Abstract
SARS-CoV-2 is a novel coronavirus responsible for the COVID-19 pandemic, in which acute respiratory infections are associated with high socio-economic burden. We applied high-content screening to a well-defined collection of 5632 compounds including 3488 that have undergone previous clinical investigations across 600 indications. The compounds were screened by microscopy for their ability to inhibit SARS-CoV-2 cytopathicity in the human epithelial colorectal adenocarcinoma cell line, Caco-2. The primary screen identified 258 hits that inhibited cytopathicity by more than 75%, most of which were not previously known to be active against SARS-CoV-2 in vitro. These compounds were tested in an eight-point dose response screen using the same image-based cytopathicity readout. For the 67 most active molecules, cytotoxicity data were generated to confirm activity against SARS-CoV-2. We verified the ability of known inhibitors camostat, nafamostat, lopinavir, mefloquine, papaverine and cetylpyridinium to reduce the cytopathic effects of SARS-CoV-2, providing confidence in the validity of the assay. The high-content screening data are suitable for reanalysis across numerous drug classes and indications and may yield additional insights into SARS-CoV-2 mechanisms and potential therapeutic strategies.
Measurement(s) | cytotoxicity • killing by virus of host cell |
Technology Type(s) | bright-field microscopy |
Factor Type(s) | concentration • incubation time |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.13562570
Similar content being viewed by others
Background & Summary
Coronaviridae is a family of encapsulated single-stranded positive-sense RNA viruses that typically cause mild respiratory diseases such as the common cold in humans1. However, several β coronavirus strains have emerged over the last two decades that cause acute respiratory infections associated with high mortality, particularly in individuals with underlying health conditions. Three major outbreaks have occurred, the first caused by Severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002/20032, the second caused by Middle East respiratory syndrome coronavirus (MERS-CoV) in 20123, and the latest caused by Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which was first recorded in December 2019 and was declared pandemic in March 20204. At the time of writing (9th December 2020), more than 69 million confirmed SARS-CoV-2 infections have been reported worldwide and more than 1.6 million people have died from the associated disease, COVID-195.
As a complement to the safe and effective vaccines against SARS-CoV-2, the repurposing of existing drugs represents a pragmatic strategy for the treatment of COVID-19 patients6. Drug repurposing (drug repositioning) is advantageous in the face of rapidly-spreading emerging diseases because the pathway to approval is facilitated by pre-existing preclinical (and often clinical or post-marketing) data, existing production capacity and supply chains can be used, and new mechanisms of action can be discovered that may suggest potential drug combinations for enhanced efficacy by exploiting synergistic effects7. Furthermore, bioinformatics analysis has identified 66 druggable proteins based on the SARS-CoV-2/human host cell interactome, which will allow the selection of compounds that interfere with interactions essential in the viral replication cycle8. Regardless of the origin of drugs against SARS-CoV-2, properly controlled clinical safety and efficacy studies are still needed before the approval of pharmacological treatments for COVID-19.
Multiple interventional clinical trials have been initiated in the search for effective treatments against SARS-CoV-29. The individual drugs or combination treatments for these studies have often been selected based on known activities against SARS-CoV, Ebola virus, HIV or Plasmodium spp., hence the drugs under investigation include remdesivir, interferon β and ribavirin and lopinavir/ritonavir10. However, the search for effective drugs against SARS-CoV-2 could extend beyond known antivirals and anti-infectives if suitable high-throughput assays are used to identify candidates.
Here we describe a high-content imaging dataset generated by screening a well-defined collection of 5632 compounds including 3488 with clinical or post-marketing data across 600 indications. The compounds were screened for their ability to inhibit the cytopathic effect of SARS-CoV-2 in the human epithelial colorectal adenocarcinoma cell line Caco-2 using the assay design shown in Fig. 1. The compounds we used are well annotated in terms of primary and secondary targets and we therefore hope the data and meta-data presented herein will be combined with our results and the findings of other researchers to identify additional treatment options for COVID-19, including drug combinations. One important avenue for reuse is to determine whether any of these clinical-stage compounds or related molecules could safely achieve active concentrations at the principal SARS-CoV-2 infection site, the human lung epithelium. In this manner, the combination of our in vitro activity data with information about tissue distribution may help to determine the most promising avenues for future COVID-19 preclinical and clinical studies.
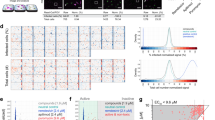
Cytopathicity assay workflow. To test for compound cytotoxicity, Caco-2 cells were grown to confluence and incubated with test compounds for 48 h. To test whether the compounds influenced the cytopathic effect of SARS-CoV-2, Caco-2 cells were grown to confluence and incubated with the test compounds and SARS-CoV-2 for 48 h. For image acquisition, the cells were fixed to comply with BSL1 requirements and imaged by digital phase contrast (DPC) microscopy. Upper image set shows the primary screen to determine cytotoxicity and cytopathic effect. Top two rows are representative wells of the compound area including three active molecules. Bottom row shows four positive control wells (virus treated, no compounds) and four negative control wells (no virus, no compounds). Lower image set shows the dose-response screen used to measure the anti-cytopathic effects of each compound at eight concentrations, allowing the calculation of IC50 values. Scale bar = 500 µm.
Methods
Compound collection
The collection of screening compounds was assembled by an external partner (SPECS) to mirror the set established by the Broad Institute11. The compounds were purchased from the same set of >70 high-quality suppliers identified by the Broad Institute (Dr Joshua Bitker, personal communication) and were quality controlled by liquid chromatography/mass spectrometry (LC/MS) for purity and identity (minimum purity >90%). The compounds were stored at a concentration of 10 mM in 100% DMSO at -20 °C. A curated database is available listing the compounds, indications, known primary targets and known mechanisms of action, as well as analysis tools that can help to determine the targets and mechanisms of action if unknown11.
Cell culture
Human epithelial colorectal adenocarcinoma cell line Caco-2 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Cells were grown in minimal essential medium (MEM) supplemented with 10% foetal bovine serum (FBS), 100 IU/ml penicillin and 100 µg/ml streptomycin at 37 °C. All culture reagents were obtained from Sigma-Aldrich.
Virus culture
SARS-CoV-2 was isolated from samples of travellers returning from Wuhan, China to Frankfurt, Germany12. SARS-CoV-2 stocks used in the experiments were passaged once on Caco-2 cells and were stored at -80 °C. Virus titres were determined as TCID50/ml using confluent cells in 96-well microtiter plates.
Cytopathicity assays for primary screening and concentration-response studies
The inhibition of the SARS-CoV-2 cytopathic effect was measured using the assay workflow summarized in Fig. 112. Compounds were added to confluent layers of Caco-2 cells in MEM supplemented with 1% FBS in 96-well plates. For the primary screen, the final compound concentration was 10 µM (in 0.1% DMSO) and each compound was tested in singlicate. Concentration-response profiling of primary hits was performed in triplicate with eight concentrations ranging from 20 µM to 20 nM (half-log dilution factor, 0.1% DMSO final). For initial dose response studies, compounds were tested on three different plates to minimise the influence of plate effects. Later dose response studies at lower concentrations were also done on the same plate. Following the addition of compounds, cells were immediately infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01. Positive controls were prepared with the virus but no compounds, and negative controls were prepared without the virus. After 48 h, the medium was removed and cells were fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS). The plates were sealed and disinfected to inactivate the virus13 then screened by high-content imaging using the Operetta CLS platform (PerkinElmer). The image data were processed as described below and deposited on the Image Data Resource14.
Image acquisition and analysis
Images were acquired using a 10× objective with nine imaged fields per well in label-free mode by digital phase contrast (DPC). In DPC two brightfield images are acquired at different Z positions and then an overlay image is calculated showing superior contrast and intensity properties. This method improves the signal-to-noise ratio and allows for image segmentation and single-cell analysis15. The images were analysed using PerkinElmer Columbus v2.9.0. The analysis sequence started with cell detection (method c, common threshold 0.05, area >100 µm2, splitting coefficient 6.5, individual threshold 0.05, contrast >0.05) and was followed by calculating morphology, intensity and position properties as well as cell confluence (Fig. 2). The data were deposited on the Image Data Resource14. Although we used cell confluence as a read out parameter additional parameters were calculated and deposited as well. These parameters can be used for further analysis to include parameters related to cellular stress or cytoskeletal impairment, both critical for viral replication. Well-level data were analysed in ActivityBase (IDBS) and R v3.6.1, using cell confluence as final read out parameter. Test results were normalized to the corresponding intra-plate virus-free negative control (assigned as 100% inhibition of cytopathicity) and virus-plus positive control (assigned as 0% inhibition of cytopathicity). Outliers were eliminated according to the three-sigma method. Plate-level statistical performance was assessed using the Z’factor calculation with values between 0.35 and 1.0 representing an acceptable assay16. Data for the primary screen were deposited on ChEMBL17.Concentration-response curves were fitted in GraphPad Prism v8.0.0 (GraphPad Software) using a four-parameter logarithmic (least squares fit). Data for the dose-response screen were deposited on ChEMBL18.
Analysis workflow. The analysis workflow is shown using mock and virus control. Images are magnified from representative fields of view. Digital phase contrast images were acquired using 10x and nine fields of view per well and subjected to image analysis using Columbus v2.9.0 (PerkinElmer). Calculated parameters offered are derived from the wells the pictures presented were taken and are given as mean with standard deviation except Confluent Cell Area which is given as sum of all nine fields of view per well. Additionally, the parameters refer only to cells recognized by the software. Confluent Cell Area (Sum) was used for the calculation of % effect, but other parameters were deposited as well and images can be downloaded and reanalysed from Image Data Resource at (https://idr.openmicroscopy.org under accession number idr0094).
Analysis of compound toxicity
Cellular toxicity was determined by measuring intracellular ATP concentrations. Caco-2 cells were harvested using Acutase followed by resuspension to the required number of 2000 cells/well in Eagle’s minimal essential medium (EMEM) supplemented with 10% FCS, 1% non-essential amino acids, 2 mM l-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 1 mM sodium pyruvate. We transferred 20 µl of the cell suspension from each well to white solid 384-well CellStar plates (GreinerBio) and incubated the cells for 20 h at 37 °C in a 5% CO2 atmosphere. We then added compounds to the cells using an Echo 550 R Liquid Handler. Assay plates were incubated for an additional 48 h at 37 °C in a 5% CO2 atmosphere. We then prepared the CellTiter-Glo Luminescent Cell Viability Assay reagent (Promega) according to the manufacturer’s protocol. The assay plates were equilibrated at room temperature for 10 min then supplemented with 10 µl/well of the CellTiter-Glo reagent. The assay plates were centrifuged for 1 min at 700 × g and incubated for 10 min in the dark at room temperature before luminescence measurements were recorded on an EnVision plate reader (Perkin Elmer), with a measurement time of 0.05 s per sample. The image data were analysed using ActivityBase, outliers were eliminated according to the two-sigma method, and the data relating to each compound were normalized to the high (0.5% v/v DMSO, 100% viability) and low (25 µM sodium selenite, 0% viability) controls, 16 wells each, of the corresponding plates19. All data were generated as triplicates. The cytotoxicity data were deposited on ChEMBL20. The combination of cytotoxicity and cytopathic effect data allowed us to calculate the cytotoxic index (the gap between the cytopathic effect and cytotoxicity) which was also deposited on ChEMBL21.
Hit selection and definition of hit
Cytopathicity assays were conducted in 96 well Cellstar plates (Greiner bio-one) using 4 wells with cells without virus (negative control) and 4 wells with cells and with virus (positive control). A three-sigma invalidation was performed on the raw data of the controls on each plate, resulting in maximal one invalidated well per plate during primary screening. During hit confirmation and dose-response studies no wells were eliminated as all wells were within three-sigma. After invalidation the plate specific z’factor was calculated and the plate was removed if quality criteria (z’factor > 0.35) were not met. Plates passing quality control were analysed and normalized as specified above. Hits were identified by having at least 75% efficiency in blocking viral cytopathicity at 10 µM. These data are available as stated in in data records section. All hit compounds were subjected to dose response studies and the 67 most active ones were followed up in dose-response studies. These data were also made available as stated in in data records section.
Data Records
Data structure and repositories
Five datasets were generated during this study. The first contains all the microscopy images and raw data acquired during primary screening and dose response studies in the context of the cytopathicity assay and was deposited on the Image Data Resource (https://idr.openmicroscopy.org) under https://doi.org/10.17867/10000148 and accession number idr009414. Digital object identifier for primary screening data and the associated raw data at IDR is https://doi.org/10.17867/10000148a. Digital object identifier for dose response screening data and the associated raw data at IDR is https://doi.org/10.17867/10000148b. These datasets contain the variables listed in Table 1. The four other datasets describe calculated compound effects and were deposited on ChEMBL (https://www.ebi.ac.uk/chembl/) under accession numbers CHEMBL4303805 (cytopathicity, primary screen)17, CHEMBL4303806 (dose-response)18, CHEMBL4303807 (cytotoxicity data)20 and CHEMBL4303808 (cytotoxicity index – the ratio between the cytopathic effect and cytotoxicity)21. These accession numbers are aggregated using the Document ChEMBL ID CHEMBL4303101. The variables of these ChEMBL entries are explained in Table 2.
Data sets and file types
The image data are stored as 16-bit TIFF files with a resolution of 1080 × 1080. The dataset features one channel for DPC acquired at 740 nm. All image data can be downloaded (for details see: https://idr.openmicroscopy.org/about/download.html) or inspected using an online web client at https://idr.openmicroscopy.org.
The activity data are stored as CSV files in ChEMBL and can be downloaded or analysed in combination with other datasets using the variables given in Table 2 as connectors between the datasets.
Technical Validation
Primary screening was validated by calculating Z’factor values and all 64 plates passed the quality control threshold (Z’factor > 0.35). The average Z’factor value in the primary screen was 0.72. Pharmacological validation of the assay was carried out using four compounds previously reported to inhibit the cytopathic effects of SARS-CoV-2, SARS-CoV or MERS-CoV in phenotypic readouts using the African green monkey kidney epithelial cell line Vero-E6, namely cycloheximide, emetine, remdesivir and chloroquine. Our data are compared to the results from previous reports in Table 3. The IC50 values for emetine (0.52 ± 0.09 µM) and cycloheximide (0.58 ± 0.02 µM) were higher than the reported activities of these compounds against MERS-CoV, SARS-CoV and SARS-CoV-2 (range 0.04–0.18 µM). The IC50 of the RNA polymerase inhibitor remdesivir (0.76 ± 0.18 µM) was similar to the recently reported value of 0.77 µM22. Chloroquine showed no activity in the Caco-2 cell assay (IC50 > 20 µM) thus contrasting with its reported micromolar activity in Vero-E6 cell assays. This may reflect the use of different (human vs monkey) cell lines or different viral strains, and was confirmed in the primary screen, where chloroquine and related compounds were mostly inactive.
The confirmed hits included six compounds (camostat, nafamostat, lopinavir, mefloquine, papaverine and cetylpyridinium) that have already been shown to inhibit SARS-CoV-2, SARS-CoV or MERS-CoV in phenotypic assays using Vero-E6, Calu-3 (human lung) or BHK-21 (hamster kidney fibroblast) cells. Our data are compared to the results from previous reports in Table 4. Camostat and nafamostat are inhibitors of the viral entry protease TMPRSS-2. The potency of camostat against SARS-CoV-2 was ~100-fold greater in our Caco-2 cell assay than in a previous study using Vero-E6 cells23 but similar to the results of another study with the same virus and cell line24, as well as studies of MERS-CoV and SARS-CoV using Calu-3 cells25,26. Likewise, the potency of nafamostat against SARS-CoV-2 in our Caco-2 cell assay was >500-fold greater than in previous studies using Vero-E6 cells27,28 but similar to the results of a previous study of MERS-CoV cytopathicity against Calu-3 cells26. Papaverine was eight-fold more potent against SARS-CoV-2 (in Caco-2 cells) than against MERS-CoV (in BHK-21 cells)29 whereas cetylpyridinium showed similar potency in both assays29. These data confirm that the Caco-2 assay is pharmacologically relevant and suitable for the identification of anti-cytopathic compounds with diverse mechanisms of action. Our results using Caco-2 cells are more similar to previous studies with the human cell line Calu-3 than studies using non-human cells (Vero-E6 or BHK-21), particularly for viral entry inhibitors.
Concentration-dependent anti-cytopathic effects were not observed with the reference compound chloroquine although the related antimalarial mefloquine was identified in the primary screen and was moderately active in the subsequent concentration-response screen (IC50 = 14.1 µM). The anti-malarial drugs tafenoquine and pyronaridine were inactive as they showed < 50% inhibition in the concentration-response screen (Hit criteria was at > 75% maximum inhibition) and, along with hydroxychloroquine, were also inactive in the primary screen. This discrepancy may reflect the greater sensitivity of Vero-E6 cells to this class of compounds, or their limited solubility.
Finally, we compared our hits with the 69 compounds in clinical or pre-clinical development recently predicted to interact with druggable targets in the SARS CoV-2/human host cell interactome screen8. We identified 52 compounds or close analogues (Tanimoto > 0.9) in our compound library, seven of which (14%) were classified as active in our primary screen and three of which (nafamostat, pevonedistat and camostat) displayed an IC50 value < 1 µM.
Usage Notes
In this study, we screened a drug library of 5632 compounds for antiviral activity against SARS-CoV-2, a recently emerged virus responsible for COVID-19. A total of 117 compounds belonging to multiple drug classes showed the ability to inhibit viral cytopathicity to at least 50% at compound concentrations of up to 20 µM. Four of these hits had already been shown to inhibit SARS-CoV-2, providing validation for our choice of assay. In addition, we confirmed the antiviral activity of remdesivir (IC50 = 0.76 µM) during assay validation, a drug targeting the viral nsp12 RNA-dependent RNA polymerase30 that has been authorized for COVID-19 treatment under emergency use protocols (9). We also confirmed the antiviral activity of nafamostat (IC50 = 0.04 µM), which inhibits membrane fusion, and camostat (IC50 = 0.64 µM), which blocks viral entry by inhibiting the serine protease TMPRSS2 (11). Finally, thioguanine (IC50 = 1.71 µM) is reported as a competitive inhibitor of the protease PL-Pro in MERS-CoV and SARS-CoV31. Interestingly, the related glycoside analog thioguanosine (IC50 = 0.78 µM) was also active in our study. These results confirm that the assay can correctly select clinically relevant compounds with evidence for activity against viral entry and/or replication-dependent mechanisms.
Among the first group of drugs initially reported to show activity against SARS-CoV were anti-malarial compounds such as chloroquine and its close relative hydroxychloroquine28 although larger randomized controlled trials indicated they were largely ineffective32. We found that chloroquine diphosphate and other anti-malarial compounds (tafenoquine, amodiaquine, lumefantrine and primaquine) were inactive in the primary assay against Caco-2 cells whereas the chloroquine analog mefloquine showed concentration-dependent activity (IC50 = 14.1 µM) at the highest compound concentrations. Our hits also included the antifungal compounds posaconazole, ravuconazole, chlormidazole, and ketoconazole, all showing IC50 values of ~2 µM, as well as sulconazole and cloconazole, showing IC50 values ≥ 20 µM. Antiviral activity is rarely reported for this class of compounds, but the exceptions include posaconazole, which is active against Parechovirus A333. The effective concentrations are an order of magnitude higher than those reported for antifungal activity in vitro. Similar membrane-interacting molecules have been reported to interfere with viral fusion to the host membrane, potentially explaining the mechanism of action for this class of compounds34. Interestingly, a recently preliminary analysis of historical electronic health records for SARS-CoV-2 positive patients presenting with COVID-19 symptoms identified ketoconazole as a medication associated with reduced likelihood of hospital admission35. In terms of possible mechanisms of action, in silico studies predict that posaconazole binds the SARS-CoV-2 spike protein, supporting the case for more detailed epidemiological and mechanistic analysis to determine the role of these antifungal compounds in a SARS-CoV-2 setting36.
The crystallization of the SARS-CoV-2 spike protein complexed to angiotensin-converting enzyme 2 (ACE2), its main receptor37, provides insight into the viral entry mechanism. ACE2 is expressed on the apical membrane domains of Caco-2, Calu-3 and Vero-E6 cells38. ACE2 expression levels can be enhanced by modulating the ACE/ACE2/AT1R/TMPRSS2 system using sartans such as candesartan and telmisartan, or ACE inhibitors such as captopril and the pro-drug enalapril. Such inhibitors could either benefit or increase the risk for COVID-19 patients because AT1R inhibitors may induce ACE2 beneficially but at the expense of facilitating viral entry39. Our primary screen did not identify either classical ACE inhibitors such as captopril or classical AT1R inhibitor sartans.
Several drugs showing antiviral activity in our primary screen have not been shown to influence SARS-CoV-2 cytopathicity in previous studies but have been shown to inhibit other viruses. Dapivirine (IC50 = 0.73 µM) is a non-nucleoside reverse transcriptase inhibitor developed for the treatment of HIV, which was recently shown to possess broad antiviral activity with micromolar IC50 values against influenza viruses A and B in vitro40. The orally available farnesyl transferase inhibitor lonafarnib (IC50 = 5.68 µM) has been considered as a treatment for renal carcinoma41 and showed activity against hepatitis delta virus42. The moderately potent pre-clinical compound NSC319276 elevates intracellular Zn2+ levels, thus modulating the folding of p5343. This compound also interferes with transition metal metabolism, inducing oxidative stress and the depletion of nucleotide reserves, suggesting a possible mechanism for inhibiting viral replication.
Papaverine (IC50 = 1.1. µM) is a non-narcotic alkaloid phosphodiesterase (PDE) inhibitor indicated for heart disease, impotence, and psychosis, which also inhibits multiple strains of influenza virus (25). Similarly, ethaverine (IC50 = 2.15 µM) and drotaverine (IC50 = 6.07 µM) inhibit PDE-444. Hematoporphyrin (IC50 = 1.85 µM) is a photodynamic activated DNA intercalator and single strand breaker with anticancer activity45, which also shows antiviral effects via DNA polymerase inactivation46. SB-612111 (IC50 = 0.77 µM) has been developed as a potent and selective nociceptin opioid peptide receptor antagonist47 and binds the opiate and adenosine receptor proteins47. Overall, this diverse group of compounds shares the common feature of moderate antiviral efficacy in previous assays in addition to their primary indication. Additional studies using analog, or structurally unrelated inhibitors of the same primary targets will be useful to determine whether the SARS-CoV-2 effects we observed are dependent on the compound class or target class.
We also identified an extended group of kinase inhibitors with activity against SARS-CoV-2. There are several potential explanations for the representation of multiple inhibitors from the same class, but kinase inhibitors are usually developed as oral drugs for the treatment of cancer or inflammation. They are intrinsically optimized to enter cells and are likely to act on host target pathways to exert anti-cytopathic effects. Indeed, some inhibitors developed for cancer treatment showed lower CC50 values than MAPKx and JNK inhibitors, which are mainly developed as anti-inflammatory drugs. For example, LY2228820, (IC50 = 0.87 µM) also known as ralimetinib, is a potent and selective inhibitor of the α and β isoforms of p38 MAPK in vitro48. Other MAPK kinase inhibitors, such as bentamapimod, talmapimod and RO5126766, showed IC50 values of 5.9 µM, 9.6 µM and 18.2 µM, respectively, but exhibited lower cytotoxicity (CC50 > 20 µM). Amuvatinib was one of the most potent compounds we identified (IC50 = 0.02 µM). It was unsuccessful in trials for the treatment of solid tumors and small cell lung carcinoma, although it was well tolerated49. Amuvatinib is a promiscuous tyrosine kinase inhibitor, with activity against c-MET, c-RET and the mutant forms of c-KIT, PDGFR and FLT3. Other tyrosine kinase inhibitors we identified included sorafenib (IC50 = 1.55 µM), regorafenib (IC50 = 1.67 µM), pexidartinib (5.43 µM) and vatalanib (IC50 = 18.27 µM) suggesting a role for kinase signaling in the receptor mediated host response to SARS-Cov-2. As well as kinase pathway modulators, we also identified a set of additional anti-cancer compounds with well-defined anti-proliferative mechanisms of action, as might be anticipated in an anti-cytopathicity assay. These compounds included the alkaloid homo-harringtonine, as well as LDE225 and LY335979 (zosuquidar).
A comparison of our primary hits to the recently proposed list of 69 interactome-targeting compounds8 revealed an overlapping set of 40 identical and 12 similar compounds. Four of these compounds were confirmed in the second screen with IC50 values < 20 µM and all but one (loratadine, IC50 = 15.13 µM) were among the most promising candidates with IC50 values < 1 µM (nafamostat, IC50 = 0.04 µM; pevonedistat, IC50 = 0.63 µM; and camostat, IC50 = 0.64 µM). The corresponding biochemical targets were serine proteases (nafamostat and camostat), the histamine 1 receptor (loratadine), and NEDD-8 activating enzyme (pevonedistat). The lack of activity of the predicted compound ligands, for the majority of the originally identified pathways, may reflect low target expression levels, compound solubility, or chemical/metabolic stability.
Interestingly, cyclosporine (IC50 = 16.7 µM) showed moderate activity in the assay, consistent with its reported activity against SARS-CoV50,51. Intriguingly, cyclosporine is also proposed for the treatment of COVID-19 due to its ability to downregulate host immune responses52. A set of apparently nonspecific antiseptic agents was identified, which is thought to act via general membrane disruption. These compounds all showed IC50 values < 2 µM and included cetylpyridinium, octenidine and AC1NDSS5, a sulfonium salt.
Finally, we tested several drugs indicated for the treatment of HIV infections. Among the HIV aspartyl protease inhibitors currently undergoing clinical testing for the treatment of SARS-CoV-2 infections, only lopinavir was active at the highest concentration in the concentration-response screen whereas ritonavir and darunavir were inactive in the primary screen, suggesting this enzyme class may not play a role in viral processing. Although several of the more active compounds showed a relatively small difference between cytotoxic and antiviral IC50 values, with cytotoxicity indices (CI) < 3 (e.g., thioguanosine, amuvatinib, cetylpyridinium, and sorafenib), detailed pharmacokinetic and human safety data are available for these compounds. Similarly, cycloheximide (IC50 = 0.58 µM) showed low toxicity toward Caco-2 cells (CI ~ 10) but has significant reported side effects, including DNA damage and reproductive toxicity, and is not a clinically relevant molecule. Therefore, it will be important to use all information on the safety and toxicity of compounds to establish whether a sufficiently high concentration can be achieved safely at the site of action, presumably the lung epithelium, before proposing follow-on clinical studies.
We would like make readers aware that other parallel data sets are available from the COVID-19 portal (https://www.covid19dataportal.org/) and the NCATS databases (https://ncats.nih.gov/expertise/covid19-open-data-portal). Our data should be analysed in conjunction with these complementary data sources.
Code availability
Images were acquired using an Operetta CLS system combined with Harmony v4.9 followed by image analysis with Columbus v2.9.0 (all from PerkinElmer). Cytotoxicity data based on the high-content images were analysed using ActivityBase v8.0.5.4 (IDBS). Data were visualised in Prism8 for Windows 64-bit version 8.0.0 (GraphPad Software) and TIBCO Spotfire Analyst v7.11.2 (PerkinElmer). A collection of R scripts and KNIME workflows for the analysis of our datasets have been made available (https://github.com/agiani99/KNIME_Screen) with no usage restrictions.
Change history
09 April 2021
Open Access funding information has been added to this article.
References
Coronaviridae. Virus Taxonomy 2012, 806–828, https://doi.org/10.1016/B978-0-12-384684-6.00068-9 (2011).
Sørensen, M. D. et al. Severe acute respiratory syndrome (SARS): development of diagnostics and antivirals. Annals of the New York Academy of Sciences 1067, 500–505, https://doi.org/10.1196/annals.1354.072 (2006).
Groot, R. Jde et al. Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. Journal of virology 87, 7790–7792, https://doi.org/10.1128/JVI.01244-13 (2013).
WHO. WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19–11-march-2020 (2020).
Johns Hopkins University. Coronavirus Resource Center. https://coronavirus.jhu.edu/ (2020).
Harrison, C. Coronavirus puts drug repurposing on the fast track. Nature biotechnology 38, 379–381, https://doi.org/10.1038/d41587-020-00003-1 (2020).
Oprea, T. I. et al. Drug Repurposing from an Academic Perspective. Drug discovery today. Therapeutic strategies 8, 61–69, https://doi.org/10.1016/j.ddstr.2011 (2011).
Gordon, D. E. et al. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. Nature 583, 459–468 (2020).
Kupferschmidt, K. & Cohen, J. Race to find COVID-19 treatments accelerates. Science (New York, N.Y.) 367, 1412–1413, https://doi.org/10.1126/science.367.6485.1412 (2020).
Li, G. & Clercq, E. de. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature reviews. Drug discovery 19, 149–150, https://doi.org/10.1038/d41573-020-00016-0 (2020).
Corsello, S. M. et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nature medicine 23, 405–408, https://doi.org/10.1038/nm.4306 (2017).
Bojkova, D. et al. SARS-CoV-2 infected host cell proteomics reveal potential therapy targets. Nature 583, 469–472 (2020).
Kampf, G., Todt, D., Pfaender, S. & Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. The Journal of hospital infection 104, 246–251, https://doi.org/10.1016/j.jhin.2020.01.022 (2020).
Ellinger, B. et al. A SARS-CoV-2 cytopathicity dataset generated by high-content screening of a large drug repurposing collection. Image Data Resource https://doi.org/10.17867/10000148 (2020).
Vicar, T. et al. Cell segmentation methods for label-free contrast microscopy: review and comprehensive comparison. BMC bioinformatics 20, 360, https://doi.org/10.1186/s12859-019-2880-8 (2019).
Zhang, Chung & Oldenburg. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of biomolecular screening 4, 67–73, https://doi.org/10.1177/108705719900400206 (1999).
Ellinger, B. et al. Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of Caco-2 cells at 10 uM after 48 hours by high content imaging. CHEMBL https://www.ebi.ac.uk/chembl/assay_report_card/CHEMBL4303805/ (2020).
Ellinger, B. et al. Determination of IC50 values for inhibition of SARS-CoV-2 induced cytotoxicity of Caco-2 cells after 48 hours by high content imaging. CHEMBL https://www.ebi.ac.uk/chembl/assay_report_card/CHEMBL4303806/ (2020).
Borsari, C. et al. SAR Studies and Biological Characterization of a Chromen-4-one Derivative as an Anti-Trypanosoma brucei Agent. ACS medicinal chemistry letters 10, 528–533, https://doi.org/10.1021/acsmedchemlett.8b00565 (2019).
Ellinger, B. et al. Toxicity against Caco-2 cells determined at 48 hours by intracellular ATP concentration using the CellTiter-Glo Luminescent Cell Viability Assay. CHEMBL https://www.ebi.ac.uk/chembl/assay_report_card/CHEMBL4303807/ (2020).
Ellinger, B. et al. Cytotoxicity index (CI): selectivity for inhibition of SARS-CoV-2 induced cytotoxicity of Caco-2 cells versus toxicity against Caco-2 cells (CC50/IC50). CHEMBL https://www.ebi.ac.uk/chembl/assay_report_card/CHEMBL4303808/ (2020).
Sheahan, T. P. et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Science translational medicine 9, https://doi.org/10.1126/scitranslmed.aal3653 (2017).
Sangeun J., et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrobial Agents Chemotherapy 64, e00819-20, https://doi.org/10.1101/2020.03.20.999730 (2020).
Hoffmann, M. et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280, https://doi.org/10.1016/j.cell.2020.02.052 (2020).
Kawase, M., Shirato, K., van der Hoek, L., Taguchi, F. & Matsuyama, S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. Journal of virology 86, 6537–6545, https://doi.org/10.1128/JVI.00094-12 (2012).
Yamamoto, M. et al. Identification of Nafamostat as a Potent Inhibitor of Middle East Respiratory Syndrome Coronavirus S Protein-Mediated Membrane Fusion Using the Split-Protein-Based Cell-Cell Fusion Assay. Antimicrobial agents and chemotherapy 60, 6532–6539, https://doi.org/10.1128/AAC.01043-16 (2016).
Danesh, F. & Ghavidel, S. Coronavirus: Scientometrics of 50 Years of Global Scientific Productions. International Journal of Medical Microbiology 14, 1–16, https://doi.org/10.30699/ijmm.14.1.1 (2020).
Wang, M. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell research 30, 269–271, https://doi.org/10.1038/s41422-020-0282-0 (2020).
Shen, L. et al. High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. Journal of virology 93, https://doi.org/10.1128/JVI.00023-19 (2019).
Agostini, M. L. et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 9, e00221–18, https://doi.org/10.1128/mBio.00221-18 (2018).
Cheng, K.-W. et al. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antiviral research 115, 9–16, https://doi.org/10.1016/j.antiviral.2014.12.011 (2015).
Statement from the Chief Investigators of the Randomised Evaluation of COVid-19 thERapY (RECOVERY) Trial on hydroxychloroquine (University of Oxford, 2020).
Rhoden, E., Nix, W. A., Weldon, W. C. & Selvarangan, R. Antifungal azoles itraconazole and posaconazole exhibit potent in vitro antiviral activity against clinical isolates of parechovirus A3 (Picornaviridae). Antiviral research 149, 75–77, https://doi.org/10.1016/j.antiviral.2017.11.011 (2018).
Speerstra, S. et al. Antivirals acting on viral envelopes via biophysical mechanisms of action. Antiviral research 149, 164–173, https://doi.org/10.1016/j.antiviral.2017.11.018 (2018).
Victor, M. C., Rachel, A. R., Sean, M. J. M. & Roy, H. P. Identifying common pharmacotherapies associated with reduced COVID-19 morbidity using electronic health records. Preprint at https://doi.org/10.1101/2020.04.11.20061994 (2020).
Wu, C. et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 10, 766–788, https://doi.org/10.1016/j.apsb.2020.02.008 (2020).
Yan, R. et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.) 367, 1444–1448, https://doi.org/10.1126/science.abb2762 (2020).
Ren, X. et al. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. The Journal of general virology 87, 1691–1695, https://doi.org/10.1099/vir.0.81749-0 (2006).
Gurwitz, D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug development research 81, 537–540, https://doi.org/10.1002/ddr.21656 (2020).
Hu, Y. et al. Discovery of dapivirine, a nonnucleoside HIV-1 reverse transcriptase inhibitor, as a broad-spectrum antiviral against both influenza A and B viruses. Antiviral research 145, 103–113, https://doi.org/10.1016/j.antiviral.2017.07.016 (2017).
Wang, J. et al. Synergistic effect of farnesyl transferase inhibitor lonafarnib combined with chemotherapeutic agents against the growth of hepatocellular carcinoma cells. Oncotarget 8, 105047–105060, https://doi.org/10.18632/oncotarget.22086 (2017).
Rizzetto, M. & Ciancio, A. The prenylation inhibitor, lonafarnib: a new therapeutic strategy against hepatitis delta. The Lancet Infectious Diseases 15, 1119–1120, https://doi.org/10.1016/S1473-3099(15)00155-3 (2015).
Nguyen, D., Liao, W., Zeng, S. X. & Lu, H. Reviving the guardian of the genome: Small molecule activators of p53. Pharmacology & therapeutics 178, 92–108, https://doi.org/10.1016/j.pharmthera.2017.03.013 (2017).
Muravyov, A. V. et al. Hemorheological efficiency of drugs, targeting on intracellular phosphodiesterase activity: In vitro study. Clinical Hemorheology and Microcirculation 36, 327–334 (2007).
Boegheim, J. P., Dubbelman, T. M., Mullenders, L. H. & van Steveninck, J. Photodynamic effects of haematoporphyrin derivative on DNA repair in murine L929 fibroblasts. The Biochemical journal 244, 711–715, https://doi.org/10.1042/bj2440711 (1987).
Munson, B. R. & Fiel, R. J. Hematoporphyrin-sensitized photodynamic inactivation of viral RNA-dependent DNA polymerase. Res Commun Chem Pathol Pharmacol 16, 175–178 (1977).
Tosh, D. K. et al. Repurposing of a Nucleoside Scaffold from Adenosine Receptor Agonists to Opioid Receptor Antagonists. ACS omega 3, 12658–12678, https://doi.org/10.1021/acsomega.8b01237 (2018).
Campbell, R. M. et al. Characterization of LY2228820 dimesylate, a potent and selective inhibitor of p38 MAPK with antitumor activity. Molecular cancer therapeutics 13, 364–374, https://doi.org/10.1158/1535-7163.MCT-13-0513 (2014).
Byers, L. A. et al. A phase 2, open-label, multi-center study of amuvatinib in combination with platinum etoposide chemotherapy in platinum-refractory small cell lung cancer patients. Oncotarget 8, 81441–81454, https://doi.org/10.18632/oncotarget.19888 (2017).
Wilde, A. H. D., Pham, U., Posthuma, C. C. & Snijder, E. J. Cyclophilins and cyclophilin inhibitors in nidovirus replication. Virology 522, 46–55, https://doi.org/10.1016/j.virol.2018.06.011 (2018).
Pfefferle, S. et al. The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors. PLoS pathogens 7, https://doi.org/10.1371/journal.ppat.1002331 (2011).
Cour, M., Ovize, M. & Argaud, L. Cyclosporine A: a valid candidate to treat COVID-19 patients with acute respiratory failure? Critical Care 24, https://doi.org/10.1186/s13054-020-03014-1 (2020).
Dyall, J. et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrobial agents and chemotherapy 58, 4885–4893, https://doi.org/10.1128/AAC.03036-14 (2014).
Meehyun Ko, So Young Chang, Soo Young Byun, Inhee Choi, Anne-Laure Pham Hung d’Alexandry d’Orengiani, David Shum, Ji-Young Min, Marc P. Windisch. Screening of FDA-approved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19. Preprint at https://doi.org/10.1101/2020.02.25.965582 (2020).
Stuart Weston, Rob Haupt, James Logue, Krystal Matthews, Matthew B. Frieman. FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2 in vitro. Preprint at https://doi.org/10.1101/2020.03.25.008482 (2020).
Vincent, M. J. et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology journal 2, 69, https://doi.org/10.1186/1743-422X-2-69 (2005).
Chan, J. F. W. et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. The Journal of infection 67, 606–616, https://doi.org/10.1016/j.jinf.2013.09.029 (2013).
Wilde, A. H. d. et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrobial agents and chemotherapy 58, 4875–4884, https://doi.org/10.1128/AAC.03011-14 (2014).
Acknowledgements
We thank Timothy Wells of the Medicine for Malaria Venture for critical reading of the manuscript and providing valuable insight into repurposing approaches. We acknowledge funding from the Johanna Quandt Universitäts-Stiftung. This work has been supported, in part, by the LOEWE Center for Translational Medicine and Pharmacology (TMP) and the Fraunhofer cluster of excellence for immune mediated diseases (CIMD). FAIRification of data for community use was supported by funding from the EU Horizon 2020 project EOSC-LIFE (grant no. 824087). We thank Andrew Leach, Anna Gaulton, Anne Hersey, Eloy Felix, Emma Manners and Patricio Bento of the European Bioinformatics Institute (Hinxton) for preparing FAIR data sets. We thank Yulia Gerhardt and Peter Maas (SPECS) and Joshua Bitker (formerly Broad Institute) for input into the selection and quality control of the compound library. We thank Giuseppe Vangelista for infrastructural support.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
B.E. conceived the study, carried out the tissue culture work, cytotoxicity assays, analysed the data and prepared the initial manuscript. D.B. carried out the tissue culture work, virus experiments and prepared the initial manuscript. A.Z. conceived the study, analysed the data and prepared the initial manuscript. J.C. conceived the study, carried out the tissue culture work and virus experiments. C.C. conceived the study and supervised the work. S.W. carried out the tissue culture work, virus experiments and cytotoxicity assays. O.K. carried out the tissue culture work, cytotoxicity assays and analysed the data. J.R. analysed the data. M.K. carried out the tissue culture work, cytotoxicity assays and analysed the data. M.W. carried out the tissue culture work, virus experiments and cytotoxicity assays. G.G. conceived the study. P.G. conceived the study and prepared the initial manuscript. S.C. conceived the study and supervised the work. All authors contributed to the text of the article and read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Ellinger, B., Bojkova, D., Zaliani, A. et al. A SARS-CoV-2 cytopathicity dataset generated by high-content screening of a large drug repurposing collection. Sci Data 8, 70 (2021). https://doi.org/10.1038/s41597-021-00848-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00848-4
This article is cited by
-
SARS-CoV-2 and human retroelements: a case for molecular mimicry?
BMC Genomic Data (2022)
-
Nafamostat Mesylate for Treatment of COVID-19 in Hospitalised Patients: A Structured, Narrative Review
Clinical Pharmacokinetics (2022)
-
An update on novel approaches for diagnosis and treatment of SARS-CoV-2 infection
Cell & Bioscience (2021)
-
A machine learning platform to estimate anti-SARS-CoV-2 activities
Nature Machine Intelligence (2021)
-
Strategies to identify candidate repurposable drugs: COVID-19 treatment as a case example
Translational Psychiatry (2021)