Abstract
We provide the raw and processed data produced during the genome sequencing of isolates from six species of parasites from the sub-family Leishmaniinae: Leishmania martiniquensis (Thailand), Leishmania orientalis (Thailand), Leishmania enriettii (Brazil), Leishmania sp. Ghana, Leishmania sp. Namibia and Porcisia hertigi (Panama). De novo assembly was performed using Nanopore long reads to construct chromosome backbone scaffolds. We then corrected erroneous base calling by mapping short Illumina paired-end reads onto the initial assembly. Data has been deposited at NCBI as follows: raw sequencing output in the Sequence Read Archive, finished genomes in GenBank, and ancillary data in BioSample and BioProject. Derived data such as quality scoring, SAM files, genome annotations and repeat sequence lists have been deposited in Lancaster University’s electronic data archive with DOIs provided for each item. Our coding workflow has been deposited in GitHub and Zenodo repositories. This data constitutes a resource for the comparative genomics of parasites and for further applications in general and clinical parasitology.
Measurement(s) | DNA • genome • sequence_assembly • sequence feature annotation |
Technology Type(s) | DNA sequencing • Oxford Nanopore Sequencing • Illumina sequencing • sequence assembly process • sequence annotation |
Sample Characteristic - Organism | Leishmaniinae |
Sample Characteristic - Location | Namibia • Thailand • Ghana • Brazil |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.15134085
Similar content being viewed by others
Background & Summary
Leishmaniasis is a neglected tropical disease. It is considered to be a disease of poverty, primarily affecting low and middle-income countries (LMICs). Leishmaniasis is caused by parasites of the genus Leishmania and 18 different species are known to infect humans1. 98 sandfly species are suspected or confirmed vectors of Leishmania2. There are three major types of leishmaniasis: visceral, also known as kala-azar, is fatal if left untreated in over 95% of cases; cutaneous, the most common form, causes skin lesions leaving life-long scars and serious disability or stigma; mucocutaneous, leads to partial or total destruction of mucous membranes of the nose, mouth and throat3. Over one billion people live in endemic areas and are at risk of leishmaniasis. It is estimated that each year, globally, new cases of cutaneous leishmaniasis occur at an incidence of 700,000 to 1.2 million or more in over 100 countries4. Additionally, up to 300,000 visceral leishmaniasis cases cause more than 200,000 deaths annually5.
The genus Leishmania is divided into four subgenera: L. Leishmania, L. Viannia, L. Sauroleishmania and the newest subgenus L. Mundinia, the latter now accommodating several species from the L. enriettii complex and others, from five continents6,7,8,9,10,11,12. In 1994, the Leishmania Genome Network was initiated13 and announced, ten years later, the assembly of the Leishmania major Friedlin strain as the first Leishmania reference genome14. Since then, a total of 58 genomes have become available publicly, assembled at a variety of levels of completeness ranging from contigs to chromosome level. Prior to our project, only two L. Mundinia subgenus genomes have been sequenced and assembled: Leishmania enriettii, strain LEM3045 (GCA_000410755) and Leishmania sp. MAR, strain LEM2494 (GCA_000410755). The genus Porcisia is a sister genus of Leishmania within the sub-family Leishmaniinae. Prior to the release of our genome, there were no genome sequences for genus Porcisia. Subsequently, the partial genome of P. deanei was released and published15.
We assembled and annotated the genomes of five L. Mundinia species – those of L. martiniquensis, L. orientalis, L. enriettii, L. sp. Ghana and L. sp. Namibia - and one genome in the genus Porcisia – that of P. hertigi, formerly known as L. hertigi16 - using Illumina and Nanopore sequencing. The two isolates from Ghana and Namibia are from new species that have not yet been formally named. The World Health Organization (WHO) codes for the six isolates are: L. martiniquensis MHOM/TH/2012/LSCM1;LV760; L. orientalis MHOM/TH/2014/LSCM4;LV768; L. enriettii MCAV/BR/2001/CUR178;LV673; L. sp. Ghana MHOM/GH/2012/GH5;LV757; L. sp. Namibia MPRO/NA/1975/252;LV425; and P. hertigi MCOE/PA/1965/C119;LV43. Nanopore long reads were used for the initial scaffolding assemblies, followed by mapping of the Illumina short reads onto these scaffolds, thus increasing quality of the assembled sequence while preserving whole chromosome integrity. Final polishing, reordering and reorienting of chromosomes, along with masking and classifying of repeat regions, was guided by the most closely related reference genome for each species. Finished genome annotation was both evidence-based and ab initio.
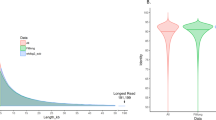
Figure 1 summarises data sizes and total yield per sample. The total sequencing data file size for all samples was 139.33 Gigabytes, yielding 58.70 GigaBases of sequence data from 23.71 GigaReads. Figure 2 summarises our analysis workflow. This workflow generated four main outputs for each assembly: genome, proteome, and transcriptome files in FASTA format, and a General Feature Format file (GFF) that contains the coordinates for all proteins and transcripts in the assembly.
Methods
Sample collection, sequencing and software
From the parasite cryobank at Lancaster University, we selected six samples of the species listed above without publicly available reference genomes. Table 1 gives details for strains, isolates, BioSample and BioProject accessions17,18,19,20,21,22,23,24,25,26,27,28. Illumina HiSeq 4000 and MiSeq sequencing was contracted to BGI Genomics and Aberystwyth University. Nanopore sequencing was performed in-house using MinION FLO-MIN106 flow cells with SQK-LSK109 ligation sequencing protocol. Throughout the text we provide literature citations to software where available. Links to both published and unpublished software used are provided in Table 2. We created public GitHub and Zenodo repositories for the analysis pipeline29,30.
Genome assembly
De novo assemblies were performed with Nanopore MinION long reads using Flye31. Due to the low quality scores in Nanopore long reads, we mapped high quality Illumina short reads onto the assemblies and created corrected consensus sequences using minimap232 and SAMtools33. The consensus sequence was scanned for any contamination or any sequence of vector origin by BLAST+34 on the UniVec database35. Finally, a polishing step was done to minimise gaps using Pilon36.
Chromosome verification
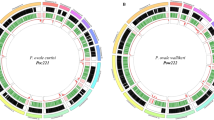
For all chromosomes of each polished genome, we then ran BLAST + (parameters: -max_target_seqs. 1 -max_hsps 1) against all TriTrypDB37 release-47 genomes. The output for each genome was then visualized using wordcloud to suggest the closest relative among TriTrypDB genomes38. Then, synteny was plotted for each genome by aligning each of its chromosomes with the corresponding chromosomes of its wordcloud-predicted closest relative, using MUMmer39 (Fig. 3). This confirmed that the order and orientation of the chromosomes of each genome was equivalent to those of its closest TriTrypDB genome. Completion was then achieved by sorting and removing any duplicate scaffolds or contigs using funannotate40, followed by a final quality check using Genome Assembly Annotation Service (GAAS).
Repetitive element annotation
We identified and classified repeat regions in the polished assemblies using RepeatModeller and TEclass41. Then, we generated a stratified genome-wide repeat plot for each assembly38 (see also L. martiniquensis example in Fig. 4) to assist the decision of which repeats to mask, using RepeatMasker.
Example genome-wide repeat plot for L. martiniquensis, stratified: simple (micro-satellites), low complexity, DNA, long terminal repeats (LTRs), long interspersed nuclear elements (LINEs), RNA, rolling circle (RC), satellites, short interspersed nuclear elements (SINEs) and retroposons. The middle pie chart represent the proportion of each repeat class in the genome: none (94.4%), simple (micro-satellites) (4.11%), low complexity (0.655%), DNA (0.419%), unknown (0.161%), LTRs (0.110%), LINEs (0.052%), RNA (0.027%), RC (0.019%), satellites (0.010%), retroposons (0.005%), SINEs (0.004%).
Gene prediction and functional annotation
After repeat masking, we annotated the assemblies using the MAKER242 annotation pipeline over two rounds: 1) an evidence-based annotation round using EST, mRNA-seq and protein homology evidence from TriTrypDB release-47 along with our repeat-masking output, 2) an ab initio round using AUGUSTUS43, with the pre-trained L. tarentolae as the model organism. After each round, Annotation Edit Distance (AED) scores were calculated and plotted (Fig. 5). We calculated brief statistics for each round, e.g. the number of genes and other features, using Genometools44 and AGAT45. After completion of all annotation rounds, we assigned functional annotations from the Uniprot46 and Pfam47 databases using BLAST + and InterProScan48.
Analysis pipeline
To make sure that all assemblies and annotations are reproducible by future investigators, the entire process from obtaining the SRAs49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91 to the annotation assignments92,93,94,95,96,97 has been made available29 using Snakemake98. This Snakemake pipeline ought to be easily adaptable to the sequencing of further similar parasite genomes, throughout the parasitology community30.
Data Records
Table 3 details the sequencing output. Short and long reads were deposited in the NCBI Sequence Read Archive (SRA)49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91. Six BioProjects23,24,25,26,27,28 and six BioSamples17,18,19,20,21,22 were also created at NCBI. The assembled genomes were deposited at NCBI Assembly99,100,101,102,103,104. Additional files containing raw reads quality reports105,106,107,108,109,110, mapped reads111,112,113,114,115,116, classified repeated sequences117,118,119,120,121,122 and functional annotations92,93,94,95,96,97 were deposited at Lancaster University electronic data archive.
Technical Validation
Genomic DNA integrity
Genomic DNA was extracted using Trizol (Invitrogen) and quantified using Qubit® dsDNA HS Assay Kits (ThermoFisher Scientific) prior to sequencing. Concentrations ranged between 68.2 and 120 ng/µL. For consistency, we used the same extracted DNA for all three sequencing platforms (Nanopore MinION, Illumina HiSeq 4000 and MiSeq). Furthermore, we assessed the gDNA high molecular weight using N50 estimates of MinION long reads which were ranged between 12.07 and 22.92 kilobases.
Contamination screening
We scanned all assemblies for any contamination or any sequence of vector origin by first building a UniVec Database and then using BLAST+ . All contaminants were found either at the beginning or at the end of contigs and then deleted. No contaminants affected assembly integrity.
Quality of short and long raw sequence reads
We used FastQC to check the sequence quality of Illumina short reads sequences and pycoQC to check the Nanopore long reads sequence quality. We used MultiQC123 to output all sequence quality scores in one interactive report105,106,107,108,109,110.
Assembly validation
Since the analysis took many steps to finish, quality checks were introduced between each step. Some checks were focused on completeness, for instance using BUSCO124 as a benchmark for the presence of expected universal single-copy orthologues. Other checks focussed on the correct order and orientation of the chromosomes, for instance MUMmer alignment to find synteny between assemblies and other Leishmania genomes. Yet further checks focussed on the accuracy and precision of annotation, for instance using Annotation Edit Distance score (AED) in MAKER2 (Fig. 5). We checked reproducibility of the assemblies and annotations using Snakemake.
References
Steverding, D. The history of leishmaniasis. Parasit Vectors 10, 82–91 (2017).
Maroli, M., Feliciangeli, M. D., Bichaud, L., Charrel, R. N. & Gradoni, L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med Vet Entomol 27, 123–147 (2013).
Zijlstra, E. E. PKDL and other dermal lesions in HIV co-infected patients with Leishmaniasis: review of clinical presentation in relation to immune responses. PLoS Negl Trop Dis 8, e3258 (2014).
Al-Salem, W., Herricks, J. R. & Hotez, P. J. A review of visceral leishmaniasis during the conflict in South Sudan and the consequences for East African countries. Parasit Vectors 9, 460–470 (2016).
Burza, S., Croft, S. L. & Boelaert, M. Leishmaniasis. Lancet 392, 951–970 (2018).
Desbois, N., Pratlong, F., Quist, D. & Dedet, J. P. Leishmania (Leishmania) martiniquensis n. sp. (Kinetoplastida: Trypanosomatidae), description of the parasite responsible for cutaneous leishmaniasis in Martinique Island (French West Indies). Parasite 21, 12–15 (2014).
Jariyapan, N. et al. Leishmania (Mundinia) orientalis n. sp. (Trypanosomatidae), a parasite from Thailand responsible for localised cutaneous leishmaniasis. Parasit Vectors 11, 351–359 (2018).
Kwakye-Nuako, G. et al. First isolation of a new species of Leishmania responsible for human cutaneous leishmaniasis in Ghana and classification in the Leishmania enriettii complex. Int J Parasitol 45, 679–684 (2015).
Lobsiger, L. et al. An autochthonous case of cutaneous bovine leishmaniasis in Switzerland. Vet Parasitol 169, 408–414 (2010).
Muller, N. et al. Occurrence of Leishmania sp. in cutaneous lesions of horses in Central Europe. Vet Parasitol 166, 346–351 (2009).
Reuss, S. M. et al. Autochthonous Leishmania siamensis in horse, Florida, USA. Emerg Infect Dis 18, 1545–1547 (2012).
Rose, K. et al. Cutaneous leishmaniasis in red kangaroos: isolation and characterisation of the causative organisms. Int J Parasitol 34, 655–664 (2004).
Ivens, A. C. & Blackwell, J. M. The Leishmania genome comes of age. Parasitol Today 15, 225–231 (1999).
Ivens, A. C. et al. The genome of the kinetoplastid parasite, Leishmania major. Science 309, 436–442 (2005).
Albanaz, A. T. S. et al. Genome analysis of Endotrypanum and Porcisia spp., closest phylogenetic relatives of Leishmania, highlights the role of amastins in shaping pathogenicity. Genes (Basel) 12, 444–463 (2021).
Espinosa, O. A., Serrano, M. G., Camargo, E. P., Teixeira, M. M. G. & Shaw, J. J. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology 145, 430–442 (2018).
NCBI BioSample https://identifiers.org/ncbi/biosample:SAMN17294109 (2021).
NCBI BioSample https://identifiers.org/ncbi/biosample:SAMN17294111 (2021).
NCBI BioSample https://identifiers.org/ncbi/biosample:SAMN17294112 (2021).
NCBI BioSample https://identifiers.org/ncbi/biosample:SAMN17294115 (2021).
NCBI BioSample https://identifiers.org/ncbi/biosample:SAMN17294129 (2021).
NCBI BioSample https://identifiers.org/ncbi/biosample:SAMN17294121 (2021).
NCBI BioProject https://identifiers.org/ncbi/bioproject:PRJNA691531 (2021).
NCBI BioProject https://identifiers.org/ncbi/bioproject:PRJNA691532 (2021).
NCBI BioProject https://identifiers.org/ncbi/bioproject:PRJNA691534 (2021).
NCBI BioProject https://identifiers.org/ncbi/bioproject:PRJNA691536 (2021).
NCBI BioProject https://identifiers.org/ncbi/bioproject:PRJNA689706 (2021).
NCBI BioProject https://identifiers.org/ncbi/bioproject:PRJNA691541 (2021).
Almutairi, H. hatimalmutairi/LGAAP. https://doi.org/10.5281/zenodo.4663265 (2021).
Almutairi, H. et al. LGAAP: Leishmaniinae Genome Assembly and Annotation Pipeline. Microbiol Resour Announc 10, e0043921 (2021).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37, 540–546 (2019).
Li, H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008. https://doi.org/10.1093/gigascience/giab008 (2021).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421–428 (2009).
NCBI. The UniVec Database. https://www.ncbi.nlm.nih.gov/tools/vecscreen/univec/ (2016).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014).
Aslett, M. et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38, D457–462 (2010).
Almutairi, H. Supplementary materials for chromosome-scale genome sequencing, assembly and annotation of six genomes from subfamily Leishmaniinae. Lancaster University https://doi.org/10.17635/lancaster/researchdata/474 (2021).
Delcher, A. L., Salzberg, S. L. & Phillippy, A. M. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics Chapter 10: Unit 10.3. https://doi.org/10.1002/0471250953.bi1003s00 (2003).
Palmer, J. & Stajich, J. nextgenusfs/funannotate: funannotate v1.5.3 (Version 1.5.3). Zenodo. https://doi.org/10.5281/zenodo.2604804 (2019).
Abrusan, G., Grundmann, N., DeMester, L. & Makalowski, W. TEclass–a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25, 1329–1330 (2009).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011).
Hoff, K. J. & Stanke, M. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinformatics 65, e57 (2019).
Gremme, G., Steinbiss, S. & Kurtz, S. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform 10, 645–656 (2013).
Dainat, J., Hereñú, D., & Pucholt, P. NBISweden/AGAT: AGAT-v0.7.0 (v0.7.0). Zenodo. https://doi.org/10.5281/zenodo.5036996 (2021).
UniProt, C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49, D480–D489 (2021).
Mistry, J. et al. Pfam: The protein families database in 2021. Nucleic Acids Res 49, D412–D419 (2021).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957074 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957073 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957072 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957071 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957070 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957069 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957068 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957067 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957066 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957065 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957064 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957063 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957062 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957061 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957060 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957059 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957058 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957057 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957056 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957055 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957054 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957079 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957078 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957077 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957076 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957075 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957086 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957085 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957084 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957083 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957082 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957081 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957080 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957038 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957037 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957036 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957035 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957034 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957048 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957047 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957046 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957045 (2021).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRX9957044 (2021).
Almutairi, H. L. (Mundinia) martiniquensis: functional annotations. Lancaster University https://doi.org/10.17635/lancaster/researchdata/446 (2021).
Almutairi, H. L. (Mundinia) orientalis: functional annotations. Lancaster University https://doi.org/10.17635/lancaster/researchdata/449 (2021).
Almutairi, H. L. (Mundinia) enriettii: functional annotations. Lancaster University https://doi.org/10.17635/lancaster/researchdata/452 (2021).
Almutairi, H. L. (Mundinia) sp. Ghana: functional annotations. Lancaster University https://doi.org/10.17635/lancaster/researchdata/455 (2021).
Almutairi, H. L. (Mundinia) sp. Namibia: functional annotations. Lancaster University https://doi.org/10.17635/lancaster/researchdata/458 (2021).
Almutairi, H. Porcisia hertigi: functional annotations. Lancaster University https://doi.org/10.17635/lancaster/researchdata/461 (2021).
Mölder, F. et al. Sustainable data analysis with Snakemake. F1000Research 10, https://f1000research.com/articles/10-33/v2 (2021).
NCBI Assembly https://identifiers.org/insdc.gca:GCA_017916325.1 (2021).
NCBI Assembly https://identifiers.org/insdc.gca:GCA_017916335.1 (2021).
NCBI Assembly https://identifiers.org/insdc.gca:GCA_017916305.1 (2021).
NCBI Assembly https://identifiers.org/insdc.gca:GCA_017918215.1 (2021).
NCBI Assembly https://identifiers.org/insdc.gca:GCA_017918225.1 (2021).
NCBI Assembly https://identifiers.org/insdc.gca:GCA_017918235.1 (2021).
Almutairi, H. L. (Mundinia) martiniquensis raw reads quality reports. Lancaster University https://doi.org/10.17635/lancaster/researchdata/437 (2021).
Almutairi, H. Leishmania (Mundinia) orientalis raw reads quality reports. Lancaster University https://doi.org/10.17635/lancaster/researchdata/438 (2021).
Almutairi, H. Leishmania (Mundinia) enriettii raw reads quality reports. Lancaster University https://doi.org/10.17635/lancaster/researchdata/439 (2021).
Almutairi, H. Leishmania (Mundinia) sp. Ghana raw reads quality reports. Lancaster University https://doi.org/10.17635/lancaster/researchdata/440 (2021).
Almutairi, H. Leishmania (Mundinia) sp. Namibia raw reads quality reports. Lancaster University https://doi.org/10.17635/lancaster/researchdata/441 (2021).
Almutairi, H. Porcisia hertigi raw reads quality reports. Lancaster University https://doi.org/10.17635/lancaster/researchdata/442 (2021).
Almutairi, H. L. (Mundinia) martiniquensis: mapped reads in SAM and BAM format. Lancaster University https://doi.org/10.17635/lancaster/researchdata/444 (2021).
Almutairi, H. L. (Mundinia) orientalis: mapped reads in SAM and BAM format. Lancaster University https://doi.org/10.17635/lancaster/researchdata/447 (2021).
Almutairi, H. L. (Mundinia) enriettii: mapped reads in SAM and BAM format. Lancaster University https://doi.org/10.17635/lancaster/researchdata/450 (2021).
Almutairi, H. L. (Mundinia) sp. Ghana: mapped reads in SAM and BAM format. Lancaster University https://doi.org/10.17635/lancaster/researchdata/453 (2021).
Almutairi, H. L. (Mundinia) sp. Namibia: mapped reads in SAM and BAM format. Lancaster University https://doi.org/10.17635/lancaster/researchdata/456 (2021).
Almutairi, H. Porcisia hertigi: mapped reads in SAM and BAM format. Lancaster University https://doi.org/10.17635/lancaster/researchdata/459 (2021).
Almutairi, H. L. (Mundinia) martiniquensis: classified repeated sequences. Lancaster University https://doi.org/10.17635/lancaster/researchdata/445 (2021).
Almutairi, H. L. (Mundinia) orientalis: classified repeated sequences. Lancaster University https://doi.org/10.17635/lancaster/researchdata/448 (2021).
Almutairi, H. L. (Mundinia) enriettii: classified repeated sequences. Lancaster University https://doi.org/10.17635/lancaster/researchdata/451 (2021).
Almutairi, H. L. (Mundinia) sp. Ghana: classified repeated sequences. Lancaster University https://doi.org/10.17635/lancaster/researchdata/454 (2021).
Almutairi, H. L. (Mundinia) sp. Namibia: classified repeated sequences. Lancaster University https://doi.org/10.17635/lancaster/researchdata/457 (2021).
Almutairi, H. Porcisia hertigi: classified repeated sequences. Lancaster University https://doi.org/10.17635/lancaster/researchdata/460 (2021).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Seppey, M., Manni, M. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol 1962, 227–245 (2019).
Acknowledgements
We thank the Ministry of Health and Public Health Authority of Saudi Arabia for funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Almutairi, H., Urbaniak, M.D., Bates, M.D. et al. Chromosome-scale genome sequencing, assembly and annotation of six genomes from subfamily Leishmaniinae. Sci Data 8, 234 (2021). https://doi.org/10.1038/s41597-021-01017-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-01017-3