black background. The checkerboard flickered (i.e., alternated black and white fields), at rate of 8 Hz. During the 16 s ‘off’ periods, participants rested with eyes open while viewing a white fixation cross (size 3 cm, visual angle 0°45′) presented on a black background. Following the 10 min stimulation block, a 15 min eyes closed rest period followed, then 5 min of left hemifield stimulation (150 sec on, then 32/16 s on/off), 5 min eyes closed rest, and 5 min of right hemifield stimulation. Note that participants were instructed to open both eyes during hemifield stimulation, and so, activity is likely to be obtained in both hemispheres of the visual cortex during this condition. Following 20 mins of eyes closed rest, a final 10 min full checkerboard (parameters identical to the first 10 min block) was presented. Due to short delays between MR scans, we found that the 90 min duration of the PET scan did not always conclude with the end of the MR sequences. Therefore, we increased the duration of the PET scan for subjects acquired later in the study. PET scan duration for each participant is noted in Table 1. Participants were directed to close their eyes and rest following the cessation of the checkerboard block.
Procedure
Participants were cannulated in the vein of each forearm with a minimum size 22-gauge cannula, and a 10 mL baseline blood sample was taken. For all participants, the left cannula was used for FDG infusion, and the right cannula was used for blood sampling. Primed extension tubing was connected to the right cannula for blood sampling, via a 3-way tap.
The 90-min simultaneous PET-MR scan was performed using a Siemens Biograph 3 T molecular MR (mMR) scanner (Erlangen, Germany). Participants were positioned supine in the scanner bore with head in a 16ch radiofrequency (RF) head-neck coil. Visual stimuli were viewed via a mirror placed on the RF coil, which reflected a 32-inch Cambridge Research Systems (UK) BOLDscreen MR-compatible LCD screen. [18 F]-fluorodeoxyglucose (FDG; average dose 97.0Mbq) was infused over the course of the scan at a rate of 36 mL/hr using a Caesarea Medical Electronics (Israel) BodyGuard 323 MR-compatible infusion pump. One participant received a dose of 39MBq due to technical error (Table 1). Infusion onset was locked to the onset of the PET scan (see PET-MR Protocol, below).
Plasma radioactivity levels were manually obtained throughout the duration of the scan. At 10-min post-infusion onset, a 10 mL blood sample was taken from the right forearm using a vacutainer; the time of the 5 mL mark was noted for subsequent decay correction. Subsequent blood samples were taken at 10 min intervals for a total of ten samples for the duration of the scan. The cannula line was flushed with 10 mL of saline after every sample to minimise line clotting. As noted in Table 1, blood samples from one subject were unable to be obtained, and samples for one additional subject were unable to be obtained after timepoint 4, due to difficulties drawing blood.
PET-MR Protocol
PET data was acquired in list-mode. PET data acquisition and infusion of FDG started with the ultrashort TE (UTE) MRI for PET attenuation correction. Non-functional MRI scans were acquired in the first 20-mins, and included T1 3D MPRAGE (TA = 7.01 mins, TR = 1640 ms, TE = 2.34 ms, flip angle = 8°, FOV = 256 × 256 mm2, voxel size = 1 × 1 × 1 mm3, 176 slices; sagittal acquisition), T2 FLAIR (TA = 5.52 mins), gradient field map (TA = 1.02 mins, TR = 466 ms, TE1 = 4.92 ms, TE2 = 7.38 ms, voxel size = 3 × 3 × 3mm3, FOV = 190 mm, 44 slices), and ASL (TA = 3.52 min, data not released). For the remainder of the scan, seven consecutive blocks of T2*-weighted echo planar images (EPIs) were acquired (TR = 2450 ms, TE = 30 ms, FOV = 190 mm, 3 × 3 × 3 mm3 voxels, 44 slices, ascending interleaved axial acquisition). The duration of each EPI block was determined by the stimulus block duration (Fig. 1a): block 1 full checkerboard (TA = 10.02 min, 242 volumes), block 2 eyes closed (TA = 15.01 min, 364 volumes), block 3 half checkerboard (TA = 5.05 min, 121 volumes), block 4 eyes closed (TA = 5.05 min, 121 volumes), block 5 half checkerboard (TA = 5.05 min, 121 volumes), block 6 eyes closed (TA = 20.02, 487 volumes), block 7 full checkerboard (TA = 10.02 min, 242 volumes).
Data Records
Online-only Table 1 defines the data available for each subject on OpenNeuro. The dataset containing the demographic and anthropometric data, T2* EPI fMR images, PET images reconstructed in 1-min bins, unreconstructed PET list-mode data, T1 structural images and gradient field maps is freely available in BIDS format24 from the OpenNeuro repository (http://openneuro.org) with the accession number ds003382 (https://doi.org/10.18112/openneuro.ds003382.v1.0.0)25.
Participants.tsv is a text file reporting the demographic and anthropometric data for each subject ordered by subject ID. Plasma_radioactivity.tsv is a text file reporting the plasma radioactivity counts and measurement times for each subject, ordered by subject ID.
The Monash vis-fMPRI-fPET dataset contains both raw (unprocessed) images and source data (i.e. unconstructed PET listmode data). Both are organized in sub-directories that are corresponding to the subjects, according to the BIDS specification. For each subject, T1-weighted MPRAGE images, fMRI images and gradient field maps are in the anat (anatomical data), func (functional data) and fmap (field map) sub-directories respectively, along with metadata in the json sidecar. Dixon and UTE scans are also added to the dataset for reconstructing the PET source data into images, which are organized in the ‘dixon’ and ‘ute’ sub-directories. At least two UTE echos are included (sub-*/ute/*ute.nii.gz), and the computed attenuation correction maps are also shared (sub-*/ute/*ute-umap.nii.gz). The UTE sequence parameters are included in the json sidecar, and can be found under the ‘global’ tag, where the values of ‘EchoNumbers’ are corresponding to the first and second UTE echoes. MR coil attenuation has been corrected during PET image reconstruction, however the attenuation from the mirror used for visual stimulation was not corrected due to practical limitations. Although there is not currently a BIDS-compliant format, the same structure is followed with a json sidecar along with each of the image data. Both static and dynamic reconstructed PET images are contained in the ‘pet’ directory. The static PET images (1.5 zooming) are reconstructed, using the Ordered Subset Expectation Maximization (OSEM) (Burgos et al., 2014) algorithm with 21 subsets and 3 sub-iterations, and smoothed with 2 mm FWHM Gaussian spatial filter, are stored as sub-*/pet/*static*.nii.gz files The dynamic reconstruction was performed for each 1-min acquisition time interval to produce 90–95 mins of sequential fPET images with a 4 mm Gaussian spatial filter applied. The images are store in the same directory as sub-*/pet/*dynamic_1min-ac.nii.gz files. Following the current working copy of the proposed BIDS Extension for PET (BEP009), blood data (i.e. discrete plasma measurements of radioactivity) are also included in the ‘pet’ directory, which report the plasma radioactivity counts and measurement times for the subject. Data in sub-*/dixon, sub-*/ute and sub-*/pet are ignored in the BIDS validation process, as they are not officially supported by the current BIDS specification.
The ‘sourcedata’ directory contains the raw, non-reconstructed PET source data that was directly exported from Siemens scanner console. The source data includes PET listmode data, normalization data, sinogram data and physiology data. The raw PET data are in the form of a file pair (one DICOM header and one raw binary file) with the two paired files having the same file name but different filename extensions (.dcm for the DICOM file and.bf for raw binary file). A json metadata sidecar file was added for each subject’s raw dataset, similar to the other image types officially supported by BIDS specification. The blood data containing the blood plasma measurements is included identically as for the reconstructed PET image data. The ‘sourcedata’ directory is also excluded in the BIDS validation process.
To prepare the BIDS dataset, the open source conversion tool, Heudiconv (https://github.com/nipy/heudiconv, version 0.8.0) was used to organize the imaging data into structured directory layouts, and the Dcm2niix convertor (https://github.com/rordenlab/dcm2niix, version 1.0.20200427) was used to convert the image data from DICOM to NifTI format. Customized scripts that were written to (i) remove personal identifiable information (PII) from the raw PET dicom header; (ii) add custom json sidecar files to the PET raw data and reconstructed image data; (iii) generate plasma radioactivity files; are available on a github repository (https://github.com/szho42). Defacing was applied to T1-weighted images, Dixon and UTE images, using the tool, pydeface (https://github.com/poldracklab/pydeface, version 2.0.0). The original umaps are shared, as defaced umaps will impact on the reconstruction results, due to losing bone information. The uMaps contain little facing information, which does not violate the OpenNeuro privacy policy. The reconstructed PET images and PET raw data were not defaced as the subjects are not able to be visually identified from the PET images. The scripts used in the BIDS dataset conversion are publicly available (https://github.com/szho42/bids-pet-conv, version 2.0.1).
Technical Validation
Whilst the Monash vis-fPET-fMRI dataset consists of simultaneous FDG-fPET and BOLD-fMRI datasets, this section primarily focuses on the validation of the fPET raw data as similar fMRI image data has previously been reported10,13. The unreconstructed PET listmode data is only useful as a publicly accessible dataset if tools are available for offline binning of the listmode data into sinogram data, and reconstruction of the sinogram data into PET images. Proprietary tools exist to perform offline image reconstructions but these are not publicly available. Technical validation of the images reconstructed using the open source reconstruction frameworks compared to the proprietary tools is required to facilitate meaningful sharing of the PET listmode data. However, the current limitations of the open source frameworks do not enable a direct quantitative comparison between the reconstruction methods. Nevertheless, a qualitative comparison has been undertaken as described below.
PET Raw Data Image Reconstruction with SIRF
The PET listmode data was reconstructed using the SIRF21 framework (with STIR26 as the backend, https://github.com/UCL/STIR, version 4.0.2) to demonstrate the technical feasibility of reconstructing the listmode data using open source PET reconstruction tools. The process required use of several open source tools including: (i) pet-rd-tools (https://github.com/UCL/pet-rd-tools, version 2.0.1) for converting the PET raw data from the Siemens-specific format to Interfile format used by the STIR reconstruction toolkit; (ii) the STIR reconstruction toolkit (https://github.com/UCL/STIR) that is a framework for iterative image reconstruction for PET and SPECT raw data; and (iii) the SIRF framework (https://github.com/SyneRBI/SIRF, version 2.2.0) that provides a user-friendly interface to write scripts in Python and Matlab. The SIRF built-in template image for ‘Siemens mMR’ was used, with a span of 11, maximal ring difference of 60 and mashing factor of 2. To reconstruct the fPET raw data dynamically, flexible binning patterns were customized in SIRF by setting specific listmode date start times and the time interval for conversion of the listmode data to sinogram data. In some cases, the first time tag is not zero (which is known as a STIR issue in the current version), in which a small threshold was set to identify the true start time. The corresponding sinogram was generated with the estimated random coincidences map, which were fed into the iterative methods for reconstruction. In our case, UTE-based attenuation maps were used for attenuation correction, which was converted by pet-rd-tool with the orientation of RAS. The acquisition sensitivity model was set by chaining the normalization and attenuation map, and background term was set by combining the randoms map and the estimated scatter map. The Ordered Subsets Maximum A Posteriori One Step Late algorithm (OSMAPOSL) was used to iteratively reconstruct the PET images, using the Poisson log-likelihood objective function. The SIRF reconstruction uses 21 subsets which is identical to the setting on the vendor console, however 40 sub-iterations are applied on the SIRF reconstruction pipeline for achieving reasonable quality images. A 4 mm FWHM Gaussian spatial filter was applied to the PET images after reconstruction. To fairly compare the results, we also reconstruct images on Siemens console, with the same setting, by only enabling 4 mm Gaussian smoothing for both static and dynamic reconstructions.
Qualitative comparison between the reconstructed PET images
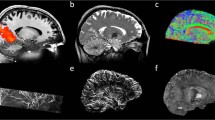
An example of the dynamically reconstructed PET images for 5-minute of the sinogram data are shown in Fig. 2. The reconstructed PET image using (a) the Siemens reconstruction pipeline on the scanner console and (b) the open source framework, SIRF with scatter and UTE-based attenuation correction applied are shown in Fig. 2(a,b) respectively. Both images were reconstructed using the last 5-minute of list mode data for subject 005 in the dataset. It can be seen from Fig. 2 that the listmode data can be reconstructed using open source tools like SIRF, and have very similar contrasts, compared to the images from the Vendor console in a dynamic setup. It is worth mentioning that the image quality using SIRF is similar but still different from the vendor’s reconstruction pipeline. The functionalities in STIR/SIRF are fast developing and we expect further improvements in later releases. Further quantitative validation of the open source PET reconstruction tools will be required as the STIR/SIRF framework is further developed.
Example of a dynamic reconstructed PET image for subject 005 using the last 5-min of the acquisition data. (a) The reconstructed PET images using the vendor reconstruction pipeline. (b) The reconstructed PET images using the SIRF21 open source framework with random, scatter and attenuation correction applied.
For reference, the static PET image reconstructed for one subject (subject 005 using the SIRF opensource framework with random, scatter and UTE-based attenuation correction applied) using the entire listmode acquisition is shown in Fig. 3. As expected, the image demonstrates significantly higher signal to noise in comparison to the PET images from the last 5-min of the acquisition as shown in Fig. 2, as well as superior anatomical localisation of the FDG signal. The scripts used to reconstruct the dynamic and static PET images are publicly available at (https://github.com/szho42/pet-image-recon).
The static reconstructed PET images for subject 005, using (a) vendor reconstruction pipeline and (b) the SIRF21 open source frame with random, scatter and attenuation corrections applied.
In summary, the STIR/SIRF reconstructed PET images demonstrate that the PET list mode data can be successfully reconstructed, but further validation of the open source reconstruction framework still remains to be undertaken. Future validation work could for example involve reconstruction of the PET list mode data with smaller time intervals (e.g. 30secs) for comparison with comparable images reconstructed using the proprietary Siemens framework. This would have the advantage of increasing the temporal resolution of the fPET image dataset that could potentially increase the sensitivity for the functional PET method to detect visually evoked brain activity.
Usage Notes
With the release of unprocessed fMR image data and unreconstructed PET list mode data, the Monash vis-fPET-fMRI dataset1 has significant re-use value. The primary motivation for the public release of the dataset is to facilitate the development of new processing pipelines, signal optimisation methods, and stimulate new hypotheses concerning the relationship between glucose uptake and cerebral haemodynamics in the human brain. The task-related design is particularly powerful, as the visual stimulation task robustly activates a well localised region-of-interest of visual cortex with a known time-course. The experimental design facilitates functional PET imaging methods development and evaluation of statistical analytical techniques for fPET data23. Some examples of re-use would be to explicitly test whether the 5 min on/off hemifield alternation is sufficient to distinguish between stimulation and rest period in the fPET data; or alternatively whether the fast 32/16 sec on/off in the checkerboard stimulation could be differentiated (Fig. 1). Furthermore, the release of unreconstructed PET list-mode data is particularly significant. We believe that this dataset represents the first PET list-mode dataset available through the OpenNeuro platform, and probably the first publicly available list-mode dataset simultaneously acquired with BOLD-fMRI data.
Access to unreconstructed PET list-mode data facilitates the development of novel image reconstruction17,18 and attenuation correction methods22, including synergistic PET-MR image reconstruction21. For researchers interested in standard reconstructions with differing parameters (e.g., frame lengths), data can be reconstructed using Siemens e7tools software, or using open-source reconstruction methods such as STIR26. For researchers interested in using reconstructed data, we have additionally provided PET data reconstructed into 1-min bins using the OP-OSEM reconstruction software. The reconstructed image data is useful for developing post-processing pipelines27, and for exploring physiological and neuroscientific questions about glucose uptake in the brain12,14. Thus, the Monash vis-fPET-fMRI dataset is a uniquely powerful and flexible resource for the neuroimaging community.
In this data release, MR images, unreconstructed list-mode PET data and reconstructed PET images were converted to BIDS format prior to upload. While the BIDS specification for MR is mature, the BIDS specification for PET is still under development (https://github.com/ohbm/osr2020/issues/57)28, and does not include specifications for standardisation of list-mode data. In fact, the definition of ‘raw’ data for BIDS does not currently appear to extend to list-mode data; rather, the term refers to unprocessed reconstructed data (https://bids-specification.readthedocs.io/en/stable/02-common-principles.html#source-vs-raw-vs-derived-data). As such, we provide these currently unspecified data types using BIDS-style naming conventions and structures. Future BIDS releases may standardise structures and naming conventions for these currently unspecified data types, at which time we will endeavour to update the OpenNeuro data repository to reflect these new standards.
Simultaneous PET-MRI data is costly and difficult to acquire, requiring access to facilities that a small (but growing) number of biomedical imaging facilities worldwide currently possess. It is expected that release of this data in both raw and reconstructed formats will provide opportunities for further work validating synergistic multimodal reconstruction routines, algorithms for data preparation and signal detection optimisation; as well as new fundamental discoveries on the dynamic use of energy in the human brain.
Code availability
Scripts used to insert required metadata into the published BIDS dataset are freely available at https://github.com/BioMedAnalysis/petmr-bids under Apache License 2.0.
References
Attwell, D. & Laughlin, S. B. An energy budget for signaling in the grey matter of the brain. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 21, 1133–1145 (2001).
Raichle, M. E. & Mintun, M. A. Brain Work and Brain Imaging. Annu. Rev. Neurosci. 29, 449–476 (2006).
Harris, J. J., Jolivet, R. & Attwell, D. Synaptic Energy Use and Supply. Neuron 75, 762–777 (2012).
Chen, Z. et al. From simultaneous to synergistic MR‐PET brain imaging: A review of hybrid MR‐PET imaging methodologies. Hum. Brain Mapp. 39, 5126–5144 (2018).
Villien, M. et al. Dynamic Functional Imaging of Brain Glucose Utilization using fPET-FDG. NeuroImage 100, 192–199 (2014).
Carson, R. E. Pet physiological measurements using constant infusion. Nucl. Med. Biol. 27, 657–660 (2000).
Hahn, A. et al. Quantification of Task-Specific Glucose Metabolism with Constant Infusion of 18F-FDG. J. Nucl. Med. 57, 1933–1940 (2016).
Hahn, A. et al. Task-relevant brain networks identified with simultaneous PET/MR imaging of metabolism and connectivity. Brain Struct. Funct. 1–10, https://doi.org/10.1007/s00429-017-1558-0 (2017).
Rischka, L. et al. Reduced task durations in functional PET imaging with [18F]FDG approaching that of functional MRI. NeuroImage 181, 323–330 (2018).
Jamadar, S. D. et al. Simultaneous task-based BOLD-fMRI and [18-F] FDG functional PET for measurement of neuronal metabolism in the human visual cortex. NeuroImage 189, 258–266 (2019).
Jamadar, S. D. et al. Radiotracer Administration for High Temporal Resolution Positron Emission Tomography of the Human Brain: Application to FDG-fPET. J. Vis. Exp. 60259, https://doi.org/10.3791/60259 (2019).
Hahn, A. et al. Reconfiguration of functional brain networks and metabolic cost converge during task performance. eLife 9, e52443 (2020).
Jamadar, S. D. et al. Simultaneous BOLD-fMRI and constant infusion FDG-PET data of the resting human brain. Sci. Data 7, 363 (2020).
Jamadar, S. D. et al. Metabolic and Hemodynamic Resting-State Connectivity of the Human Brain: A High-Temporal Resolution Simultaneous BOLD-fMRI and FDG-fPET Multimodality Study. Cereb. Cortex https://doi.org/10.1093/cercor/bhaa393 (2021).
Catana, C. The Dawn of a New Era in Low-Dose PET Imaging. Radiology 290, 657–658 (2019).
Chen, K. T. et al. Ultra–Low-Dose 18 F-Florbetaben Amyloid PET Imaging Using Deep Learning with Multi-Contrast MRI Inputs. Radiology 290, 649–656 (2019).
Sudarshan, V. P. et al. Incorporaion of anatomical MRI knowledge for enhanced mapping of brain metabolism using functional PET. NeuroImage (2021).
Sudarshan, V. P., Egan, G. F., Chen, Z. & Awate, S. P. Joint PET-MRI image reconstruction using a patch-based joint-dictionary prior. Med. Image Anal. 62, 101669 (2020).
Li, S., Jamadar, S. D., Ward, P. G. D., Egan, G. F. & Chen, Z. Estimation of simultaneous BOLD and dynamic FDG metabolic brain activations using a multimodality concatenated ICA (mcICA) method. NeuroImage 226, 117603 (2021).
Shokri-Kojori, E. et al. Correspondence between cerebral glucose metabolism and BOLD reveals relative power and cost in human brain. Nat. Commun. 10, 690 (2019).
Ovtchinnikov, E. et al. SIRF: Synergistic Image Reconstruction Framework. Comput. Phys. Commun. 249, 107087 (2020).
Baran, J. et al. Accurate hybrid template–based and MR-based attenuation correction using UTE images for simultaneous PET/MR brain imaging applications. BMC Med. Imaging 18, 41 (2018).
Li, S. et al. Analysis of continuous infusion functional PET (fPET) in the human brain. NeuroImage 213, 116720 (2020).
Gorgolewski, K. J. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044 (2016).
Jamadar, S. D. et al. Monash vis-fPET-fMRI. OpenNeuro https://doi.org/10.18112/openneuro.ds003382.v1.3.1 (2020).
Thielemans, K. et al. STIR: software for tomographic image reconstruction release 2. Phys. Med. Biol. 57, 867–883 (2012).
Chen, Z. et al. MR-PET head motion correction based on co-registration of multicontrast MR images. Hum. Brain Mapp. https://doi.org/10.1002/hbm.24497 (2019).
Knudsen, G. M. et al. Guidelines for the content and format of PET brain data in publications and archives: A consensus paper. J. Cereb. Blood Flow Metab. 40, 1576–1585, https://doi.org/10.1177/0271678X20905433 (2020).
Acknowledgements
This work was supported by an Australian Research Council (ARC) Linkage Project (LP170100494) that includes financial support from Siemens Healthineers. SDJ, PGDW & GFE are supported by the ARC Centre of Excellence for Integrative Brain Function (CE140100007). SDJ is supported by an ARC Discovery Early Career Researcher Award (DE150100406) and National Health and Medical Research Council Fellowship (APP1174164). The authors acknowledge Winnie Orchard, Irene Graafsma and Disha Sasan for their assistance in acquiring data; and Lauren Hudswell and Cameron Pain for assistance with data preparation.
Author information
Authors and Affiliations
Contributions
S.D.J., G.F.E., P.G.D.W., Z.C., N.J.S., M.P., A.F. designed the research question. Data was analysed by S.Z. Data was prepared for release by S.Z., S.D.J., A.C. & R.M.c. Funding was obtained by G.F.E., S.D.J., Z.C., A.F., M.P., K.O’.B., D.S. & N.J.S. Manuscript first draft was written by S.D.J., S.Z., & G.F.E. All authors have contributed to manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
Siemens Healthineers contributed financial support to the ARC Linkage project held by GFE, SDJ, ZC, AF, MP & NJS. KO’B and DS are employees and shareholders of Siemens Healthcare Pty Ltd. SZ, PGDW, AC & RMc have no competing or conflicting interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Table
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Jamadar, S.D., Zhong, S., Carey, A. et al. Task-evoked simultaneous FDG-PET and fMRI data for measurement of neural metabolism in the human visual cortex. Sci Data 8, 267 (2021). https://doi.org/10.1038/s41597-021-01042-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-01042-2