Abstract
Brain computer interfaces (BCIs) are valuable tools that expand the nature of communication through bypassing traditional neuromuscular pathways. The non-invasive, intuitive, and continuous nature of sensorimotor rhythm (SMR) based BCIs enables individuals to control computers, robotic arms, wheel-chairs, and even drones by decoding motor imagination from electroencephalography (EEG). Large and uniform datasets are needed to design, evaluate, and improve the BCI algorithms. In this work, we release a large and longitudinal dataset collected during a study that examined how individuals learn to control SMR-BCIs. The dataset contains over 600 hours of EEG recordings collected during online and continuous BCI control from 62 healthy adults, (mostly) right hand dominant participants, across (up to) 11 training sessions per participant. The data record consists of 598 recording sessions, and over 250,000 trials of 4 different motor-imagery-based BCI tasks. The current dataset presents one of the largest and most complex SMR-BCI datasets publicly available to date and should be useful for the development of improved algorithms for BCI control.
Measurement(s) | brain measurement |
Technology Type(s) | electroencephalography (EEG) |
Factor Type(s) | task • mindfulness • brain computer interface • alpha waves |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.14153504
Similar content being viewed by others
Background & Summary
Millions of individuals live with paralysis1. The emerging field of neural prosthetics seeks to provide relief to these individuals by forging new pathways of communication and control2,3,4,5,6. Invasive techniques directly record neural activity from the cortex and translate neural spiking into actionable commands such as moving a robotic arm or even individuals’ own muscles7,8,9,10,11,12,13. However, one major limitation of this approach is that roughly half of all implantations fail within the first year14.
A promising alternative to invasive techniques uses the electroencephalogram, or EEG, to provide brain recordings. One popular approach, the sensorimotor rhythm (SMR) based brain computer interface (BCI) detects characteristic changes in the SMR in response to motor imagery3,4,5,6,15,16. The mu rhythm, one of the most prominent SMRs, is an oscillation in the alpha band and reduces in strength when we move (or think about moving), which is called event-related desynchronization (ERD)17,18. The reliable detection of ERD, and its converse event-related synchronization (ERS), enables the intuitive and continuous control of a BCI19,20, which has been used to control computer cursors, wheelchairs, drones, and robotic arms21,22,23,24,25,26,27,28.
However, mastery of SMR-BCIs requires extensive training, and even after training, 15–20% of the population remain unable to control these devices27,29. While the reasons for this remain obscure, the strength of the resting SMR has been found to predict BCI proficiency, and evidence suggests that the resting SMR can be enhanced through behavioral interventions such as mindfulness training30,31,32. Alternatively, better decoding strategies can also improve BCI performance33,34.
One encouraging trend in BCI is to use artificial neural networks to decode brain states13,35,36,37. Critically, progress in creating robust and generalizable BCI decoding systems is currently hindered by the limited data available to train these decoding models. Most deep learning BCI studies perform training and testing on the BCI Competition IV datasets38,39,40,41,42. While these datasets enable benchmarking performance, the BCI Competition IV datasets 2a and 2b are small (9 subjects, 2–5 sessions) and simple (2a—4 class, no online feedback, 2b—2 class with online feedback, but only 3 motor electrodes)43. Most other datasets available online (e.g., those that can be found at http://www.brainsignals.de, or http://bnci-horizon-2020.eu) share similar limitations. Fortunately, two recently published datasets have attempted to address these concerns44,45. Cho et al. provide a large EEG BCI dataset with full scalp coverage (64 electrodes) and 52 participants, but only 36 minutes and 240 samples of 2-class motor imagery (i.e., left/right hand) per subject. Kaya et al. present a larger dataset (~60 hours of EEG recordings from 13 participants over 6 sessions, ~4.8 hours and 4600 trials per participant) with a more complicated design consisting of simple 2-classs motor imagery (left/right hand), 6-class motor imagery (left/right hand, left/right leg, tongue, rest), and motor imagery of individual fingers. However, the motor imaginations were only performed once, which is not suitable for continuous control, and scalp coverage is limited (19 electrodes). Further, neither study provided continuous online feedback.
We recently collected, to our knowledge, the largest SMR-BCI dataset to date30. In total, this dataset comprises 62 participants, 598 individual BCI sessions, and over 600 hours of high-density EEG recordings (64 channels) from 269,099 trials. Each individual participant completed 7–11 online BCI training sessions, and the dataset includes on average 4,340 trials and 9.9 hours’ worth of EEG data per participant. This dataset contains roughly 4.5 times as many trials and 10 times as much data as the next largest dataset currently available for public use44.
We believe this dataset should be of particular value to the field for four reasons: (1) the amount of EEG data is sufficient to train large decoding models, (2) the sample size permits tests of how well decoding models and signal processing techniques will generalize, (3) the BCI decoding tasks are challenging (e.g., up to 4-class continuous 2D control with online feedback), and (4) the longitudinal study design enables tests of how well decoding models and signal processing techniques adapt to session by session changes. This dataset may additionally provide new insights into how individuals control a SMR-BCI, respond to feedback, and learn to modulate their brain rhythms.
Methods
Participants and Experimental procedure
The main goals of our original study were to characterize how individuals learn to control SMR-BCIs and to test whether this learning can be improved through behavioral interventions such as mindfulness training30. Participants were initially assessed for baseline BCI proficiency and then randomly assigned to an 8-week mindfulness intervention (Mindfulness-based stress reduction), or waitlist control condition where participants waited for the same duration as the MBSR class before starting BCI training, but were offered a comparable MBSR course after completing all experimental requirements46. Following the 8-weeks, participants returned to the lab for 6–10 sessions of BCI training.
All experiments were approved by the institutional review boards of the University of Minnesota and Carnegie Mellon University. Informed consents were obtained from all subjects. In total, 144 participants were enrolled in the study and 76 participants completed all experimental requirements. Seventy-two participants were assigned to each intervention by block randomization, with 42 participants completing all sessions in the experimental group (MBSR before BCI training; MBSR subjects) and 34 completing experimentation in the control group. Four subjects were excluded from the analysis due to non-compliance with the task demands and one was excluded due to experimenter error. We were primarily interested in how individuals learn to control BCIs, therefore analysis focused on those that did not demonstrate ceiling performance in the baseline BCI assessment (accuracy above 90% in 1D control). The dataset descriptor presented here describes data collected from 62 participants: 33 MBSR participants (Age = 42+/−15, (F)emale = 26) and 29 controls (Age = 36+/−13, F = 23). In the United States, women are twice as likely to practice meditation compared to men47,48. Therefore, the gender imbalance in our study may result from a greater likelihood of women to respond to flyers offering a meditation class in exchange for participating in our study.
For all BCI sessions, participants were seated comfortably in a chair and faced a computer monitor that was placed approximately 65 cm in front of them. After the EEG capping procedure (see data acquisition), the BCI tasks began. Before each task, participants received the appropriate instructions. During the BCI tasks, users attempted to steer a virtual cursor from the center of the screen out to one of four targets. Participants initially received the following instructions: “Imagine your left (right) hand opening and closing to move the cursor left (right). Imagine both hands opening and closing to move the cursor up. Finally, to move the cursor down, voluntarily rest; in other words, clear your mind.” In separate blocks of trials, participants directed the cursor toward a target that required left/right (LR) movement only, up/down (UD) only, and combined 2D movement (2D)30. Each experimental block (LR, UD, 2D) consisted of 3 runs, where each run was composed of 25 trials. After the first three blocks, participants were given a short break (5–10 minutes) that required rating comics by preference. The break task was chosen to standardize subject experience over the break interval. Following the break, participants competed the same 3 blocks as before. In total, each session consisted of 2 blocks of each task (6 runs total of LR, UD, and 2D control), which culminated in 450 trials performed each day.
Data acquisition
Researchers applied a 64-channel EEG cap to each subject according to the international 10–10 system. The distances between nasion, inion, preauricular points and the cap’s Cz electrode were measured using a measuring tape to ensure the correct positioning of the EEG cap to within ±0.25 cm. The impedance at each electrode was monitored and the capping procedure ensured that the electrodes’ impedance (excluding the rare dead electrode; see artifacts below) remained below 5 kΩ. EEG was acquired using SynAmps RT amplifiers and Neuroscan acquisition software (Compumedics Neuroscan, VA). The scalp-recorded EEG signals were digitized at 1000 Hz and filtered between 0.1 to 200 Hz with an additional notch filter at 60 Hz and then stored for offline analysis. EEG electrode locations were recorded using a FASTRAK digitizer (Polhemus, Colchester, Vermont).
BCI recordings
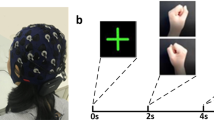
Each trial is composed of 3 parts: the inter-trial interval, target presentation, and feedback control. Participants initially saw a blank screen (2 s). They were then shown a vertical or horizontal yellow bar (the target) that appeared in one of the cardinal directions at the edge of the screen for 2 s. After the target presentation, a cursor (i.e., a pink ball) appeared in the center of the screen. The participants were then given up to 6 s to contact the target by moving the cursor in the correct direction by modulating their SMRs. Trials were classified as “hits” when the cursor contacted the correct target and “misses” when the cursor happened to contact one of the other 3 edges of the screen. “Timeouts” occurred when 6 s elapsed without selecting any target. An inter-trial interval of 2 s followed the end of a trial, and then a new trial began. Performance was quantified by a percent valid correct (PVC) metric and was defined as hits/(hits + misses). PVC was averaged across runs for each BCI session. Participants were considered proficient in a given task if their average block or session PVC crossed a given threshold (70% for 1D tasks [LR,UD] and 40% for the 2D task)49.
BCI experiments were conducted in BCI200020. Online control of the cursor proceeded in a series of steps. The first step, feature extraction, consisted of spatial filtering and spectrum estimation. During spatial filtering, the average signal of the 4 electrodes surrounding the hand knob of the motor cortex was subtracted from electrodes C3 and C4 to reduce the spatial noise. Following spatial filtering, the power spectrum was estimated by fitting an autoregressive model of order 16 to the most recent 160 ms of data using the maximum entropy method. The goal of this method is to find the coefficients of a linear all-pole filter that, when applied to white noise, reproduces the data’s spectrum. The main advantage of this method is that it produces high frequency resolution estimates for short segments of data. The parameters are found by minimizing (through least squares) the forward and backward prediction errors on the input data subject to the constraint that the filter used for estimation shares the same autocorrelation sequence as the input data. Thus, the estimated power spectrum directly corresponds to this filter’s transfer function divided by the signal’s total power. Numerical integration was then used to find the power within a 3 Hz bin centered within the alpha rhythm (12 Hz).
The translation algorithm, the next step in the pipeline, then translated the user’s alpha power into cursor movement. Horizontal motion was controlled by lateralized alpha power (C4 - C3) and vertical motion was controlled by up and down regulating total alpha power (C4 + C3). These control signals were normalized to zero mean and unit variance across time by subtracting the signals’ mean and dividing by its standard deviation. A balanced estimate of the mean and standard deviation of the horizontal and vertical control signals was calculated by estimating these values across time from data derived from 30 s buffers of individual trial type (e.g., the normalized control signal should be positive for right trials and negative for left trials, but the average of left and right trials should be zero). Finally, the normalized control signals were used to update the position of the cursor every 40 ms.
Data Records
Distribution for use
The data files for the large electroencephalographic motor imagery dataset for EEG BCI have been uploaded to the figshare repository50. These files can be accessed at https://doi.org/10.6084/m9.figshare.13123148.
EEG data organization for BCI tasks
This dataset (Citation 51) consists of 598 data files, each of which contains the complete data record of one BCI training session. Each file holds approximately 60 minutes of EEG data recorded during the 3 BCI tasks mentioned above and comprises 450 trials of online BCI control. Each datafile is from one subject and one session and is identified by their subject number and BCI session number.
All data files are shared in .mat format and contain MATLAB-readable records of the raw EEG data and the recording session’s metadata described below. The data in each file are represented as an instance of a Matlab structure named “BCI” having the following key fields “data,” “time,” “positionx,” “positiony,” “SRATE,” “TrialData,” “metadata,” and “chaninfo” (detailed in Table 1). The file naming system is designed to facilitate batch processing and scripted data analysis. The file names are initially grouped by subject number and then session number and can be easily accessed through nested for loops. For example, SX_Session_Y.mat is the filename for subject X’s data record of their Yth BCI training session. Participants are numbered 1 through 62 and sessions are numbered 1 (i.e., the baseline BCI session that occurs 8 weeks prior to the main BCI training) through 11 (or 7 if the participant only completed 6 post intervention training sessions).
The fields of the structure “BCI” comprising the data record of each file are as follows. The main field is “data,” where the EEG traces of each trial of the session are stored. Since each trial varies in length, the “time” data field contains a vector of the trial time (in ms) relative to target presentation. The two position fields, “positionx” and “positiony,” contain a vector documenting the cursor position on the screen for each trial. “SRATE” is a constant representing the sampling rate of the EEG recording, which was always 1000 Hz in these experiments. “TrialData” is a structure containing all the relevant descriptive information about a given trial (detailed in Table 2). The field “metadata” contains participant specific demographic information (detailed in Table 3). Finally, “chaninfo” is a data structure containing relevant information about individual EEG channels (detailed in Table 4).
The “data” field contains the recording session’s EEG data in the format of a 1 × nTrials cell array, where each entry in the cell array is a 2D Matlab array of size nChannels × nTime. The number of trials per session, nTrials, is nearly always 450. Each row of a trial’s 2D data matrix is the time-series of voltage measurements (in μV) from a single EEG input lead such as C3 or C4. The “time” field contains the time data in the format of a 1 × nTrials cell array, where each entry is a 1 × nTime vector of the time index of the trial referenced to target presentation. Each trial begins with a 2 s inter-trial interval (index 1, t = −2000ms), followed by target presentation for 2 s (index 2001, t = 0). Feedback begins after target presentation (index 4001, t = 2000ms) and continues until the end of the trial. The end of the trial can be found in the “resultind” subfield of “TrialData” (see below). A post-trial interval of 1 s (1000 samples) follows target selection or the timeout signal.
While the length of the trial may vary, nTime (the number of time samples of the trial), the number of channels, nChannels, is always 62. The ordering of the channels can be found in the “chaninfo” structure’s subfield “label”, which lists the channel names of the 62 electrodes. If individual electrode positions were recorded, the subfield “positionsrecorded” will be set to 1 and the “electrodes,” “fiducials,” and “shape” subfields will be populated according to this information. “Electrodes” contains the electrode labels and their X, Y, and Z coordinates. The information in the “fiducials” subfield contains the X, Y, and Z locations of the nasion as well as the left and right preauricular points. The shape of the face was recorded by sweeping from ear to ear under the chin and then drawing arcs horizontally across the brow and vertically from the forehead to chin. This information is contained in the “shape” subfield. If the session specific individual electrode positions are not available, “positionsrecorded” will be set to zero, the “fiducials” and “shape” subfields will be empty, and the “electrodes” subfield will include generic electrode locations for the 10–10 system positioning of the Neuroscan Quik-Cap. Finally, the subfield “noisechan” identifies which channels were found to be particularly noisy during the BCI session (see Technical Validation) and can be used for easy exclusion of this data or channel interpolation.
The position subfields of the “BCI” structure, “positionx” and “positiony,” provide the cursor positions throughout the trial in the format of a 1 × nTrials cell array. Each entry of this cell array is a 1 × nTime vector of the horizontal or vertical position of the cursor throughout the trial. Positions are provided by BCI2000 starting with feedback control (index 4001) and continue through the end of the trial (TrialData(trial).resultind). The inter-trial intervals before and after feedback are padded with NaNs. The positional information ranges from 0 (left/bottom edge of the screen) to 1 (right/top edge of the screen), with the center of the screen at 0.5. During 1D tasks (LR/UD), the orthogonal position remains constant (e.g., in the LR task, positiony = 0.5 throughout the trial).
The “TrialData” structure subfield of “BCI” provides valuable information for analysis and supervised machine learning (detailed in Table 2). This structure contains the subfields “tasknumber,” “runnumber,” “trialnumber,” “targetnumber,” “triallength,” “targethitnumber,” “resultind,” “result,” “forcedresult,” and “artifact.” Tasknumber is used to identify the individual BCI tasks (1 = ‘LR’, 2 = ‘UD’, 3 = ‘2D’). Targetnumber is used to identify which target was presented to the participants (1 = right, 2 = left, 3 = up, 4 = down). “Triallength” is the length of the feedback control period in seconds, with the maximum length of 6.04 s occurring in timeout trials. “Resultind” is the time index that indicates when the feedback ended (when a target was selected or the trial was considered a timeout). To easily extract the feedback portion of the EEG data, you simply define an iterator variable (e.g., trial = 5) and index into the data subfield [e.g., trial_feedback = BCI.data{trial}(4001:BCI.TrialData(trial).resultind)]. “Targethitnumber” identifies which target was selected during feedback and takes the same values as “targetnumber” mentioned above. These values will be identical to “targetnumber” when the trial is a hit, different when the trial is a miss, and NaN when a trial is a timeout. “Result” is a label for the outcome of the trial which takes values of 1 when the correct target was selected, 0 when an incorrect target is selected and NaN if the trial was labeled as a timeout. “Forcedresult” takes the same values as before, however in timeout trials, the final cursor position is used to select the target closest to the cursor thereby imposing the BCI system’s best guess onto timeout trials. Finally the “artifact” subfield of the “TrialData” structure is a bool indicating whether an artifact was present in the EEG recording (see Technical Validation).
Lastly, the “metadata” field of the “BCI” data structure includes participant and session level demographic information (detailed in Table 3). “Metadata” contains information related to the participant as an individual, factors that could influence motor imagery, information regarding the mindfulness intervention, and individual session factors. Participant demographic information includes “age” (in years), “gender” (‘M’, ‘F’ NaN if not available), and “handedness” (‘R’, ‘L’, NaN). Motor imagery factors include “instrument”, “athlete”, “handsport”, and “hobby.” These factors are reported as ‘Y’ for yes, ‘N’ for no, ‘U’ for used to, and NaN if not available. Subfields for the mindfulness intervention include “MBSRsubject” “meditationpractice”. Finally, session factors included “date, “day”, and “time”.
While the data is formatted for ease of use with Matlab, this software is not required. The data structure is compatible with Octave. However, to open the data in python, the fields of the BCI structure should be saved in separate variables. For example, the cell BCI.data can be saved as ‘data’ in a.mat file and then loaded into python as an ndarray with the scypy.io function loadmat. The TrialData structure can also be converted to a matrix and cell of column names then converted to a pandas dataframe.
Technical Validation
Automatic artifact detection was used to demonstrate the high quality of the individual data records45,51. First, the EEG data were bandpass filtered between 8 Hz and 30 Hz. If the variance (across time) of the bandpass filtered signals exceeded a z-score threshold of 5 (z-scored between electrodes), these electrodes were labeled as artifact channels for a given session. This information can be found in BCI.chaninfo.noisechan. Overall, only 3% of sessions contained more than 4 artifact channels (Fig. 1a). These channels were excluded from the subsequent artifact detection step and their values were interpolated spatially with splines for the ERD topographies shown below52. Then, if the bandpass filtered data from any remaining electrode crossed a threshold of ±100 μV throughout a trial, this trial was labeled as containing an artifact. Artifact trials are labeled in the TrialData structure (BCI.TrialData.artifact). Again, only 3% of sessions had more than 5% of trials labeled as containing artifacts (Fig. 1b).
More research is needed to understand how individuals learn to control brain computer interfaces27, and this dataset provides a unique opportunity to study this process. Not only does the online BCI performance demonstrate that motor imagery can be decoded from the EEG data records, it also shows that the decidability of the participants’ EEG rhythms improves as individuals learn to control the device (Fig. 2a). BCI proficiency is also an active area of study53, and, by the end of training, roughly 90% of all the participants in our study were considered proficient in each task (Fig. 2b). However, the sample size of this dataset is large enough that 7–8 participants can still be examined for BCI inefficiency in each task. This dataset additionally enables the study of the transition from BCI inefficiency to proficiency.
Online BCI performance across sessions (a) Following the baseline assessment, the average online BCI performance exceeds chance level across all tasks suggesting decodable motor imagery patterns are present in the EEG data records. As BCI training continues, participants produce better linearly classifiable signals in the alpha band. The shaded area represents ±1 standard error of the mean (SEM). (b) Group-level BCI proficiency throughout training. This curve shows the percentage of participants that are proficient in BCI control as a function of BCI training session. At the end of training, approximately 90% of all subjects were considered proficient in each task of BCI control.
Event-Related Desynchronization (ERD) was calculated to verify the quality of the EEG signals collected during BCI control17,45. To calculate ERD for each channel, the EEG data were first highpass filtered above 1 Hz to remove drifts, then bandpass filtered between 8 Hz and 14 Hz52. The Hilbert transform was then applied to all of the trials and the absolute magnitude was taken for each complex value of each trial. The magnitudes of the Hilbert-transformed samples were averaged across all trials. Finally, these averages were baseline corrected to obtain a percentage value for ERD/ERS according to the formula \({\rm{ERD}} \% =\frac{A-R}{R}\ast 100 \% \), where A is each time sample and R is the mean value of the baseline period (−1 s to 0 s). The participant averaged ERD signal for channels C3 and C4, and for each trial type, are shown in Fig. 3a. ERD values were averaged over the feedback portion of trials to create the topography images in Fig. 3b. The ERD curves and scalp topographies show the expected responses to motor imagery such as stronger desynchronization in the contralateral motor cortex during motor imagery and stronger synchronization during rest.
Event-related desynchronization during BCI control (a) ERD averaged across participants and training sessions. These curves show the change in alpha power (8–14 Hz) over the left (C3; red) and right (C4; blue) motor cortex relative to the power in the inter-trial interval. The ERD curves show the expected responses to motor imagery such as stronger desynchronization (negative values) in the contralateral motor cortex during motor imagery and stronger synchronization during rest (positive values). The rectangles at the bottom of each plot display the different phases of each trial with red, yellow, and green representing the inter-trial interval, target presentation, and feedback control periods, respectively. The shaded are represents ± 1 SEM. (b) Topographies of ERD values across the cortex for each trial type. T-values were found by testing the ERD values across the population in our study. Cooler colors represent more desynchronization during motor imagery and warmer colors represent more synchronization.
Code availability
The code used to produce the figures in this manuscript is available at https://github.com/bfinl/BCI_Data_Paper.
References
Armour, B. S., Courtney-Long, E. A., Fox, M. H., Fredine, H. & Cahill, A. Prevalence and Causes of Paralysis-United States, 2013. Am. J. Public Health 106, 1855–7 (2016).
Chaudhary, U., Birbaumer, N. & Ramos-Murguialday, A. Brain-computer interfaces for communication and rehabilitation. Nature Reviews Neurology 12, 513–525 (2016).
Wolpaw, J. R., Birbaumer, N., McFarland, D. J., Pfurtscheller, G. & Vaughan, T. M. Brain-computer interfaces for communication and control. Clinical Neurophysiology 113, 767–791 (2002).
He, B., Baxter, B., Edelman, B. J., Cline, C. C. & Ye, W. W. Noninvasive brain-computer interfaces based on sensorimotor rhythms. Proc. IEEE 103, 907–925 (2015).
Vallabhaneni, A., Wang, T. & He, B. Brain Computer Interface. in Neural Engineering (ed. He, B.) 85–122 (Kluwer Academic/Plenum Publishers, 2005).
Yuan, H. & He, B. Brain-computer interfaces using sensorimotor rhythms: Current state and future perspectives. IEEE Transactions on Biomedical Engineering 61, 1425–1435 (2014).
Taylor, D. M., Tillery, S. I. H. & Schwartz, A. B. Direct cortical control of 3D neuroprosthetic devices. Science (80-.). 296, 1829–1832 (2002).
Serruya, M. D., Hatsopoulos, N. G., Paninski, L., Fellows, M. R. & Donoghue, J. P. Instant neural control of a movement signal. Nature 416, 141–142 (2002).
Musallam, S., Corneil, B. D., Greger, B., Scherberger, H. & Andersen, R. A. Cognitive control signals for neural prosthetics. Science (80-.). 305, 258–262 (2004).
Carmena, J. M. et al. Learning to Control a Brain–Machine Interface for Reaching and Grasping by Primates. PLoS Biol. 1, e42 (2003).
Velliste, M., Perel, S., Spalding, M. C., Whitford, A. S. & Schwartz, A. B. Cortical control of a prosthetic arm for self-feeding. Nature 453, 1098–1101 (2008).
Hochberg, L. R. et al. Reach and grasp by people with tetraplegia using a neurally controlled robotic arm. Nature 485, 372–375 (2012).
Schwemmer, M. A. et al. Meeting brain–computer interface user performance expectations using a deep neural network decoding framework. Nat. Med. 24, 1669–1676 (2018).
Barrese, J. C. et al. Failure mode analysis of silicon-based intracortical microelectrode arrays in non-human primates. J. Neural Eng. 10, 066014 (2013).
He, B., Yuan, H., Meng, J. & Gao, S. Brain-Computer Interfaces. in Neural Engineering (ed. He, B.) 131–183, https://doi.org/10.1007/978-1-4614-5227-0 (Springer, 2020).
Wang, T., Deng, J. & He, B. Classifying EEG-based motor imagery tasks by means of time–frequency synthesized spatial patterns. Clin. Neurophysiol. 115, 2744–2753 (2004).
Pfurtscheller, G. & Lopes Da Silva, F. H. Event-related EEG/MEG synchronization and desynchronization: Basic principles. Clinical Neurophysiology 110, 1842–1857 (1999).
Yuan, H. et al. Negative covariation between task-related responses in alpha/beta-band activity and BOLD in human sensorimotor cortex: An EEG and fMRI study of motor imagery and movements. Neuroimage 49, 2596–2606 (2009).
Pfurtscheller, G. & Neuper, C. Motor imagery and direct brain-computer communication. Proc. IEEE 89, 1123–1134 (2001).
Schalk, G., McFarland, D. J., Hinterberger, T., Birbaumer, N. & Wolpaw, J. R. BCI2000: A general-purpose brain-computer interface (BCI) system. IEEE Trans. Biomed. Eng. 51, 1034–1043 (2004).
Huang, D. et al. Electroencephalography (EEG)-based brain-computer interface (BCI): A 2-D virtual wheelchair control based on event-related desynchronization/synchronization and state control. IEEE Trans. Neural Syst. Rehabil. Eng. 20, 379–388 (2012).
Rebsamen, B. et al. A brain controlled wheelchair to navigate in familiar environments. IEEE Trans. Neural Syst. Rehabil. Eng. 18, 590–598 (2010).
Edelman, B. J. et al. Noninvasive neuroimaging enhances continuous neural tracking for robotic device control. Sci. Robot. 4, 1–13 (2019).
Lafleur, K. et al. Quadcopter control in three-dimensional space using a noninvasive motor imagery-based brain–computer interface. J. Neural Eng 10, 46003–15 (2013).
Wolpaw, J. R. & McFarland, D. J. Control of a two-dimensional movement signal by a noninvasive brain-computer interface in humans. Proc. Natl. Acad. Sci. USA 101, 17849–17854 (2004).
Luu, T. P., Nakagome, S., He, Y. & Contreras-Vidal, J. L. Real-time EEG-based brain-computer interface to a virtual avatar enhances cortical involvement in human treadmill walking. Sci. Rep. 7, 1–12 (2017).
Perdikis, S. et al. The Cybathon race: Successful longitudinal mutual learning with two tetraplegic users. PLoS Biol. 16, e2003787, https://doi.org/10.1371/journal.pbio.200 (2018).
Li, Y. et al. An EEG-based BCI system for 2-D cursor control by combining Mu/Beta rhythm and P300 potential. IEEE Trans. Biomed. Eng. 57, 2495–2505 (2010).
Guger, C., Edlinger, G., Harkam, W., Niedermayer, I. & Pfurtscheller, G. How many people are able to operate an EEG-based brain-computer interface (BCI)? IEEE Trans. Neural Syst. Rehabil. Eng. 11, 145–147 (2003).
Stieger, J. R. et al. Mindfulness Improves Brain–Computer Interface Performance by Increasing Control Over Neural Activity in the Alpha Band. Cereb. Cortex 31, 426–438 (2021).
Ahn, M., Cho, H., Ahn, S. & Jun, S. C. High theta and low alpha powers may be indicative of BCI-illiteracy in motor imagery. PLoS One 8(11), e80 (2013).
Blankertz, B. et al. Neurophysiological predictor of SMR-based BCI performance. Neuroimage 51, 1303–1309 (2010).
Guger, C. et al. Complete Locked-in and Locked-in Patients: Command Following Assessment and Communication with Vibro-Tactile P300 and Motor Imagery Brain-Computer Interface Tools. Front. Neurosci. 11, 251 (2017).
Blankertz, B., Tomioka, R., Lemm, S., Kawanabe, M. & Müller, K. R. Optimizing spatial filters for robust EEG single-trial analysis. IEEE Signal Process. Mag. 25, 41–56 (2008).
Craik, A., He, Y. & Contreras-Vidal, J. L. Deep learning for electroencephalogram (EEG) classification tasks: A review. Journal of Neural Engineering 16, 031001, https://doi.org/10.1088/1741-2552/ab0ab5 (2019).
Schirrmeister, R. T. et al. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 38, 5391–5420 (2017).
Jiang, X., Lopez, E., Stieger, J. R., Greco, C. M. & He, B. Effects of Long-Term Meditation Practices on Sensorimotor Rhythm-Based Brain-Computer Interface Learning. Front. Neurosci. 14, 1443 (2021).
Lu, N., Li, T., Ren, X. & Miao, H. A Deep Learning Scheme for Motor Imagery Classification based on Restricted Boltzmann Machines. IEEE Trans. Neural Syst. Rehabil. Eng. 25, 566–576 (2017).
Lawhern, V. J. et al. EEGNet: A compact convolutional neural network for EEG-based brain-computer interfaces. J. Neural Eng. 15, 56013–56030 (2018).
Sakhavi, S., Guan, C. & Yan, S. Learning Temporal Information for Brain-Computer Interface Using Convolutional Neural Networks. IEEE Trans. Neural Networks Learn. Syst. 29, 5619–5629 (2018).
Zhang, Z. et al. A Novel Deep Learning Approach with Data Augmentation to Classify Motor Imagery Signals. IEEE Access 7, 15945–15954 (2019).
Wang, P., Jiang, A., Liu, X., Shang, J. & Zhang, L. LSTM-based EEG classification in motor imagery tasks. IEEE Trans. Neural Syst. Rehabil. Eng. 26, 2086–2095 (2018).
Tangermann, M. et al. Review of the BCI competition IV. Frontiers in Neuroscience 6, 55 (2012).
Kaya, M., Binli, M. K., Ozbay, E., Yanar, H. & Mishchenko, Y. Data descriptor: A large electroencephalographic motor imagery dataset for electroencephalographic brain computer interfaces. Sci. Data 5, 180211 (2018).
Cho, H., Ahn, M., Ahn, S., Kwon, M. & Jun, S. C. EEG datasets for motor imagery brain-computer interface. GigaScience 6, 1–8 (2017).
Kabat-Zinn, J. An outpatient program in behavioral medicine for chronic pain patients based on the practice of mindfulness meditation: Theoretical considerations and preliminary results. Gen. Hosp. Psychiatry 4, 33–47 (1982).
Cramer, H. et al. Prevalence, patterns, and predictors of meditation use among US adults: A nationally representative survey. Scientific Reports 6, 1–9 (2016).
Upchurch, D. M. & Johnson, P. J. Gender differences in prevalence, patterns, purposes, and perceived benefits of meditation practices in the United States. J. Women’s Heal. 28, 135–142 (2019).
Combrisson, E. & Jerbi, K. Exceeding chance level by chance: The caveat of theoretical chance levels in brain signal classification and statistical assessment of decoding accuracy. J. Neurosci. Methods 250, 126–136 (2015).
Stieger, J. R., Engel, S. A. & He, B. Human EEG Dataset for Brain-Computer Interface and Meditation. figshare https://doi.org/10.6084/m9.figshare.13123148 (2021).
Muthukumaraswamy, S. D. High-frequency brain activity and muscle artifacts in MEG/EEG: A review and recommendations. Frontiers in Human Neuroscience 7 (2013).
Oostenveld, R., Fries, P., Maris, E. & Schoffelen, J. M. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput. Intell. Neurosci. 2011, 156869 (2011).
Alkoby, O., Abu-Rmileh, A., Shriki, O. & Todder, D. Can We Predict Who Will Respond to Neurofeedback? A Review of the Inefficacy Problem and Existing Predictors for Successful EEG Neurofeedback Learning. Neuroscience 378, 155–164 (2018).
Acknowledgements
The authors would like to thank the following individuals for useful discussions and assistance in the data collection: Drs. Mary Jo Kreitzer and Chris Cline. This work was supported in part by NIH under grants AT009263, EB021027, NS096761, MH114233, EB029354.
Author information
Authors and Affiliations
Contributions
B.H. conceptualized and designed the study; and supervised the project. J.S. conducted the experiments, collected all data, and validated the data. S.E. contributed to the experimental design of data collection.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Stieger, J.R., Engel, S.A. & He, B. Continuous sensorimotor rhythm based brain computer interface learning in a large population. Sci Data 8, 98 (2021). https://doi.org/10.1038/s41597-021-00883-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00883-1
This article is cited by
-
A large EEG database with users’ profile information for motor imagery brain-computer interface research
Scientific Data (2023)
-
Neural correlates of user learning during long-term BCI training for the Cybathlon competition
Journal of NeuroEngineering and Rehabilitation (2022)
-
Open multimodal iEEG-fMRI dataset from naturalistic stimulation with a short audiovisual film
Scientific Data (2022)