Abstract
Recent advancements in magnetoencephalography (MEG)-based brain-computer interfaces (BCIs) have shown great potential. However, the performance of current MEG-BCI systems is still inadequate and one of the main reasons for this is the unavailability of open-source MEG-BCI datasets. MEG systems are expensive and hence MEG datasets are not readily available for researchers to develop effective and efficient BCI-related signal processing algorithms. In this work, we release a 306-channel MEG-BCI data recorded at 1KHz sampling frequency during four mental imagery tasks (i.e. hand imagery, feet imagery, subtraction imagery, and word generation imagery). The dataset contains two sessions of MEG recordings performed on separate days from 17 healthy participants using a typical BCI imagery paradigm. The current dataset will be the only publicly available MEG imagery BCI dataset as per our knowledge. The dataset can be used by the scientific community towards the development of novel pattern recognition machine learning methods to detect brain activities related to motor imagery and cognitive imagery tasks using MEG signals.
Measurement(s) | brain physiology trait |
Technology Type(s) | Magnetoencephalography |
Factor Type(s) | age group • sex |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.13561976
Similar content being viewed by others
Background & Summary
Mental imagery activities such as imagination of limb movement or mathematical calculation induce explicit and predictive patterns of brain activity that can be detected using electroencephalography (EEG) or magnetoencephalography (MEG)1. One of the most prominent brain patterns is event-related desynchronization/synchronization (ERD/ERS) of brainwaves in the alpha (8–13 Hz) and beta (16–30 Hz) frequency bands during motor imagery tasks. Brain-computer interfaces (BCIs) can detect and translate these patterns into actions and thus, provide a potential medium for communication and rehabilitation for patients with severe neuromuscular impairment2,3,4. MI-based BCIs employed with neurofeedback training paradigms can induce brain plasticity and possibly contribute to the enhancement of motor rehabilitation for stoke patients5,6,7, thus, may provide an alternative to conventional recovery methods e.g. physical practice8 for these patients.
While majority of the research to date has focused on EEG modality, MEG can also be useful towards developing effective BCI systems9,10. MEG has the advantage of recording brain activity across the whole scalp while maintaining much higher spatial and temporal resolution. In addition, compared to EEG, MEG allows detection of higher frequencies as magnetic fields are less attenuated by the head bone and tissue as compared to electric fields11. Though not portable, MEG-based BCIs are relevant for rehabilitation interventions.
Regardless of great potential, MEG-based BCI systems still need significant improvement in terms of robust and efficient signal processing algorithms. A big constraint towards the development of novel algorithms or validating currently available BCI signal processing pipelines is lack of open source MEG-BCI datasets. As per our knowledge, there are no sizable datasets available currently. In this work, we publish an MEG-based BCI dataset recorded using a conventional BCI paradigm involving MI and cognitive imagery (CI) tasks. The dataset contains 1134 minutes of MEG recordings across 34 recording sessions of 17 healthy participants (two sessions for each participant recorded on different days), and 6,800 imagery trials. BCI interactions involved two MI (both hands and both feet imagination) and two CI (word generations and mathematical subtraction) states. On average, 66 minutes of MEG recordings and 400 imagery trials are available per participant. The dataset is one of the first MI- and CI-related MEG-based BCI datasets published to date and presents a significant step from existing datasets in terms of uniformity, state-of-the-art MEG system, number of participants and MEG channels.
Methods
Participants
The study involved recruitment of 20 healthy participants. However, data of three participants are excluded from the dataset due to quality issues. Thus, the current dataset consists of 17 participants including 14 males (82.35%) and 3 females (17.64%), wherein median age of participants is 28 years with minimum age 22 years and maximum age 40 years. Out of 17, 15 participants are right-handed and 2 participants are left-handed (by self reporting). Table 1 provides the demographic information of all the participants. The names of all participants have been hereby anonymised. The participants are identified only by their participant Ids i.e. ‘sub-1’ through ‘sub-20’. Ids of excluded participants are ‘sub-5’, ‘sub-8’ and ‘sub-10’.
The experimental procedures were approved by the University Research Ethics Committee of the Ulster University, Northern Ireland, UK. All research procedures were carried out in accordance with approved institutional guidelines and regulations and guidelines of the Helsinki declaration. Prior to the data acquisition process, all participants were informed about the purpose and the procedures of the experiments and informed consenting procedure was followed wherein participants provided written consent to allow usage of their anonymised data for research purposes by other researchers. The participants had been screened for the absence of any psychiatric condition, any medications taken, and contraindications to MEG. Inclusion criteria were as follows: healthy individuals, age between 18 to 80 years (both inclusive), and no history of neurological, developmental or language deficits. Exclusion criteria were as follows: claustrophobic, pregnant or breastfeeding, body tattoos, metal or active body implants and on-going medications.
MEG data acquisition
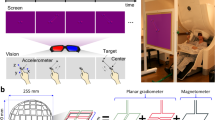
MEG data were recorded with a 306-channel (102 magnetometers and 204 planar gradiometers) Elekta NeuromagTM system (Elekta Oy, Helsinki, Finland) located at the Northern Ireland Functional Brain Mapping (NIFBM) Facility of the Intelligent Systems Research Centre, Ulster University. Elekta NeuromagTM system (Elekta Oy, Helsinki, Finland) is installed with MaxShieldTM system which is a high-performance magnetic shielding system designed and optimised for bioelectromagnetic measurements using Elekta NeuromagTM. The system consists of structurally optimal magnetically shielded room with internal active shielding. All the participants were screened for any metallic foreign substance e.g. jewelry, coins, keys or any other ferromagnetic material before entering the magnetically shielded room. The standard fiducial landmarks (left and right pre – auricular points and Nasion), five head position indicator (HPI) coils (placed over scalp), and the additional reference points over the scalp were digitized (Fastrak Polhemus system) to store information about the participant’s head position, orientation, and shape. In addition, ocular and cardiac activities were recorded with two sets of bipolar electro – oculogram (EOG) electrodes (horizontal – EOG and vertical – EOG) and one set of electrocardiogram (EKG) electrodes, respectively. Before starting the data acquisition, the complete procedure and the experimental paradigm were described to the participants. All recordings were made with participants seated on a comfortable chair approximately 80 cm away from the projector screen and in upright position of MEG scanner. The MEG signals were filtered at a bandwidth of 0.01–300 Hz (online) and sampled at the rate of 1 kHz during the acquisition itself. Continuous head position estimation was started after 20 s of raw data recording and kept running for rest of the acquisition period.
Experimental paradigm
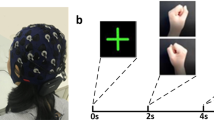
Figure 1 presents the timing diagram of the BCI paradigm used for the data acquisition. Each trial starts with a rest period of 2 s followed by 5 s of imagery task period. The cue remains visible during the whole imagery task period. During the rest period, participants were asked to fixate on a red cross presented at the center of the screen. A randomly selected inter- trial – interval (ITI) of 1.5–2 s was presented after the imagery task period. The fixation point and the cue were displayed on a Panasonic projector with a screen resolution of 1024 × 768 and refresh rate of 60 Hz. MEG data were acquired over 2 sessions (each session on different days) using the same BCI paradigm. Each session consisted of 50 trials for each of the imagery tasks, thus includes a total of 200 trials. A break of 5 minute duration was provided in each session after completion of first 100 trials. The participants were kept seated during the break and asked to relax.
The experimental paradigm was designed to cover four mental imagery tasks: two related to MI i.e. both hands movement imagery, both feet movement imagery and two related to CI i.e. mathematical subtraction imagery and word generation imagery. During the MI-related tasks, participants imagined movement of both hands/both feet when the related cue appeared at the screen (i.e. during the task period). Similarly, for CI tasks, participants either subtracted two numbers presented as cue or generated words related to the English language alphabet appeared as cue. Triggers were recorded within the .fif files (Elekta NeuromagTM system) to mark the start of imagery period for each trial.
Data processing
The original MEG dataset was acquired from all 306 sensors (204 gradiometers and 102 magnetometers) during two different sessions for each participant and recorded as .fif files. As each session consists of two data files due to session break, for better handling of the data, we have merged these files to create one single .fif file for each session. Thus, there are two raw .fif data files for each participant (i.e. one for each session). Our aim here is to provide the BCI researchers least processed data to allow them with greater flexibility towards customising the processing pipeline. However, we have also processed the .fif file format to convert the data in an epoched format (.mat file) to be compatible for BCI related analysis. Each epoch (trial) is generated with time duration of 7000 ms i.e. 2000 ms (pre-stimulus) to 5000 ms (post-stimulus). The triggers are available in both BIDS and.mat formats, where the classes are defined as follows:- Class 1: Both Hand Imagery, Class 2: Both Feet Imagery, Class 3: Word generation Imagery, and Class 4: Subtraction Imagery and their associated triggers in the STIM channels are 4, 8, 16, and 32, respectively. A detailed description of the data file structure is presented in Section ‘Data Records’. The fieldTrip Toolbox12 has been used in all data processing steps.
Data Records
The data acquired during the described experiment are freely accessible and may be downloaded from figshare13, which is a general-purpose repository that makes research outputs available in a citable, shareable, and discover-able manner. It is worth to be noted that the data is available in two data formats i.e. MEG-BIDS format14 (.fif) and MATLAB compatible (.mat) file at the repository. Figure 2 shows the structure of the data directory for MEG-BIDS format where only one participant data structure is illustrated to avoid repetition. The folder named ‘MEG_BIDS’ contain two files named ‘dataset_description.json’ and ‘participant.tsv’. Further, there are 17 sub-folders (one for each participant data), each having scan file ‘_scan.tsv’ and a sub-folder named ‘meg’. Each ‘meg’ folder contains five files i.e. ‘_coordsystem.json’, ‘_channels.tsv’, ‘_events.tsv’, ‘_meg.fif’, and ‘_meg.json’.
The structure of BIDS format data directory, where MEG_BIDS is a root folder. Under MEG_BIDS folder, each participant has its data folder (e.g. sub–1 is for participant 1), where two sub-folders are given for Session 1 and Session 2 of data recording, each sub-folder has a meg folder, where all the required information is available and ‘.fif’ files contain the MEG recording.
We have also provided data in Matlab compatible format and shared the script at GitHub as well to convert the MEG-BIDS format to .mat file format. The root database folder (MEG_mat) contains two folders, namely Session_01 and Session_02, which store datasets recorded on day 1 and day 2, respectively. Within each session folder, there are seventeen.mat files i.e. one for each participant. We have used a similar name convention for all files within the database e.g. in sub-1_ses-1_task-bcimici_meg.mat filename, ‘sub-1’ shows participant Id and ‘ses-1’ stands for session number. Each of these .mat files contains a Matlab structure with name ‘dataMAT’. Table 2 provides names, data type, data size, and description of all the fields present within the ‘dataMAT’ Matlab structure. To provide more flexibility to readers, we have provided the data in both BIDS and .mat file format, which can be downloaded from figshare13. The root database folder is (MEG_BIDS) for BIDS format and (MEG_mat) for Matlab.
Each session has 200 trials, stored in a cell array [1 × 200], named ‘data.trial’, and each trial has data from 306 channels for 7 s time duration (i.e. [306 × 7000]), where sampling frequency is 1000 Hz. The class labels are stored in ‘data.trialinfo’ which is an array of size [200 × 1].
Technical Validation
We performed a technical validation of the dataset by estimating and evaluating spatio-temporal features for six binary-classification tasks. For this analysis, MEG data from 204 gradiometer sensors were used while discarding the data from 102 magnetometers, as former provide higher sensor-to-noise ratio and are more sensitive to cortical activations. It is well known that SMRs are more prominent in cortical brain regions. Further, we have selected data for a 3 s time duration i.e. from 0.5 s to 3.5 s after the onset of imagery task. To generate spatio-temporal features, one of the state-of-the-art methods (i.e. filter-bank common spatial pattern (FBCSP)) was employed. This method involves two main steps i.e. band-pass filtering within different frequency ranges (creating a filter-bank) and estimation of CSP features using the band-pass filtered data from previous step15. To explore the effect of selecting different combinations of frequency ranges, two filter-banks, namely FB1 and FB2, were created and CSP features were generated for both filter-banks separately. FB1 consisted of two frequency ranges i.e. 8–12 Hz – alpha (α) band and 14–30 Hz– beta (β) band. FB2 consisted of ten overlapping frequency ranges i.e. 8–12 Hz, 10–14 Hz, 12–16 Hz, 14–18 Hz, 16–20 Hz, 18–22 Hz, 20–24 Hz, 22–26 Hz, 24–28 Hz, and 26–30 Hz.
To evaluate the BCI performance, classification accuracies (CAs) were estimated by using a support vector machine (SVM) classifier for six binary classification tasks, i.e. hand versus feet (H-F), hand versus word generation (H-W), hand versus subtraction (H-S), feet versus word generation (F-W), feet versus subtraction (F-S), and word generation versus subtraction (W-S). This evaluation was performed for both intra-session condition (i.e. 10-fold cross-validation using Session 1 data) and inter-session condition (i.e. training of classifier with feature set of Session 1 data and evaluation on feature set of Session 2 data). The main reason for using 10-fold cross-validation estimator is that is has a lower variance than a single hold-out set estimator, which can be important if the amount of available data is limited as in our case we have 200 trials in each session.
The 10-fold cross-validation (intra-session condition) performance is reported using box plot in Fig. 3 with both filter-bank combinations (i.e. FB1 and FB2) for 6 different binary tasks comparisons (i.e. H-F, H-W, H-S, F-W, F-S, and W-S). The CA for FB1 ranged from 96.29% to 98.29% and for FB2 range from 99% to 99.94%. The overall results showed a high separability between the feature sets of different classes. The results for inter-session condition are reported in Tables 3 and 4 for FB1 and FB2, respectively. For FB1 which includes α and β frequency bands, H-W (i.e. hand vs word) class pair has achieved maximum average (over 17 participants) classification accuracy (i.e. 69.35%), wherein participant sub-3 performed best with 94% and sub-4 has the lowest accuracy of 50%. In FB2, H-S (i.e. hand vs subtraction) class pair has achieved maximum average classification accuracy (i.e. 66.65%), wherein participant sub-20 performed best with 93% and sub-4 has the lowest accuracy 50%. Figure 4 shows comparison between average classification accuracies of FB1 and FB2 for six binary classification tasks in inter-session condition. Here, FB1 performed better than FB2 for majority of class pairs.
Notably, the CAs for inter-session condition is significantly lower than the intra-session condition for all binary classification tasks. Importantly, most of the machine learning methods in BCI are facing an issue of low performance in terms of classification accuracy, which may be due to the presence of non-stationarity in the data recorded over multiple sessions16,17. According to the literature, there are several reasons for the presence of non-stationarity in the data such as head movement, user fatigue, change in mood, or external noise interfering the MEG system18. We believe that the low CAs in the inter-session condition may be due to the presence of high non-stationarity (i.e. covariate shift) between MEG data of Session 1 and Session 2. The covariate shift is a case, where the input distribution of the data shifts (i.e. (Ptrain(x) ≠ Ptest(x))), whereas the conditional probability remains the same, while transitioning from the training to testing stage, which in our case is from Session 1 to Session 219,20,21). The covariate shift between Session 1 and Session 2 is a challenging issue, as demonstrated by a large difference between the performances of single-trial classification, wherein 10-fold cross-validation average accuracy on Session 1 data is significantly higher than evaluation average accuracy on Session 2 data. We have examined input data distribution between Session 1 and Session 2 for all participants and found that all the participants’ data had some form of covariate shift. Figures 5 and 6 illustrate the presence of covariate shift in the feature set of the participant sub-20 for of α and β frequency bands, respectively. It is to be noted that the sub-20 data provided highest inter-session classification accuracy. Each figure consists of six sub-figures representing distribution between class pairs of six binary classification tasks. In each sub-figure, two ellipses with blue dashed line show the training distribution (Ptrain(x)) for the two participating classes (e.g. two classes for top-left sub-figure in Fig. 5 are Hand and Foot imagery) and black dashed line presents the decision hyper-plane for the training dataset. Similarly, the ellipses with red points boundary show the test data distribution Ptest(x) for the same classes and the red dash line presents the decision hyper-plane for the test dataset. A clear shift pattern for the datasets can be seen within both Figs. 5 and 6, i.e. for majority of the class pairs, the training data has high separability as compared to the test data and there are large shifts in decision hyper-planes in most cases. This variation in inter-class separability may explain the low classification accuracies while evaluating the trained classifier with Session 2 data.
Usage Notes
There are several potential uses for this database. Firstly, it can be used to test the effectiveness of already developed EEG-BCI data analysis pipelines using this MEG dataset. Secondly, we encourage any use that can contribute towards development of novel pattern recognition and machine learning methods to detect brain activities related to MI and CI tasks using MEG signals. Thirdly, since we have performed a basic analysis and single-trial classification of tasks using the raw data, future work may involve exploring impact of various MEG pre-processing methods e.g. head movement correction and maxfiltering22. Additionally, as the dataset contains two sessions that were recorded on different days for each participant, robustness of analysis pipelines towards inter-session non-stationarity can be assessed using this dataset. More importantly very high spatial resolution of MEG facilitates much enhanced source-level analysis. The data-sets can used for investigating source level features in accounting for inherent non-stationarity present in MEG neuro-imaging modality primarily due to head movements.
Code availability
The pre-processing and feature extraction of the MEG data, as well as the single-trial classification were performed using custom Matlab codes based on Fieldtrip toolbox12 functions. All codes are available at our GitHub repository https://github.com/sagihaider/MEGBCI2020.git.
References
Rathee, D., Cecotti, H. & Prasad, G. Single-trial effective brain connectivity patterns enhance discriminability of mental imagery tasks. Journal of neural engineering 14, 056005 (2017).
Birbaumer, N. & Cohen, L. G. Brain–computer interfaces: communication and restoration of movement in paralysis. The Journal of physiology 579, 621–636 (2007).
Daly, J. J. & Wolpaw, J. R. Brain–computer interfaces in neurological rehabilitation. The Lancet Neurology 7, 1032–1043 (2008).
Rathee, D. et al. Brain–machine interface-driven post-stroke upper-limb functional recovery correlates with beta-band mediated cortical networks. IEEE Transactions on Neural Systems and Rehabilitation Engineering 27, 1020–1031 (2019).
Prasad, G., Herman, P., Coyle, D., McDonough, S. & Crosbie, J. Applying a brain-computer interface to support motor imagery practice in people with stroke for upper limb recovery: a feasibility study. Journal of neuroengineering and rehabilitation 7, 60 (2010).
Chowdhury, A., Raza, H., Meena, Y. K., Dutta, A. & Prasad, G. Online covariate shift detection-based adaptive brain–computer interface to trigger hand exoskeleton feedback for neuro-rehabilitation. IEEE Transactions on Cognitive and Developmental Systems 10, 1070–1080 (2017).
Chowdhury, A. et al. Active physical practice followed by mental practice using BCI-driven hand exoskeleton: a pilot trial for clinical effectiveness and usability. IEEE journal of biomedical and health informatics 22, 1786–1795 (2018).
Wriessnegger, S. C., Steyrl, D., Koschutnig, K. & Müller-Putz, G. R. Short time sports exercise boosts motor imagery patterns: implications of mental practice in rehabilitation programs. Frontiers in human neuroscience 8, 469 (2014).
Mellinger, J. et al. An meg-based brain–computer interface (BCI). Neuroimage 36, 581–593 (2007).
Halme, H.-L. & Parkkonen, L. Comparing features for classification of MEG responses to motor imagery. PLOS ONE 11, 1–21 (2016).
Hämäläinen, M. S. Magnetoencephalography: a tool for functional brain imaging. Brain topography 5, 95–102 (1992).
Oostenveld, R., Fries, P., Maris, E. & Schoffelen, J.-M. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Intell. Neuroscience 2011, 1:1–1:9 (2011).
Rathee, D., Raza, H., Roy, S. & Prasad, G. A magnetoencephalography dataset for motor and cognitive imagery BCI, https://doi.org/10.6084/m9.figshare.c.5101544 (2021).
Niso, G. et al. MEG-BIDS, the brain imaging data structure extended to magnetoencephalography. Scientific data 5, 1–5 (2018).
Ang, K. K., Chin, Z. Y., Zhang, H. & Guan, C. Filter bank common spatial pattern (FBCSP) in brain-computer interface. In 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), 2390–2397 (IEEE, 2008).
Raza, H., Prasad, G. & Li, Y. Adaptive learning with covariate shift-detection for non-stationary environments. In 2014 14th UK Workshop on Computational Intelligence (UKCI), 1–8 (IEEE, 2014).
Raza, H., Cecotti, H., Li, Y. & Prasad, G. Learning with covariate shift-detection and adaptation in non-stationary environments: Application to brain-computer interface. In 2015 International Joint Conference on Neural Networks (IJCNN), 1–8 (IEEE, 2015).
Okazaki, Y. O. et al. Real-time MEG neurofeedback training of posterior alpha activity modulates subsequent visual detection performance. NeuroImage 107, 323–332 (2015).
Raza, H., Cecotti, H., Li, Y. & Prasad, G. Adaptive learning with covariate shift-detection for motor imagery-based brain–computer interface. Soft Computing 20, 3085–3096 (2016).
Raza, H., Rathee, D., Zhou, S.-M., Cecotti, H. & Prasad, G. Covariate shift estimation based adaptive ensemble learning for handling non-stationarity in motor imagery related EEG-based brain-computer interface. Neurocomputing 343, 154–166 (2019).
Raza, H., Prasad, G. & Li, Y. EWMA model based shift-detection methods for detecting covariate shifts in non-stationary environments. Pattern Recognition 48, 659–669 (2015).
Roy, S., Rathee, D., Chowdhury, A., McCreadie, K. & Prasad, G. Assessing impact of channel selection on decoding of motor and cognitive imagery from MEG data. Journal of Neural Engineering 17, 056037 (2020).
Acknowledgements
H.R. was supported by the Economic and Social Research Council (ESRC) funded Business and Local Government Data Research Centre (grant number ES/S007156/1). G. P. was supported in part by the Northern Ireland Functional Brain Mapping Facility Project through InvestNI and the Ulster University under Grant 1303/101154803 and in part by the UKIERI DST Thematic Partnership Project under Grant DST-UKIERI-2016-17-0128.
Author information
Authors and Affiliations
Contributions
D.R. prepared the experimental paradigm, recruited participants, recorded MEG data, processed the database, wrote and revised the manuscript and managed the whole process. H.R. advised data recording and wrote and revised the manuscript. S.R. recruited participants, recorded MEG data, processed the database, and revised the manuscript. G.P. advised and supervised the project and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Rathee, D., Raza, H., Roy, S. et al. A magnetoencephalography dataset for motor and cognitive imagery-based brain-computer interface. Sci Data 8, 120 (2021). https://doi.org/10.1038/s41597-021-00899-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00899-7
This article is cited by
-
Domain-independent short-term calibration based hybrid approach for motor imagery electroencephalograph classification: a comprehensive review
Multimedia Tools and Applications (2024)
-
Coherence-based channel selection and Riemannian geometry features for magnetoencephalography decoding
Cognitive Neurodynamics (2024)
-
A magnetoencephalography dataset during three-dimensional reaching movements for brain-computer interfaces
Scientific Data (2023)