Abstract
Field studies have been performed for decades to analyze effects of different management practices on agricultural soils and crop yields, but these data have never been combined together in a way that can inform current and future cropland management. Here, we collected, extracted, and integrated a database of soil health measurements conducted in the field from sites across the globe. The database, named SoilHealthDB, currently focuses on four main conservation management methods: cover crops, no-tillage, agro-forestry systems, and organic farming. These studies represent 354 geographic sites (i.e., locations with unique latitudes and longitudes) in 42 countries around the world. The SoilHealthDB includes 42 soil health indicators and 46 background indicators that describe factors such as climate, elevation, and soil type. A primary goal of this effort is to enable the research community to perform comprehensive analyses, e.g., meta-analyses, of soil health changes related to cropland conservation management. The database also provides a common framework for sharing soil health, and the scientific research community is encouraged to contribute their own measurements.
Measurement(s) | organic material • grain yield trait • mass density of soil • concentration of carbon atom in soil • soil organic carbon sequestration rate • concentration of nitrogen atom in soil • phosphorus • potassium • pH measurement • soil cation exchange capability • electrical conductivity • soil base saturation • aggregate stability • porosity of soil • soil penetration resistance • soil infiltration rate • field saturated hydraulic conductivity • soil erosion • flood • soil nutrient leaching • temperature of soil • soil water content • available water holding capacity • parasitic weed • diseases of the cropland • pests • Lumbricus terrestris • Arthropoda • Nematoda • Bacteria • Fungi • mycorrhiza • enzyme activity • dinitrogen oxide • carbon dioxide • methane gas emission process |
Technology Type(s) | digital curation |
Factor Type(s) | conservation management method |
Sample Characteristic - Environment | soil environment |
Sample Characteristic - Location | United States of America • Canada • Germany • Brazil • French Republic • Italy • Sweden • Kingdom of Denmark • Nigeria • Argentina • Kenya • Zimbabwe • China • South Korea • Kingdom of Spain • Kingdom of the Netherlands • Greece • Kingdom of Norway • New Zealand • Russia • Australia • Ghana • Malawi • Benin • Zambia • Cameroon • Peru • Indonesia • The Philippines • Rwanda • Uganda • Togo • Guinea • Tanzania • Turkey • Moldova • England • India • Costa Rica • Switzerland • Bangladesh • Poland |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.10748210
Similar content being viewed by others
Background & Summary
Soil health, sometimes used interchangeably with soil quality, represents the ability of soils to function as a biodiverse organism that sustains terrestrial life (USDA-NRCS, 2019), and is often assessed using a combination of physical, chemical and biological indicators1. Cropland soil degradation due to natural vegetation removal, intensive agricultural operations, and erosion are among the main factors causing declines in soil health and crop yields2,3,4. According to a recent report from the Food and Agriculture Organization of the United Nations (FAO), one-third of soils in the world are infertile due to unsustainable land-use management practices5. Cropland conservation management practices, including the use of cover crops within rotations and changes from traditional mouldboard or disk tillage to reduced or no-tillage, have been proposed as ways to increase soil carbon and soil health6,7. Many on-site experiments have been conducted to evaluate the effects of conservation management on soil properties, yet there has been little effort to evaluate which indicators should be measured to consistently quantify any resulting improvements in soil health. In addition, studies can differ in their results: as an example, using cover crops during normally fallow seasons can enhance soil organic carbon8, though many short-term studies have not found this same result9,10,11.
To better address such uncertainties, systematic reviews and meta-analyses have evaluated the effects of cover crops12, no-tillage13,14, organic farm15, and agroforestry systems16 on crop yield and soil properties. These efforts have generated new insights into soil health dynamics, yet there is still limited understanding of whether and how these findings translate to global scales. Historically and newly published data offer a wealth of information that can support global assessments of how conservation agricultural practices may influence soil health, provided that there is an effective mechanism to record and disseminate this information.
To address this gap, we collected studies that compared agricultural production and soil properties under traditional management strategies with those under conservational management practices. Publications that meet specific criteria were digitized and the data were integrated into a global soil health database that we have named SoilHealthDB. This web-based, open source dataset can be continuously updated by including newly published and even provisional data. The dataset can be used to perform statistical analyses (e.g., meta-analyses) on specific soil health indicators or agronomic responses. SoilHealthDB provides a common soil health framework for sharing and integrating field measurements and related information, and thereby offers valuable information for farmers, agency personnel, and scientists as they plan and evaluate cropland management.
Methods
Data collection
SoilHealthDB currently includes 46 background indicators (Online-only Table 1) and 42 soil health indicators (Online-only Table 2)1. To identify relevant studies, we conducted a systematic literature search for field comparisons between traditional and conservational management practices. We initially targeted four main conservational management methods: cover cropping (CC), no-tillage (NT), organic farming (OF), and agro-forestry systems (AF) (Table 1).
Publications were searched and collected from three sources: (1) an online literature search; (2) the Soil Health Institute “Research Landscape Tool”, which compiles soil health results into a searchable database and includes publication and research projects17; and (3) cited papers from previous meta-analyses or review papers12,15,18,19. For the online literature search we used the ISI Web of Science, Google Scholar, and the China National Knowledge Infrastructure (CNKI). We used the keywords “soil health” or “soil quality” and “conservation management”, “cover crop”, “no-till”, “organic farm”, or “agroforestry systems” when performing the literature search. Papers from peer-reviewed journals, conference collections, theses, and dissertations were included. No other restrictions or filtering criteria were used (e.g., we included eligible papers in all languages and with all publication dates). We collected a total of more than 500 papers; we then used the following criteria to determine whether the publication would be included in this study: (1) experiments were conducted in the field or at a research station; (2) the publications compared controls (i.e., traditional management) and treatments (i.e., conservational management); (3) publications provide at least one comparison of soil health indicators between controls and treatments (Online-only Table 2). Within these constraints, 321 papers were extracted and integrated into the SoilHealthDB.
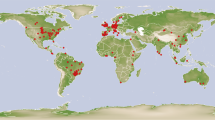
Data were digitized from tables and figures. The software Data Thief (version III)20 was used to read the data from figures. Background information was extracted from the publications and fit into 46 background indicator categories (Online-only Table 1). Whenever latitude and longitude were not reported in the literature, the site name was entered into the website (https://www.findlatitudeandlongitude.com) to estimate location. Whenever elevation was missing from the original paper, it was identified by latitude and longitude (https://www.freemaptools.com/elevation-finder.htm). In total, 5,907 comparisons were collected from across the globe (Fig. 1), for a mean of approximately 20 comparisons per study. As many studies reported multiple comparisons, we needed to identify if those comparisons were independent of one another. We therefore allocated a unique experiment ID to a comparison if the cover crop group, cash crop group, site, tillage, fertilization, soil depth, termination, or rotation were different from other comparisons (Fig. 2). This process resulted in a total of 1,407 experiments that were assumed to be independent of each other.
The spatial distribution of sites from cover cropping (CC), no-tillage (NT), organic farming (OF), and agro-forestry systems (AF) across the globe. The numbers in the parentheses represent the number of sites reporting data for each different conservation management method. Symbol sizes represent the number of comparisons in each site.
Diagram detailing the procedures for data integration, experiment ID allocation, and potential uses that the database can support. Unique experiment IDs were given to pairwise comparisons if the cash crop, site, tillage, fertilizer level, cover crop, soil sampling depth, cover crop termination, or cash crop rotation was different from other comparisons; otherwise, comparisons that had the same information for one or more of those categories received the same experiment ID (middle panel).
Data processing
After the location information was carefully checked, the climatic regions for all sites were identified according to climate Koppen classification21, using the latitude and longitude (for a detailed description please see the ‘Data Records’ section provided in the supplemental R code22). All missing MAT and MAP values were estimated using a global air temperature and precipitation dataset provided by the Center for Climate Research at the University of Delaware23. The MAP and MAT were calculated based on the monthly precipitation and temperature between 1961 and 2015. Soil texture was grouped into coarse (sand, loamy sand, and sandy loam), medium (sandy clay loam, loam, silt loam, and silt), and fine (clay, sandy clay, clay loam, silty clay, and silty clay loam) textures based on the Cornell Framework24.
The cash crops were grouped into corn, soybean, wheat, other monoculture, corn-soybean rotation (CS), corn-soybean-wheat rotation (CSW), and other rotation of more than two cash crops (ROT). The cover crops were grouped into broadleaf, grass, legume, mixture of two legumes (LL), mixture of legume and grass (LG), mixture of two cover crops other than LL or LG (MOT), and other mixtures of more than two cover crops (MTT). Soil sampling depths were grouped into 0–10 cm, 0–20 cm, 0–30 cm, and 30–100 cm (Fig. 3). It should be noted that the user can regroup the cash crop, cover crop, and soil sampling depth according their research objectives.
The number of replications and standard deviations (SD) were compiled from the publications when possible. When the studies reported standard error (SE), coefficient of variation (CV), or confidence interval (CI) rather than SD, SD was calculated using:
where n is the number of observations.
SD was calculated from CV as:
and from the CI as:
where Zα/2 is the Z score for a given level of significance, α. Zα/2 is equal to 1.96 when α = 0.05 and 1.645 when α = 0.10.
Soil organic carbon (SOC) data were reported as carbon stocks (Mg/ha). When applicable, SOC was calculated based on SOC concentrations (SOC%) and soil bulk density using:
where h represents soil sampling depth (meter), and BD represents soil bulk density (Mg/m3).
SOC sequestration rate (SOCseq) was calculated in terms of (Mg/ha/yr) using:
where SOCcc is the soil carbon stocks under CC treatments (Mg/ha), SOCbackground is the soil carbon stock either under background conditions or under the no cover crop controls (Mg/ha), and y represents years after CCs.
Data Records
The data and R code can be downloaded in figshare22; there are two folders, named data and RScripts, when ‘SoilHealthDB.zip’ is unzipped. ‘SoilHealthDB_V1.xlsx’ in the data file currently includes 5,907 rows and 268 columns, which were retrieved from 321 papers (for the detailed reference list please refer to ‘References’ under ‘SoilHealthDB_V1.xlsx’22). Each column corresponds to one data point of either background information or soil health indicator, and each row includes as many as 42 comparisons between treatments and controls (if all soil health indicators have data). The names, attributes, and descriptions of the background information and soil health indicators are presented in Online-only Tables 1 and 2. It should be noted that different measurements and/or units may be involved in the same soil health indicator (e.g., soil total nitrogen, soil organic nitrogen, or soil inorganic nitrogen are reported in different papers to represent the soil nitrogen indicator, ID 5 in Online-only Table 2); therefore, it is important that measurement objectives, units, and other detailed descriptions are recorded in the comments columns. It should also be noted that for some soil health indicators (e.g., CH4 and N2O emission), we were only able to extract limited numbers of comparisons, which may restrain the ability of those data to be used in further analyses. ‘SoilHealthDB_V1.csv’ is a simplified version of ‘SoilHealthDB_V1.xlsx’, with only soil health background and indicator information kept (e.g., all the description sheets were not kept). There are two R scripts in the ‘RScripts’ folder: the ‘SoilHealthDB_quality_check.R’ script includes code for quality check of the ‘SoilHealthDB’, and the ‘functions.R’ script defines several functions, including one to generate the location of the site in ‘SoilHealthDB’. The SoilHealthDB_V1.csv file is to be used when running the R codes.
Technical Validation
Quality control was performed to check the fidelity of the data to the original source. Each paper was carefully read at least twice, and special attention was paid to the tables, figures, and method sections, where most of the soil health indicator comparisons and background information were located. Before a new paper was extracted, we first used the bibliography database manager Mendeley to check whether it was a duplicate of previous papers (for details, please see the supplemental reference document). After the data extraction, we compared the digitized data against the tables or figures from the original paper once again to make sure the data were loaded correctly.
After the data extraction, we examined data quality using R (version 3.5.1)25. The formats of each column (numerical or string) were checked to correct any mistyping in the numerical columns (e.g., checking all soil health indicators and some background information columns like latitude and longitude). For each soil health indicator, we calculated the response ratio (RR), which is the value of treatment divided by the value of control, e.g., for cover crop studies RR = ln(xcc/xnc), where xcc is the mean parameter value under cover crops and xnc is the mean parameter value under no cover controls. We then plotted the frequency distribution of response ratio for each soil health indicator, and returned to the original articles to verify any extreme values that were identified in this process. We also visualized the data distribution for background columns that contained numeric values (e.g. latitude, elevation) and manually checked the outliers by validating them against the original papers. For the location of each site, we plotted the latitude and longitude by country and checked whether there were sites from a specific country that fell outside its border. For those sites, we checked the extracted latitude and longitude information with location information from the original paper (e.g., site name, country name). For some sites located near to coastal areas, a few sites were reported to exist in the sea, likely due to insufficient precision in reported values. For these sites, we slightly corrected the longitude and latitude to the nearest point on land.
Linkages to external data sources
The studies compiled thus far in SoilHealthDB rarely reported potentially important soil properties (e.g., cation exchange capacity, CEC) and background information (e.g., mean annual temperature, MAT, and mean annual precipitation, MAP). Similarly, some soil attributes such as soil taxonomy were classified differently between regions, making it difficult to compare this information. To resolve those issues, we associated our database with external data sources (by latitude and longitude; for details please see the code in the repository). We linked our data with Koppen21 classification (0.5° × 0.5° resolution), a global air temperature and precipitation dataset (0.5° × 0.5° resolution)23, and the Harmonized World Soil Database v1.2 (HWSD, 0.05° × 0.05° resolution)26,27. We then analysed all samples for their soil type, using the World Reference Base (WRB) classification system26,27, and for their climatic attributes (Fig. 4).
Representation of SoilHealthDB samples in different climate and soil types. Distributions of SoilHealthDB samples values across different parameters. Analyzed distributions include: (a) different climate types; (b) mean annual temperature (MAT); (c) mean annual precipitation (MAP); and (d) different WRB soil groups. Note that in (a) Equat – equatorial and Temp – temperate; in (b,c) the light blue represents samples from SoilHealthDB and gray represents global values from the Harmonized World Soil Database v1.2 (for details please see refs. 26,27).
Samples from SoilHealthDB covered all four climate types, with the majority of sites located in temperate areas and relatively few sites located in arid areas (Fig. 4a). Sites within the SoilHealthDB had somewhat different distributions for MAT and MAP as compared to global distributions (Fig. 4b,c), in part because we only included locations with MAT between −5 °C and 35 °C so as to exclude climates not conducive to crop production. The MAT from SoilHealthDB sites followed an approximately normal distribution, with the most common temperatures occurring between 5 and 20 °C. In contrast the global MAT peaked between 20 and 30 °C. The majority of sites in SoilHealthDB had MAP between 500 and 1500 mm, while global MAP followed a gamma distribution with a greater proportion of area having <500 mm MAP. SoilHealthDB sites covered 21 out of 32 soil taxonomic groups in the WRB soil classification system26,27 (Fig. 4d).
Only 11 studies reported soil CEC (thus representing approximately 4% of all studies in SoilHealthDB), for a total of 54 independent records. There thus exists a paucity of direct CEC measurements in SoilHealthDB. However, we were able to estimate CEC for all sites using the HWSD soil database (Fig. 5a). Cation exchange capacity (CEC) distributions were similar between SoilHealthDB sites and the global HWSD soil database (Fig. 5b), suggesting that samples in the SoilHealthDB properly represent soil and climatic characteristics for regions conducive to agricultural production.
Finally, because attributes such as texture and CEC are important for interpreting soil health, we encourage future submissions to record these types of information to the extent possible. We also encourage use of the WRB taxonomy for all samples, as a way to enhance the global applicability of this database.
Usage Notes
In the SoilHealthDB, the measurement objectives and units between each comparison (control vs. treatment within same row) will always be the same. However, each soil health indicator may have multiple measurement objectives and therefore involve multiple units (e.g., a researcher may measure soil total nitrogen in one site and measure organic nitrogen in another site). Detailed information about measurement objectives and units are recorded under the comments column. The user should always check the comments before data processing and analysis; otherwise, without data filtration and unit conversion only response ratios should be analysed. We recommend that users download and explore the database using the provided R code, as the code includes explanations and instructions. The user can contact the corresponding author with questions on understanding the code and using the data.
Code availability
All the data processing and data visualization were conducted using R (version 3.5.1)25. The source code is available on figshare22. The code is detailed with instructions for users. Generally, the function.R file (under RScript folder) defined several functions to obtain background information from external datasets, as well as the function to plot the samples spatial distribution (Fig. 1). The SoilHealthDB_quality_check.R file (under RScript folder) intends to check the data quality, and to explain how some soil health indicators are grouped based on the basic information. We also created a markdown file (SHDB.Rmd), which described the analysis and generated figures (Figs. 1, 4 and 5) for this study. All the code and data used are available in figshare22 and GitHub (https://github.com/jinshijian/SoilHealthDB).
References
Stewart, R. D. et al. What we talk about when we talk about soil health. Agric. Environ. Lett. 3, 5–9 (2018).
Cullum, R. F., McGregor, K. C., Mutchler, C. K., Johnson, J. R. & Boykin, D. L. Soybean yield response to tillage, fragipan depth, and slope length. Trans. ASAE 43, 563–571 (2000).
Olson, K., Ebelhar, S. A. & Lang, J. M. Long-term effects of cover crops on crop yields, soil organic carbon stocks and sequestration. Open J. Soil Sci. 04, 284–292 (2014).
Haruna, S. I., Nkongolo, N. V., Anderson, S. H., Eivazi, F. & Zaibon, S. In situ infiltration as influenced by cover crop and tillage management. J. Soil Water Conserv. 73, 164–172 (2018).
FAO. The state of the world’s land and water resources for food and agriculture (SOLAW): managing systems at risk. (The Food and Agriculture Organization of the United Nations and Earthscan, 2011).
Gardner, J. C. & Clancy, S. A. Impact of farming practices on soil quality in North Dakota. Methods Assess. Soil Qual. 49, 337–343 (1996).
Mbuthia, L. W. et al. Long term tillage, cover crop, and fertilization effects on microbial community structure, activity: implications for soil quality. Soil Biol. Biochem. 89, 24–34 (2015).
Kaye, J. P. & Quemada, M. Using cover crops to mitigate and adapt to climate change. A review. Agron. Sustain. Dev. 37, 4 (2017).
Bandick, A. K. & Dick, R. P. Field management effects on soil enzyme activities. Soil Biol. Biochem. 31, 1471–1479 (1999).
Idowu, O. J. et al. Use of an integrative soil health test for evaluation of soil management impacts. Renew. Agric. Food Syst. 24, 214–224 (2009).
Ndiaye, E. L., Sandeno, J. M., McGrath, D. & Dick, R. P. Integrative biological indicators for detecting change in soil quality. Am. J. Altern. Agric. 15, 26 (2000).
Poeplau, C. & Don, A. Carbon sequestration in agricultural soils via cultivation of cover crops - A meta-analysis. Agric. Ecosyst. Environ. 200, 33–41 (2015).
Cooper, J. et al. Shallow non-inversion tillage in organic farming maintains crop yields and increases soil C stocks: a meta-analysis. Agron. Sustain. Dev. 36, 22 (2016).
Luo, Z., Wang, E. & Sun, O. J. Can no-tillage stimulate carbon sequestration in agricultural soils? A meta-analysis of paired experiments. Agric. Ecosyst. Environ. 139, 224–231 (2010).
Gattinger, A. et al. Enhanced top soil carbon stocks under organic farming. Proc. Natl. Acad. Sci. 109, 18226–18231 (2012).
Shi, L., Feng, W., Xu, J. & Kuzyakov, Y. Agroforestry systems: Meta-analysis of soil carbon stocks, sequestration processes, and future potentials. L. Degrad. Dev., https://doi.org/10.1002/ldr.3136 (2018).
Soil Health Institude. Soil Health Research Landscape Tool, www.soilhealthinstituteresearch.org.
Alvarez, R., Steinbach, H. S. & De Paepe, J. L. Cover crop effects on soils and subsequent crops in the pampas: A meta-analysis. Soil Tillage Res. 170, 53–65 (2017).
Sileshi, G., Akinnifesi, F. K., Ajayi, O. C. & Place, F. Evidence for impact of green fertilizers on maize production in sub-Saharan Africa: a meta-analysis. ICRAF Occas. Pap. 10, 1–46 (2009).
Flower, A., McKenna, J. W. & Upreti, G. Validity and reliability of GraphClick and DataThief III for data extraction. Behav. Modif. 40, 396–413 (2016).
Kottek, M., Grieser, J., Beck, C., Rudolf, B. & Rubel, F. Updated world map of the Köppen-Geiger climate classification. Meteorol. Zeitschrift 15, 259–263 (2006).
Jian, J., Stewart, R. & Du, X. SoilHealthDB. figshare. https://doi.org/10.6084/m9.figshare.8292176.v8 (2019).
Willmott, C. & Matsuura, K. Terrestrial Air Temperature and Precipitation: Monthly and Annual Time Series 1950–1999. University of Delaware Center for Climate Research., http://climate.geog.udel.edu/~climate/html_pages/README.ghcn_ts2.html (2001).
Moebius-Clune, B. N. et al. Comprehensive Assessment of Soil Health: The Cornell Framework Manual. (Cornell University, 2016).
R, Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2013. (2014).
Nachtergaele, F. A. et al. The harmonized world soil database-Food and Agriculture Organization of the United Nations. Science, Soil Solutions for a Changing World (2010).
Wieder, W. R., Boehnert, J., Bonan, G. B. & Langseth., M. Regridded Harmonized World Soil Database v1.2. ORNL Distrubuted Active Archive Center., https://doi.org/10.3334/ORNLDAAC/1247 (2014).
Chander, K., Goyal, S., Mundra, M. C. & Kapoor, K. K. Organic matter, microbial biomass and enzyme activity of soils under different crop rotations in the tropics. Biol. Fertil. Soils 24, 306–310 (1997).
Acknowledgements
We thank the anonymous reviewers and Paolo de Souza, Editorial Board Member, for their useful comments. This work is supported by the Natural Resources Conservation Service, U.S. Department of Agriculture, under NRCS Conservation Innovation Grant 69-3A75-14-260. Funding for this work was provided in part by the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. Jinshi Jian was partially supported by the U.S. Department of Energy, Office of Science, Biological and Environmental Research as part of the Terrestrial Ecosystem Sciences Program. Xuan Du was supported by the conservation management and soil erosion project funded by the Yangling Vocational & Technical College, grant number: A2019009. Publication fees were provided by the Virginia Tech Open Access Subvention Fund. We would also like to acknowledge Christopher Poeplau for providing tabulated data from an earlier meta-analysis. The code and the SoilHealthDB are available on figshare (https://doi.org/10.6084/m9.figshare.8292176.v8) and GitHub (https://github.com/jinshijian/SoilHealthDB); those items can be used for individual, academic, research, and commercial usage, and can be repackaged or sold without written permission.
Author information
Authors and Affiliations
Contributions
Jinshi Jian and Ryan D. Stewart conceived the design of the data framework. Jinshi Jian and Xuan Du extracted and integrated the data from papers to the SoilHealthDB. Jinshi Jian drafted the manuscript, and all authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Tables
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Jian, J., Du, X. & Stewart, R.D. A database for global soil health assessment. Sci Data 7, 16 (2020). https://doi.org/10.1038/s41597-020-0356-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-020-0356-3
This article is cited by
-
Dataset of metals and metalloids in food crops and soils sampled across the mining region of Moquegua in Peru
Scientific Data (2023)
-
Opportunities and challenges of bio-based fertilizers utilization for improving soil health
Organic Agriculture (2023)
-
The concept and future prospects of soil health
Nature Reviews Earth & Environment (2020)