Abstract
Across the globe, vulture species are experiencing major population declines. A key factor for the long-term persistence of these endangered species is the maintenance of genetic diversity patterns within wild populations. The datasets presented in this descriptor includes microsatellite genotypes of 605 Cape vultures (Gyps coprotheres) drawn from across the southern African distribution of the species. Microsatellites are useful in quantifying genetic diversity at the population level. Populations of the endangered Cape vulture are currently monitored by conservation agencies and the data presented here can be used as an important baseline for future population genetic monitoring.
Measurement(s) | microsatellite |
Technology Type(s) | DNA sequencing |
Factor Type(s) | geographic location |
Sample Characteristic - Organism | Gyps coprotheres |
Sample Characteristic - Location | South Africa |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.9896372
Similar content being viewed by others
Background & Summary
In recent years, Cape vultures (Gyps coprotheres) have shown a decline in the overall number of individuals in the wild and their global range is currently undergoing a significant reduction, with most breeding colonies found in South Africa1,2,3,4,5. A more rigorous approach is required to successfully stabilize and conserve this endangered vulture4. In order to ensure the long-term conservation of vulture populations, management practices should evaluate and maintain the amount and pattern of genetic diversity within current populations6,7.
Microsatellites are useful molecular markers used to estimate the amount and pattern of genetic variation at the population-level8. These molecular markers show high levels of polymorphism within a species or among populations of the same species9. Microsatellites are sensitive to genetic changes such as, changes in effective population sizes and rates of migration among populations8 and so can be used to monitor the genetic “health” of populations.
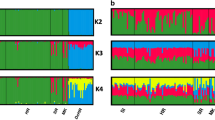
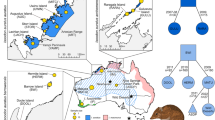
This data descriptor describes a dataset of 605 Cape vultures collected from 24 localities (Supplementary Table 1) genotyped at 13 microsatellite loci. These data were analysed in a recent study that describes the genetic diversity of South African Cape vulture populations10 and were used to estimate the regional connectivity of six Cape vulture breeding colonies in South Africa. These data represent an important baseline for future genetic monitoring of wild populations of Cape vulture.
Methods
Sampling procedure and sampling localities
A total of 605 Cape vultures from 24 localities, across the South African distribution of the species, were sampled for this study (Supplementary Table 1). This includes 266 samples collected from six breeding colonies. Samples consisted of feather, archival tissue or blood. Feather samples were collected opportunistically from feeding sites, sites of electrocutions, poisoning events and below nests at breeding colonies. Blood samples were collected when vultures were captured and fitted with global positioning system/global system for mobile transmitters11. Blood samples were stored on Whatman FTA® Elute cards (Sanford, USA). Archival museum samples (dried skin snips) were sourced from local South African museums (Supplementary Table 1).
Molecular methods
DNA extraction
The NucleoSpin® Tissue kit (Macherey-Nagel, Germany) was used for all DNA extractions. The extraction protocol was modified for feather and archival samples to improve DNA yield. Samples were incubation with proteinase K for 48 hours in a shaking water bath (56 °C), the lysate was incubated in 70 °C B3 buffer for 45 minutes, the final volume of pre-warmed elution buffer (BE) was 80 μl. During the final elution step samples were incubated at 70 °C for 20 minutes followed by centrifuging and then reapplication of the solution onto the membrane. The samples were incubated again at 70 °C for an additional five minutes followed by the final centrifuging step.
Microsatellite amplification
Thirteen microsatellite loci were selected from previous studies12,13 (Table 1). Each primer was fluorescently labeled, using three-fluorophore analogues, according to their expected allelic size and sequence motif (Table 1). Six multiplex reactions where designed according to microsatellite loci amplification, fluroscent dye and optimal annealing temperature (Table 1). All samples were amplified in six multiplex reactions using the KAPA2GTM Fast Multiplex PCR kit (KAPA Biosystems) following the manufacture’s protocol. All amplified products were analyzed using a 3130xL Genetic Analyzer housed at the Central Analytical Facility at Stellenbosch University, South Africa. The software GeneMarker v2.4.0 (Soft Genetics) was used for genotype scoring14,15.
Data Records
The datasets are available on Zenodo and include the raw fragment analysis data for the 605 Gyps coprotheres genotyped using 13 microsatellite loci14 as well as the genotype scores for the 605 Gyps coprotheres individuals15. All associated metadata (tissue type, sampling locality, and date of collection) is available in Supplementary Table 1. Multilocus microsatellite alleles are scored according to size in base pairs and missing data is encoded as “0”. Percentage of missing data included in the final dataset varied across loci (Supplementary Table 2) but was minimal (mean = 11%).
Technical Validation
To ensure genotype data quality, all archival samples were re-amplified, and each locus was genotyped multiple times (up to five times) and compared for consistency. In addition, 20% of all feather, muscle and blood samples were re-amplified multiple times (up to five times) to verify the reliability of the data. Negative controls were including in each genetic analyzer run to check for contamination of reagents. When consistent genotypes were not generated the genotype scores was inputted as missing data.
We used identity analysis in the software Cervus v3.0.716 to ensure that duplicated genotypes were not included in the final data. This is particularly important in this study as genotypes were amplified from discarded feathers collected at colonies. Null alleles can be a problem in studies that use primers not designed for the study species and this can bias population structure analysis17. Uncorrected global FST were compared to FST values corrected using the excluding null alleles (ENA) method18 using a paired t-test. The paired t-tests were not significant (p-value > 0.05) suggesting that these data are not affected by null alleles.
References
Ogada, D. L. et al. Another continental vulture crisis: Africa’s vultures collapsing toward extinction. Conservation Letters, 9, 89–97 (2016).
Pfeiffer, M. B., Venter, J. A. & Downs, C. T. Identifying anthropogenic threats to Cape Vultures Gyps coprotheres using community perceptions in communal farmland, Eastern Cape Province, South Africa. Bird Conservation International 25, 353–365 (2015).
Wolter, K., Neser, W., Hirschauer, M. T. & Camiña, A. Cape Vulture Gyps coprotheres breeding status in southern Africa: monitoring results from 2010–2014. Ostrich 87, 119–123 (2016).
BirdLife International. Gyps coprotheres (amended version of 2016 assessment). The IUCN Red List of Threatened Species, https://doi.org/10.2305/IUCN.UK.2017-3.RLTS.T22695225A118592987.en (2017).
Monadjem, A., Anderson, M. D., Piper, S. E. & Boshoff, A. F. In Proceedings of a workshop on vulture research and conservation in southern Africa. Bird of Prey Working Group, Endangered Wildlife Trust, Johannesburg, South Africa (2004).
Lerner, H. R. L. & Mindell, D. P. Phylogeny of eagles, Old World vultures, and other Accipitridae based on nuclear and mitochondrial DNA. Molecular Phylogenetics and Evolution 37, 327–346 (2005).
Arshad, M., Gonzalez, J., El-Sayed, A. A., Osborne, T. & Wink, M. Phylogeny and phylogeography of critically endangered Gyps species based on nuclear and mitochondrial markers. Journal of Ornithology 150, 419–430 (2009).
Jarne, P. & Lagoda, P. J. L. Microsatellites, from molecules to populations and back. Trends in Ecology and Evolution 11, 424–429 (1996).
Chistiakov, D. A., Hellemans, B. & Volckaert, F. A. Microsatellites and their genomic distribution, evolution, function and applications: a review with special reference to fish genetics. Aquaculture 255, 1–29 (2006).
Kleinhans, C. & Willows-Munro, S. Low genetic diversity and shallow population structure in the endangered vulture, Gyps coprotheres. Scientific Reports 9, 5536 (2019).
Pfeiffer, M. B., Venter, J. A. & Downs, C. T. Foraging range and habitat use by Cape Vulture Gyps coprotheres from the Msikaba colony, Eastern Cape province, South Africa. Koedoe 57, 1–11 (2015).
Mira, S., Billot, C., Guillemaud, T., Palma, L. & Cancela, M. L. Isolation and characterization of polymorphic microsatellite markers in Eurasian vulture Gyps fulvus. Molecular Ecology Notes 2, 557–558 (2002).
Gautschi, B., Tenzer, I., Müller, J. P. & Schmid, B. Isolation and characterization of microsatellite loci in the bearded vulture (Gypaetus barbatus) and cross‐amplification in three Old World vulture species. Molecular Ecology 9, 2193–2195 (2000).
Kleinhans, C. & Willows-Munro, S. Microsatellite fragment sizing of South African Cape vulture (Gyps coprotheres). Zenodo. https://doi.org/10.5281/zenodo.2807990 (2019).
Kleinhans, C. & Willows-Munro, S. Microsatellite genotypes of South African Cape vulture (Gyps coportheres). Zenodo. https://doi.org/10.5281/zenodo.3357761 (2019).
Kalinowski, S. T., Taper, M. L. & Marshall, T. C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology 16, 1099–1106 (2007).
Horváth, M. B., Martínez-Cruz, B., Negro, J. J., Kalmár, L. & Godoy, J. A. An overlooked DNA source for non-invasive genetic analysis in birds. Journal of Avian Biology 36, 84–88 (2005).
Chapuis, M. P. & Estoup, A. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 24, 621–631 (2007).
Acknowledgements
This study was funded by a Rated Researcher grant to awarded to S. Willows-Munro by the National Research Foundation (South Africa). The first author received a free-standing MSc bursary from the NRF. The authors wish to thank the curators of the Ditsong National Museum of Natural History (Pretoria, South Africa), Durban Natural Sciences Museum (Durban, South Africa), Iziko Museums (Cape Town, South Africa), National Museum (Bloemfontein, South Africa) and National Zoological Gardens of South Africa (Pretoria, South Africa) for providing access to archival samples. Many thanks to L. Arnot, A. Botha, M. Streicher, A. Butt, B. Coversdale, C. Downs, B. Hoffman, S. Krüger, S. McPherson, W. Neser, M. Pfeiffer, K. Shaw, L. Thompson and R. Visagie, who collected samples in the field. Ethical approval was obtained for this study from the University of KwaZulu-Natal Animal Ethics subcommittee (Reference number: 045/15/Animal). All necessary sampling permits were obtained through Ezemvelo KZN Wildlife (Permit number: OP3407/2016) and the National Department of Environmental Affairs (Permit Numbers: 05054, 27273 and 28553).
Author information
Authors and Affiliations
Contributions
C.K. conducted data collection. C.K. and S.W.-M. drafted and revised the article.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Kleinhans, C., Willows-Munro, S. Microsatellite genotypes of the South African Cape vulture, Gyps coprotheres. Sci Data 6, 200 (2019). https://doi.org/10.1038/s41597-019-0221-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-019-0221-4