Abstract
Human adenoviruses (HAdVs) are fatal to immuno-suppressed individuals, but no effective anti-HAdV therapy is available. Here, we present a novel image-based high-throughput screening (HTS) platform, which scores the full viral replication cycle from virus entry to dissemination of progeny and second-round infections. We analysed 1,280 small molecular weight compounds of the Prestwick Chemical Library (PCL) for interference with HAdV-C2 infection in a quadruplicate, blinded format, and performed robust image analyses and hit filtering. We present the entire set of the screening data including all images, image analyses and data processing pipelines. The data are made available at the Image Data Resource (IDR, idr0081). Our screen identified Nelfinavir mesylate as an inhibitor of HAdV-C2 multi-round plaque formation, but not single round infection. Nelfinavir has been FDA-approved for anti-retroviral therapy in humans. Our results underscore the power of image-based full cycle infection assays in identifying viral inhibitors with clinical potential.
Measurement(s) | nucleus • number of infected nuclei • infection index (number of infected nuclei/number of nuclei) • Plaque • fluorescent reporter intensity • percent cell viability |
Technology Type(s) | epifluorescence microscopy • compound toxicity assay |
Factor Type(s) | compound treatment |
Sample Characteristic - Organism | Human mastadenovirus C |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.12594470
Similar content being viewed by others
Background & Summary
Human adenoviruses (HAdVs) affect the respiratory, urinary and gastrointestinal tracts and the eyes. They cause morbidity and mortality, especially to immuno-compromised patients1,2 as indicated by a recent outbreak in the USA killing 12 children, or a recent case of meningoencephalitis in a middle-aged woman in the USA3. HAdVs have a high prevalence4,5,6,7 and are broadly used as gene therapy and vaccination vectors as well as oncolytic viruses8,9,10. The high seroprevalence of HAdV-C2 and C5 (species C, types 2 and 5) underlines that HAdV infections are asymptomatic in healthy individuals, but persist in mucosal lymphocytes, and thereby pose a risk for immunosuppressed patients undergoing stem cell transplantation11,12. More than 100 HAdV genotypes are grouped into seven species based on hemagglutination assays and genome sequences13,14. Types of the species A, F and G replicate in the gastrointestinal tract, B, C and E in the respiratory tract, and B and D in the conjunctiva of the eyes. Notably, species B members have a broad tropism, including kidney and hematopoietic cells6,12.
HAdV has a double-stranded DNA genome of ~36 kbp tightly packaged into an icosahedral protein capsid of about 90 nm in diameter15,16. HAdV-C2 and C5 enter cells by receptor-mediated endocytosis, shed minor capsid proteins, expose the membrane lytic protein, penetrate the endosomal membrane and are transported to the nuclear membrane, where they uncoat and release their genome to the nucleus17,18,19,20. In the nucleus, the viral genome gives rise to the immediate early viral mRNA encoding the E1A protein, which transactivates the subviral promoters, drives lytic infection and maintains genome persistence in presence of interferon21,22,23. Proteolytically matured HAdV progeny is released upon rupture of the nuclear envelope and the plasma membrane24,25,26.
Currently, there is no effective therapy available against HAdV disease. The standard of care is the nucleoside analogue Cidofovir, with poor clinical efficacy6,27. The problem is exacerbated by the shortage of a suitable small animal model for HAdV disease, although Syrian Hamsters are susceptible to HAdV-C infection and give rise to viral progeny28. Here, we developed an image-based procedure to identify novel inhibitors of HAdV infection in cell culture. We used the commercially available Prestwick Chemical Library (PCL) comprising 1,280 off-patent mostly FDA-approved small molecules (listed in PCL compounds tested in the screening procedure29). The PCL comprises compounds against diseases including infection and cancer30,31, collected at prestwickchemical.com/libraries-publications.
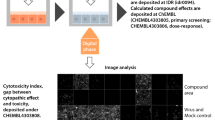
We performed a phenotypic screen against HAdV-C2 infection employing automated fluorescence microscopy and image-based scoring of the progression of multi-round infections using the Plaque2.0 software32 (Fig. 1a,b). For representative images in 384 well plates, see Fig. 1c. The procedure features robust imaging, image analysis and data processing, as concluded from two parallel workflows carried out at independent institutions, the Department of Molecular Life Sciences at University of Zurich (UZH), and the Biomolecular Screening Facility at Ecole Polytechnique Fédérale de Lausanne (EPFL).
The compound screening procedure. (a) Following assay development, stability and quality testing, the screening of the PCL against HAdV infection was performed. Imaging, image analysis and data processing were independently carried out at UZH and EPFL, before hit ranking. (b) Schematic overview of the wet-lab pipeline. PCL compounds and DFT positive control in DMSO as well as DMSO alone as negative control were pre-spotted onto 384-well imaging plates by Echo acoustic liquid handling at 10 nl corresponding to a final concentration of 1.25 µM in 80 µl assay volume/well and stored at −20 °C. Compound-blinded plates are thawed and 4,000 A549 cells/wells seeded. The following day, the cells were inoculated with HAdV-C2-dE3B at 1.77*105 genome equivalents/well. Allowing for multiple viral replication rounds, the cells were PFA-fixed at 72 hpi and the nuclei stained with Hoechst 33342. The infection phenotypes were imaged using an epifluorescence HT microscope and scored using Plaque2.0. The data of the four technical replicates were further processed in R or through EPFL-BSF LIMS. (c) Exemplary epifluorescence microscopy images of cells in 384-wells stitched to a screening plate overview of 32 replicates of negative (two most left columns) and positive control (two most right columns) and 320 blinded PCL compounds (centre 20 columns). Hoechst-stained nuclei are shown in blue, viral GFP in green. (d) Representative 384-well epifluorescence microscopy images of the DMSO negative control (most left), the DFT positive control (most right) and the top hit Nelfinavir mesylate (centre). Empty black triangle indicates a plaque (infection focus) from a productively infected cell. White arrows point out infected cells that did not form a plaque. Hoechst-stained nuclei are shown in blue, infected cells expressing GFP in green. Scale bar is 5 mm.
Five phenotypic features scored the effects of the compounds on HAdV-C2-dE3B-GFP-infected human lung cancer epithelial A549 cells – the number of infected and uninfected cell nuclei, the infection index (infected nuclei per total nuclei), the number of plaques (areas of infection foci originating from a single productively infected cell as in non-perturbed infections depicted in Fig. 1d, left image) and the integrated signal of the infection marker green fluorescence protein (GFP) encoded in the reporter virus genome. All data are available at the Image Data Resource (IDR, idr.openmicroscopy.org), IDR accession number idr008133, and can be accessed via the IDR web client. Raw and scored infection phenotypes are provided for UZH and EPFL analyses. Rigorous assay development ensured a high data quality, as indicated by mean Z’-factors of 0.52 for the plaque numbers. The screening was performed in four biological replicates at high reproducibility. Compounds that gave significant toxicity in uninfected cells were excluded during hit filtering.
Imaging, image analysis and scoring by the two independent teams yielded well correlated data and a congruent list of top hits, provided in Table 1. We identified Nelfinavir mesylate (Nelfinavir in short) as best inhibitor of HAdV infection. As evident in representative images presented in Fig. 1d, Nelfinavir abolishes the formation of plaques, but not single first round infections. We confirmed the antiviral efficacy of Nelfinavir in a follow-up study34.
Methods
Virus
HAdV-C2-dE3B-GFP was produced as described24 and fully sequenced35. In brief, the virus was generated by exchange of the viral E3B genome region with a reporter cassette harbouring the enhanced green fluorescent protein (GFP) under the immediate early Cytomegalovirus (CMV) promoter24. The virus was grown in A549 cells and purified by double caesium chloride gradient centrifugation36. Aliquots supplemented with 10% glycerol (v/v) were stored at −80 °C. HAdV-C2-dE3B-GFP was found to be homogeneous by SDS-PAGE and negative-stain analyses by transmission electron microscopy.
Cell culture
A549 (human adenocarcinomic alveolar basal epithelium) cells were obtained from the American Type Culture Collection (ATCC), Manassas, USA. The cells were maintained in full medium: high glucose Dulbecco Modified Eagle Medium (DMEM; Thermo Fisher Scientific, Waltham, USA) containing 7.5% fetal bovine serum (FBS, Invitrogen, Carlsbad, USA), 1% L-glutamine (Sigma-Aldrich, St. Louis, USA) and 1% penicillin streptomycin (Sigma-Aldrich, St. Louis, USA) and subcultured following PBS washing and trypsinisation (Trypsin-EDTA, Sigma-Aldrich, St. Louis, USA) weekly. Cells were grown at standard conditions (37 °C, 5% CO2, 95% humidity) and passage number kept below 20.
Preparation of pre-plates
Ten µl 0.0125% DMSO in PBS was spotted on all 384 wells each of imaging-compatible 384-well plates (Matrix plates #4332, Thermo Fisher Scientific, Waltham, USA) using a Matrix WellMate dispenser and normal bore Matrix WellMate tubing cartridges (Thermo Fisher Scientific, Waltham, USA). Plates were sealed and stored at −20 °C.
Blinding
The PCL compound arrangement as dispensed by EPFL in four subset plates A - D comprising each screening set replicate 1–4 was blinded and replaced by UZH with internal identifier (Raw Plaque-2.0 infection scores of the PCL screen, imaged and analysed at UZH and Processed Plaque-2.0 infection scores of the PCL screen, imaged and analysed at UZH29, compoundIdentifier 1 to 1280). The identity of the compounds was only disclosed after the screening process and hit filtering (Raw Plaque-2.0 infection scores of the PCL screen, imaged and analysed at UZH and Processed Plaque-2.0 infection scores of the PCL screen, imaged and analysed at UZH29 and Table 1, PCL_ID Prestw-1 to Prestw-1804 and compoundName).
Compounds
The PCL was obtained from Prestwick Chemical (Illkirch, France). 3′-deoxy-3′-fluorothymidine (DFT, CAS number 25526-93-6) was obtained from Toronto Research Chemical, North York, Canada. All compounds were dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich, St. Louis, USA) at a final stock concentration of 10 mM and stored at −20 °C.
PrestoBlue toxicity assay
Toxicity of the PCL compounds on uninfected A549 cells was assessed using the PrestoBlue Cell Viability reagent (Thermo Fisher Scientific, Waltham, USA). PrestoBlue37,38 is a cell viability and cytotoxicity indicator based on resazurin. Resazurin is reduced to resorufin in cellular respiration by accepting electrons from NAPDH, FADH, FMNH, NADH and cytochromes. This reduction causes PrestoBlue to change from a non-fluorescent to a strongly fluorescent form. The conversion of PrestoBlue is proportional to the number of metabolically active cells and can be evaluated quantitatively using fluorescence or absorbance measurements. Briefly, following 3.5-day continuous treatment of A549 cells with compounds at concentrations and cell densities as in the screening protocol, 10% final PrestoBlue was added to each well and incubated for 1 h at standard cell incubation conditions. Fluorescence intensity (bottom-read) was measured using a multi-well plate reader (Tecan Infinite F500, Tecan, Männedorf, Switzerland) with excitation at 560/10 nm, emission at 590/10 nm at a fixed gain. Doxorubicin hydrochloride (Prestw-438, Prestwick Chemical, Illkirch, France) was used as a positive control for cytotoxicity, at a final concentration of 10 µM, and the corresponding concentration of the drug solvent DMSO was used as a negative control. The full PCL library was tested on duplicated plates. The EPFL-BSF in-house Laboratory Information Management System (LIMS) was used for data processing and statistical validation. First, raw PrestoBlue readings were normalized per plate to negative control values at 0 and positive controls at 1. Then, the normalized values of both duplicates were averaged. Assay quality was assessed for each plate through the Z’-factor calculation. Compounds were considered toxic, when the normalized value for all replicates was higher than the average + 3σ (standard deviation, SD) of the DMSO negative control for the corresponding plate. Scores and score SD were then calculated for hit compounds by averaging normalized value for all replicates.
Preparation of plates for Z’-factor and drug screening
Ten nl of 10 mM PCL compounds, the nucleoside analogue DFT positive control (all dissolved in DMSO) and DMSO only as negative control were pre-spotted on imaging-compatible 384-well plates (Falcon plates, Corning Inc., New York, USA) using an Echo acoustic liquid handling system (Labcyte, San Jose, USA) by the EPFL-BSF, sealed and stored at −20 °C. Each Z’-factor 384-well plate consisted of 192 technical replicates of positive and negative controls, each. Each screening plate set consisted of four subset plates A to D. Each screening plate comprised 32 technical replicates of positive and negative controls, each, and 320 PCL compounds.
Wet-lab screening pipeline
The screening was performed in four independent biological replicates 1–4. Liquid handling was performed using a Matrix WellMate dispenser and Matrix WellMate tubing cartridges (Thermo Fisher Scientific, Waltham, USA). Prior to usage, tubings were rinsed with 125 ml autoclaved double-distilled (dd) H2O followed by 125 ml autoclaved PBS. Pre-spotted compound plates were thawed at room temperature (RT) for 30 min, briefly centrifuged before compounds were dissolved in 10 µl/well of PBS. 4,000 A549 cells/well in 60 µl full medium were seeded onto the compounds using standard bore tubing cartridges. Following cell adhesion over night, the cells were inoculated with 1.77*105 genome equivalents per well of HAdV-C2-dE3B-GFP in 10 µl of full medium using bovine serum albumin (BSA, cell-culture grade, Sigma-Aldrich, St. Louis, USA)-blocked small bore tubing cartridges. The final compound concentration was 1.25 µM at 0.0125% DMSO. Infection was allowed to progress over multiple infection rounds for 72 h giving rise to foci of infected cells originating from a single first round infected cell. Cells were fixed for 1 h at RT by addition of 26.6 µL 16% PFA and 4 µg/ml Hoechst 33342 (Sigma-Aldrich, St. Louis, USA) in PBS using standard bore tubing cartridges. Cells were washed three times with PBS before PBS supplemented with 0.02% N3 was added and plates were sealed for long-term storage at 4 °C. Following usage, tubings were rinsed with 125 ml autoclaved ddH2O followed by 125 ml autoclaved PBS and autoclaved for re-usage.
Imaging
Nuclei stained with Hoechst 33342 (DAPI channel) and viral GFP (FITC channel) were imaged on two devices. At UZH, plates were imaged on an IXM-C automated high-throughput fluorescence microscope (Molecular Devices, San Jose, USA) using MetaXpress (version 6.2, Molecular Devices, San Jose, USA) and a 4x air objective (Nikon S Fluor, 0.20 NA, 15.5 mm WD, Nikon Instruments, Minato, Japan) at widefield mode. Images of 2,0482 px at 1.72 µm/px resolution were acquired on an Andor sCMOS camera (Oxford Instruments, Abingdon, UK). Exposure times: DAPI 150 ms, FITC 20 ms. At EPFL, images were acquired on a IN Cell 2200 automated high-throughput fluorescence microscope (GE Healthcare, Chicago, USA) using IN Cell Analyzer (version 6.2, GE Healthcare, Chicago, USA) and a 4x air objective (Nikon Plan Apo, 0.20 NA, 15.7 mm WD, Nikon Instruments, Minato, Japan) at widefield mode. Image size 2,0482 px at 1.625 µm/px resolution acquired on an Andor sCMOS camera. Exposure times: DAPI 300 ms, FITC 40 ms.
Image analysis
The infection phenotype for each well was quantified by Plaque2.032 (https://github.com/plaque2/matlab/tree/antivir) via five main read-outs: number of nuclei, number of infected nuclei, the ratio between infected and total nuclei referred to as infection index, number of multi-round infection foci termed plaques (plaque forming unit(s), pfu) and the integrated viral transgenic GFP intensity. Plaque2.0 parameters were optimized independently at UZH and EPFL for the data acquired at the respective institution. Well- and object-based read-outs are provided in the Plaque2.0 output files.
Z’-factor calculation
The Z’-factor was computed using R version 3.3.239 according to Eq. (1)
where σ+ is the SD of the positive control, σ- is the SD of the negative control, μ+ the mean of the positive control and μ- the mean of the negative control.
Screening data processing
Plaque2.0 results were further processed and filtered. At UZH, results were processed in R version 3.3.239, EPFL used KNIME version 3.4.040 as well as the EPFL-BSF in-house LIMS. Mean infection scores over the five Plaque2.0 read-outs of the four biological replicates of each PCL compound and the 16 biological replicates containing 32 technical replicates of positive and negative control, each, were calculated. Each compound’s scores were normalized by the mean score of the DMSO negative control of the respective plate. Only non-toxic, effective PCL compounds were considered as HAdV inhibitor candidates. Non-toxic compounds were filtered by applying an inclusive μ+ (mean of the negative control) ± 2σ (SD of the negative control) threshold for number of nuclei. Efficacy was filtered by applying an excluding μ+ ± 3σ threshold for the infection scores (number of infected nuclei, infection index, number of plaques or integrated GFP intensity). Subsequently, compounds exhibiting significant toxicity to noninfected cells were excluded.
Data Records
Data structure and repositories
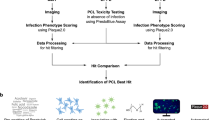
The screening data comprise the information collected during assay development, including stability, quality and screening of the PCL itself. The latter two were imaged on two different microscopes. We provide the parameters used for Plaque2.0 image analysis, and the code for the subsequent hit filtering in R. The data available for download at the IDR, accession number idr008133, are structured as outlined in Fig. 2a. For download instructions, see idr.openmicroscopy.org/about/download. Moreover, the data can be viewed conveniently on the IDR web client (idr.openmicroscopy.org/webclient), where it is structured as depicted in Fig. 2b. Additionally, an annotated list of the PCL compounds as well as raw and scored screening data are available on figshare29 as.txt files.
Project data structure available at IDR, accession number idr008133. (a) In the data provided for download, there are three sub-folders for 1-prePlates, 2-ZPlates and 3-Screen. The latter two contain both the images and analyses generated by UZH and EPFL. (b) The data provided for viewing are divided into five screens: screenA contains the pre-plates and screenB and screenC consist of the Z’-factor plates imaged and analysed at UZH and EPFL, respectively. screenD and screenE provide the screening data obtained at UZH and EPFL, respectively.
Data sets and file types
The data provided for download consists of three data sets 1 to 3 (see Fig. 2a).
- 1-prePlates contains layouts (.csv), images (.tif), Plaque2.0 image analysis parameters (.mat) and results (.csv) for the assay stability test plates performed at UZH prior to Z’-factor plates (preZ) and the screen (preScreen).
- 2-ZPlates contains layouts (.csv), images (.tif), Plaque2.0 image analysis parameters (.mat) and results (.csv) for the two Z’-factor plates a and b as imaged and analysed at UZH (Data_UZH) and EPFL (Data_EPFL).
- 3-Screen contains layouts (.csv), images (.tif), Plaque2.0 image analysis parameters (.mat) and results (.csv) for the 16 screening plates (four biological replicates 1–4, each consisting of a set of four subset plates A - D) as imaged and analysed at UZH (Data_UZH) and EPFL (Data_EPFL). Moreover, Analysis contains the Plaque2.0 batch processing (AntiVir_batchprocessing.m) and hit filtering pipeline (AntiVir_hitfiltering.R) used by UZH. Analysis also contains the PrestoBlue raw results (.csv) for toxicity in absence of infection.
The data provided for browsing via the IDR web client are assembled into five screens A to E (see Fig. 2b).
- idr0081-study.txt summarizes the overall study and the five screens that were performed.
- screenA contains the assay stability test plates performed at UZH prior to Z’-factor plates (preZ) and the screen (preScreen). idr0081-screenA-library.txt provides thorough information on the tested compounds including PubChem identifiers and their plate layout. idr0081-screenA-processed.txt presents the results of the Plaque2.0-based image analysis. idr0081-screenA-mean.txt summarises the infection scores per pre plate.
- screenB contains the assay quality test plates (Z’-factor plates a and b) performed at UZH. idr0081-screenB-library.txt provides thorough information on the tested compounds including PubChem identifiers and their plate layout. idr0081-screenB-processed.txt presents the results of the Plaque2.0-based image analysis. idr0081-screenB-mean.txt summarises the infection scores per Z’-factor plate.
- screenC contains the assay quality test plates (Z’-factor plates a and b) performed at EPFL. idr0081-screenC-library.txt provides thorough information on the tested compounds including PubChem identifiers and their plate layout. idr0081-screenC-processed.txt presents the results of the Plaque2.0-based image analysis. idr0081-screenC-mean.txt summarises the infection scores per Z’-factor plate.
- screenD contains the PCL screening plates (in replicates 1 to 4, consisting of subset plates A to D) performed at UZH. idr0081-screenD-library.txt provides thorough information on the tested compounds including PubChem identifiers and their plate layout. idr0081-screenD-processed.txt presents the results of the Plaque2.0-based image analysis. idr0081-screenB-filtered.txt summarises the infection scores per compound and indicates if it was identified as hit.
- screenE contains the PCL screening plates (in replicates 1 to 4, consisting of subsets A to D) performed at EPFL. idr0081-screenE-library.txt provides thorough information on the tested compounds including PubChem identifiers and their plate layout. idr0081-screenE-processed.txt presents the results of the Plaque2.0-based image analysis. idr0081-screenE-filtered.txt summarises the infection scores per compound and indicates, if it was identified as hit.
Technical Validation
Assay stability
The wet-lab screening pipeline was optimized regarding liquid handling, cell seeding, virus inoculum, positive and negative controls, infection time, as well as imaging and image analysis. This ensured a high assay stability and reproducibility. Furthermore, all compounds, especially media and supplements, the BSA for tubing saturation, PFA- and Hoechst-supplemented fixative were prepared as large batch from a single lot and stored as single-use aliquots. Prior to every experiment, assay stability with respect to cell and infection phenotype was tested on pre-plates according to the established wet-lab, imaging and image analysis pipeline. Since the solvent control had already been spotted in 10 µl PBS, no further PBS was added prior to cell seeding. Periodically, the virus stock dilution was tested and adjusted for experiments if necessary.
Assay quality determination: Z’-factor
The accuracy of the wet-lab, imaging and image analysis pipeline was assessed by two independently imaged and analysed Z’-factor plates (Table 2 and Fig. 3). 3σ Z’-factors of numberOfInfectedNuclei, infectionIndex and numberOfPlaques were in the range of 0.30 to 0.57, scoring good to excellent. totalVirusIntensity (Z’-factors between −0.07 to 0.08) were not suitable to identify HAdV infection inhibitors, while numberOfNuclei (Z’-factors between −1.11 to −8.10) was not a useable readout either. Additionally, the Z’-factors were determined for each of the 16 screening plates (Table 3 and Fig. 4). 3σ Z’-factors of numberOfInfectedNuclei, infectionIndex and numberOfPlaques were in the range of 0.27 to 0.57, scoring good to excellent.
Infection score density of positive and negative controls across Z’-factor plates. Distribution of (a) numberOfNuclei, (b) numberOfInfectedNuclei, (c) infectionIndex, (d) numberOfPlaques and e totalVirusIntensity in negative control (0.0125% DMSO) compared to positive control-treated (1.25 µM DFT) samples of the two Z’-factor plates. Dark green and dark grey indicate Z’-factor plate a, light green and grey show Z’-factor plate b. Dashed vertical lines mark mean of 192 technical replicates.
Infection score density of positive and negative controls across screening replicates. Distribution of (a) numberOfNuclei, (b) numberOfInfectedNuclei, (c) infectionIndex, (d) numberOfPlaques and e totalVirusIntensity in negative control (0.0125% DMSO in grey) compared to positive control-treated (1.25 µM DFT in green) samples of the screening sets. Replicates 1 to 4 indicated by colour shading are each comprised of four set plates containing 32 technical replicates per control. The dashed vertical lines indicate the corresponding mean values.
Independent analysis and filtering
Imaging, image analysis and screening data processing were performed by two independent research teams at UZH and EPFL, as depicted in Fig. 1. Raw and scored infection phenotypes are shown for UZH and EPFL analyses (Raw Plaque-2.0 infection scores of the PCL screen, imaged and analysed at UZH, Processed Plaque-2.0 infection scores of the PCL screen, imaged and analysed at UZH and Raw Plaque-2.0 infection scores of the PCL screen, imaged and analysed at EPFL, Processed Plaque-2.0 infection scores of the PCL screen, imaged and analysed at EPFL, respectively29). Both dry-lab pipelines confirmed the high assay quality (Tables 2 and 3). During hit filtering, PCL compounds that gave significant toxicity in uninfected cells were excluded during hit filtering (Fig. 5, PCL compounds excluded due to toxicity in uninfected cells29). As summarized in Fig. 6 left panel, both scores are strongly correlated with R2 between 0.6870 and 0.9870. Both approaches yielded identical top scored compounds (Fig. 6, right panel), of which Prestw-1764, Nelfinavir mesylate, was the top hit.
PCL compound toxicity in uninfected cells. Of the 1,278 PCL compounds tested, 126 PCL compounds are found to be toxic, as shown in red, and listed in PCL compounds excluded due to toxicity in uninfected cells29. A549 cells were treated with PCL compounds in duplicates according to the screening wet-lab protocol, however, in absence of HAdV infection for 3.5 days. Doxorubicin hydrochloride (Prestw-438) was used as a positive control for cytotoxicity, at a final concentration of 10 µM, and the corresponding concentration of the drug solvent DMSO was used as a negative control. Cell viability was determined by PrestoBlue assay. PrestoBlue fluorescence intensities of each well were normalized per plate to negative control values at 0 and positive controls at 1. Compounds were considered toxic, when the normalized value for all replicates was higher than the average +3σ (standard deviation, SD) of the DMSO negative control for the corresponding plate. X-axis indicates compounds by their PCL identifier (PCL ID, see PCL compounds tested in the screening procedure29). Normalized average PrestoBlue read-outs are depicted on the y-axis.
Infection scores from independent dry-lab pipelines. Imaging, image analysis and data processing were performed independently at UZH and EPFL. Infection phenotypes in PCL-treated cells of four biological replicates were averaged and normalized against the DMSO solvent control. Linear regression plots of UZH and EPFL data are shown for (a) numberOfNuclei, (b) numberOfInfectedNuclei, (c) infectionIndex, (d) numberOfPlaques and (e) totalVirusIntensity of the 1,278 tested PCL compounds from are (green line). Red dots indicate toxicity in the absence of infection. Non-toxic compounds are shown as green dots. R2 was calculated using GraphPad Prism 8.2.1. Highest scoring compounds are shown on the right, including the PCL_ID of some non-toxic compounds.
Usage Notes
Five parameters were used to score the infection phenotype of each well: the number of nuclei (numberOfNuclei), number of infected nuclei (numberOfInfectedNuclei), the ratio between number of infected and total nuclei (infectionIndex), the number of multi-round infection foci termed plaques (numberOfPlaques) and the extend of viral GFP reporter expression as integrated GFP intensity totalVirusIntensity).
Infection scoring using the Plaque2.0 GUI
A detailed manual for Plaque2.0 GUI-based infection phenotype scoring is available at plaque2.github.io/. No MATLAB license is necessary.
The following settings should be used:
Input/Output:
Processing folder: Path to folder containing the images (e.g. idr0081/3-Screen/Data_EPFL/Screen/ BSF018292_1A).
filename pattern Data_UZH:.* (? < wellName > [A-Z][0–9]*)_(? < channelName > w[0–9]*).TIF
filename pattern Data_EPFL:.* (? < wellName > [A-Z] - [0–9] + )[(]fld 1 wv (? < channel > [A-Z]{4}).*.tif
Plate name: Name of the plate to be analysed (e.g. BSF018292_1A)
Result Output Folder: Path to the results folder in the respective data folder (e.g. idr0081/3-Screen/Data_EPFL/Results).
Stitch: Stitching of the images is not necessary, since every 384-well is imaged in a single site. Do not activate the tab.
Mask:
Custom Mask File: Path to the manually defined mask file (e.g. idr0081/3-Screen/Data_UZH/Parameters). Well masking is optional and was not performed by EPFL.
Monolayer:
Channel: Nuclei were imaged in channel 1.
Plaque:
Channel: Viral GFP reporter signal was imaged in channel 2.
Infection scoring using the Plaque2.0 batch script
How to use the AntiVir_batchprocessing.m for Plaque2.0 batch processing is indicated in the comments of the code.
Code availability
Plaque2.0 batch image analysis for infection scoring. The MATLAB (version R2016b, The MathWorks, Natick, USA) script AntiVir_batchprocessing.m used by UZH for image analysis is provided for download at IDR, accession number idr0081, under idr0081/3-Screen/Analysis. It is based on the Plaque2.0 software available on GitHub under GPLv3 open source license: https://github.com/plaque2/matlab.
To batch analyse the HAdV screening data by Plaque2.0, fork or download the Plaque2.0 AntiVir code from GitHub: https://github.com/plaque2/matlab/tree/antivir. Place the AntiVir_batchprocessing.m file from idr0081/3-Screen/Analysis into the Plaque2/matlab folder and follow the instructions in AntiVir_batchprocessing.m. A MATLAB license is required.
Hit filtering using R. The R39 (version 3.6.1 (2019-07-05)) script AntiVir_hitfiltering.R used by UZH for data processing and hit filtering is provided at IDR accession number idr0081 under idr0081/3-Screen/Analysis.
References
Krilov, L. R. Adenovirus infections in the immunocompromised host. Pediatr Infect Dis J 24, 555–556 (2005).
Greber, U. F., Arnberg, N., Wadell, G., Benkő, M. & Kremer, E. J. Adenoviruses - from pathogens to therapeutics: a report on the 10th International Adenovirus Meeting. Cell Microbiol 15, 16–23 (2013).
Tunkel, A. R., Baron, E. L., Buch, K. A., Marty, F. M. & Martinez-Lage, M. Case 31-2019: A 45-Year-Old Woman with Headache and Somnolence. N Engl J Med 381, 1459–1470 (2019).
Gray, G. C. et al. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004-2006. Clin Infect Dis 45, 1120–1131 (2007).
Metzgar, D. et al. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J Infect Dis 196, 1465–1473 (2007).
Lynch, J. P. & Kajon, A. E. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin Respir Crit Care Med 37, 586–602 (2016).
Haque, E., Banik, U., Monowar, T., Anthony, L. & Adhikary, A. K. Worldwide increased prevalence of human adenovirus type 3 (HAdV-3) respiratory infections is well correlated with heterogeneous hypervariable regions (HVRs) of hexon. PLoS ONE 13, e0194516 (2018).
Jiang, H. et al. Oncolytic adenovirus research evolution: from cell-cycle checkpoints to immune checkpoints. Curr Opin Virol 13, 33–39 (2015).
Lawler, S. E., Speranza, M.-C., Cho, C.-F. & Chiocca, E. A. Oncolytic viruses in cancer treatment: A review. JAMA Oncology 3, 841–849 (2017).
Ginn, S. L., Amaya, A. K., Alexander, I. E., Edelstein, M. & Abedi, M. R. Gene therapy clinical trials worldwide to 2017: An update. J Gene Med 20, e3015 (2018).
Mennechet, F. J. D. et al. A review of 65 years of human adenovirus seroprevalence. Expert Rev Vaccines 18, 597–613 (2019).
Lion, T. Adenovirus persistence, reactivation, and clinical management. FEBS Lett, https://doi.org/10.1002/1873-3468.13576 (2019).
Ismail, A. M. et al. Genomic foundations of evolution and ocular pathogenesis in human adenovirus species D. FEBS Lett 593, 3583–3608 (2019).
Harrach, B., Tarján, Z. L. & Benkő, M. Adenoviruses across the animal kingdom: a walk in the zoo. FEBS Lett, https://doi.org/10.1002/1873-3468.13687 (2019).
Reddy, V. S., Natchiar, S. K., Stewart, P. L. & Nemerow, G. R. Crystal structure of human adenovirus at 3.5 A resolution. Science 329, 1071–1075 (2010).
Benevento, M. et al. Adenovirus composition, proteolysis, and disassembly studied by in-depth qualitative and quantitative proteomics. J Biol Chem 289, 11421–11430 (2014).
Greber, U. F. & Flatt, J. W. Adenovirus entry: from infection to immunity. Annual review of virology 6, 177–197 (2019).
Bauer, M. et al. The e3 ubiquitin ligase mind bomb 1 controls adenovirus genome release at the nuclear pore complex. Cell Rep 29, 3785–3795.e8 (2019).
Greber, U. F. Virus and host mechanics support membrane penetration and cell entry. J Virol 90, 3802–3805 (2016).
Wang, I.-H., Burckhardt, C. J., Yakimovich, A., Morf, M. K. & Greber, U. F. The nuclear export factor CRM1 controls juxta-nuclear microtubule-dependent virus transport. J Cell Sci 130, 2185–2195 (2017).
Prasad, V. et al. The UPR sensor IRE1α and the adenovirus E3-19K glycoprotein sustain persistent and lytic infections. Nat Commun 11, 1997 (2020).
King, C. R., Zhang, A., Tessier, T. M., Gameiro, S. F. & Mymryk, J. S. Hacking the cell: network intrusion and exploitation by adenovirus E1A. MBio 9, (2018).
Zheng, Y., Stamminger, T. & Hearing, P. E2f/rb family proteins mediate interferon induced repression of adenovirus immediate early transcription to promote persistent viral infection. PLoS Pathog 12, e1005415 (2016).
Yakimovich, A. et al. Cell-free transmission of human adenovirus by passive mass transfer in cell culture simulated in a computer model. J Virol 86, 10123–10137 (2012).
Tollefson, A. E. et al. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol 70, 2296–2306 (1996).
Doronin, K. et al. Overexpression of the ADP (E3-11.6K) protein increases cell lysis and spread of adenovirus. Virology 305, 378–387 (2003).
Lenaerts, L. & Naesens, L. Antiviral therapy for adenovirus infections. Antiviral Res 71, 172–180 (2006).
Wold, W. S. M., Tollefson, A. E., Ying, B., Spencer, J. F. & Toth, K. Drug development against human adenoviruses and its advancement by Syrian hamster models. FEMS Microbiol Rev 43, 380–388 (2019).
Georgi, F. et al. High-content image-based drug screen identifies a clinical compound against cell transmission of adenovirus. figshare https://doi.org/10.6084/m9.figshare.c.5052965 (2020).
Chauvin, C. et al. High-Throughput Drug Screening Identifies Pazopanib and Clofilium Tosylate as Promising Treatments for Malignant Rhabdoid Tumors. Cell Rep 21, 1737–1745 (2017).
Wall, G. et al. Screening a Repurposing Library for Inhibitors of Multidrug-Resistant Candida auris Identifies Ebselen as a Repositionable Candidate for Antifungal Drug Development. Antimicrob Agents Chemother 62 (2018).
Yakimovich, A. et al. Plaque2.0-A High-Throughput Analysis Framework to Score Virus-Cell Transmission and Clonal Cell Expansion. PLoS ONE 10, e0138760 (2015).
Georgi, F. et al. High-content image-based drug screen identifies a clinical compound against cell transmission of adenovirus. University of Dundee https://doi.org/10.17867/10000136 (2020).
Georgi, F. et al. The FDA-approved drug Nelfinavir inhibits lytic cell-free, but not cell-associated non-lytic transmission of human adenovirus. Antimicrob Agents Chemother, https://doi.org/10.1128/AAC.01002-20 (2020).
Georgi, F. et al. Mutant Human adenovirus 2 isolate HAdV-C2-dE3B-CMV-GFP, complete sequence. GenBank https://identifiers.org/ncbi/insdc:MT277585.1 (2020).
Greber, U. F., Willetts, M., Webster, P. & Helenius, A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75, 477–486 (1993).
Boncler, M., Różalski, M., Krajewska, U., Podsędek, A. & Watala, C. Comparison of PrestoBlue and MTT assays of cellular viability in the assessment of anti-proliferative effects of plant extracts on human endothelial cells. J Pharmacol Toxicol Methods 69, 9–16 (2014).
Xu, M., McCanna, D. J. & Sivak, J. G. Use of the viability reagent PrestoBlue in comparison with alamarBlue and MTT to assess the viability of human corneal epithelial cells. J Pharmacol Toxicol Methods 71, 1–7 (2015).
R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2018).
Berthold, M. R. et al. KNIME - the Konstanz information miner. ACM SIGKDD Explorations Newsletter 11, 26 (2009).
Acknowledgements
We thank the Greber lab for fruitful discussions and critical assessment of the data. We thank the IDR team for helping to make our work openly accessible. The work was supported by the Swiss National Science Foundation (SNSF) to U.F.G. (Grant numbers 316030_170799/1 and 31003A_179256/1), and the SNSF National Research Program “NCCR Chemical Biology” to G.T. and U.F.G.
Author information
Authors and Affiliations
Contributions
U.F.G., V.A., A.Y. conceived the screening idea. F.G. designed the experiments, and with U.F.G. coordinated the project. F.K. prepared the PCL-spotted plates. F.G. and R.W. performed the experiments. F.G. and F.K. acquired the data. F.G. and V.A. analysed the imaging data. L.M. and F.G. processed the data. G.T. organized and supervised the screening project at the EPFL-BSF. F.G., F.K. and U.F.G. wrote manuscript, with input from all the co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Georgi, F., Kuttler, F., Murer, L. et al. A high-content image-based drug screen of clinical compounds against cell transmission of adenovirus. Sci Data 7, 265 (2020). https://doi.org/10.1038/s41597-020-00604-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-020-00604-0