Abstract
In Amazonia, the knowledge about Fungi remains patchy and biased towards accessible sites. This is particularly the case in French Guiana where the existing collections have been confined to few coastal localities. Here, we aimed at filling the gaps of knowledge in undersampled areas of this region, particularly focusing on the Basidiomycota. From 2011, we comprehensively collected fruiting-bodies with a stratified and reproducible sampling scheme in 126 plots. Sites of sampling reflected the main forest habitats of French Guiana in terms of soil fertility and topography. The dataset of 5219 specimens gathers 245 genera belonging to 75 families, 642 specimens are barcoded. The dataset is not a checklist as only 27% of the specimens are identified at the species level but 96% are identified at the genus level. We found an extraordinary diversity distributed across forest habitats. The dataset is an unprecedented and original collection of Basidiomycota for the region, making specimens available for taxonomists and ecologists. The database is publicly available in the GBIF repository (https://doi.org/10.15468/ymvlrp).
Measurement(s) | taxonomic diversity assessment by targeted gene survey • Fungi |
Technology Type(s) | Barcode-Seq • Taxonomy |
Factor Type(s) | geographic location |
Sample Characteristic - Organism | Fungi |
Sample Characteristic - Environment | tropical lowland evergreen broadleaf rain forest |
Sample Characteristic - Location | French Guiana Region |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.9914138
Similar content being viewed by others
Background & Summary
Neotropical rainforests are poorly described when it comes to the Fungi. The distribution of the known species remains patchy, biased towards accessible sites1,2 and their ecology is still largely fragmentary3. In Amazonia, the interest in Mycology goes back to the 19th century, with Montagne and Leprieur who drew a first checklist of Fungi around Cayenne, French Guiana4 (and Berkeley5,6 for the Brazilian part). Since then, Amazon rainforests have been explored in their Brazilian part with important contributions by Hennings7 and Rick8 at the very beginning of the 20th century, and more recently by Singer9,10, Trieveiler-Pereira11, Sulzbacher12 or Ryvarden13. Great contributions have also been made by Henkel and collaborators14,15 in the Pakaraimas mountains in Guyana (www.tropicalfungi.org) or in Colombia, especially in the terra-firme and white-sand forests16,17,18. The last checklist for French Guiana (1996) listed 625 taxa19 gathered in a very limited number of coastal localities. Evidently, there is an urgent need to systematically collect and document fungi from undersampled areas to fill the knowledge gaps in a region where fungal diversity may be much higher than presently known20,21,22.
From 2011 onwards, we collected all fruiting-bodies following the same protocol in 126 plots representative of the main forest habitats of French Guiana (Fig. 1). We also gathered information on habitats, environment and first taxonomic indications. The resulting dataset provides an unprecedented collection of Basidiomycota for the region, making specimens available for taxonomists, with a molecular barcode for some of them, together with information on ecology and distribution.
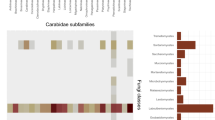
We found an extraordinary diversity across forest habitats. The dataset is not a checklist as only 27% of the specimens are determined at the species level and 96% at the genus level. However, the voucher specimens are deposited to herbaria, mainly the Fungarium of the Paris Natural History Museum, for further identification if needed. This tedious, on-going sampling increases the number of families and genera known for the territory as compared with previous collections4,19 (Table 1 and Fig. 2). The number of species reported from French Guiana increased from 62523 to 1168. The most abundant families found in French Guiana are also among the most abundant ones found in Amazonian Brazilian forests (http://splink.cria.org.br/, keywords search = Basidiomycota AND Acre, Amapa, Amazonas, Para, Roraima, 16793 records). Around one quarter of the genera we observed were also recorded in the Amazonian part of Colombia by Vasco-Palacios and Franco Molano16 (Fig. 2). They recorded 119 species belonging mainly to Tricholomataceae (Agaricales) and Coriolaceae (Polyporales). Thirteen genera of ectomycorrhizal fungi (all of which were also recorded from monodominant leguminous forests in Guyana14, e.g. Amanita, Cantharellus or Russula) were, although scarce, present in several sites, thereby confirming that ectomycorrhizal fungi can persist in hyperdiverse Neotropical forests1,22.
Successive contributions of sampling. Cladogram showing the contribution of: (from left to right) Montagne4, Courtecuisse19, Vasco-Palacios16, Species Link for Amazonia (splink.cria.org.br, 2019) and this dataset (2019) in gathering specimens. For convenience, only orders that have been more intensively sampled are displayed. We followed the classification proposed by Tedersoo et al.37.
Methods
Geographic coverage
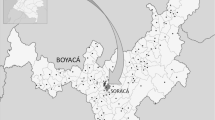
French Guiana (83,534 km2) is a French overseas region situated in South America at the eastern limit of the Guiana Shield, a mountainous tableland extending, from West to East, across Guyana, Suriname, French Guiana, as well as parts of Colombia, Venezuela and Brazil. Soils are ancient, heavily eroded and chemically poor. The country’s relief is fairly flat, rarely exceeding 200 m with three mountain chains reaching up to 830 m at Mount Itoupé24. The climate is characterized by a clear seasonal pattern: a wet season from December to July, which is normally interrupted in February or March by a short dry period, and a long dry season from August to November with monthly precipitation of less than 100 mm. Average annual precipitation is 2200 mm. Mean temperature is 25 °C with low seasonal changes25.
Study extent
From 2011, the authors collected sporocarps in French Guiana and inventoried a total of 126 1-ha plots (Fig. 1), 6 of which had previously been visited by R. Courtecuisse19. The Nouragues Ecological Research Station (4°05′N–52°41′W, www.nouragues.cnrs.fr) and the Experimental Station of Paracou (5°18′N–52°53′W, http://paracou.cirad.fr/) are research stations where permanent forest plots are monitored for their vegetation, climate and environmental data. Limonade, Itoupe, Mitaraka are part of the National Amazonian Park of French Guiana (PAG, www.pag.fr). Kaw, Laussat and Trinite are part of the Network of Natural Reserve of French Guiana on the coast (www.guyane-parcregional.fr). CSG is located within the area under the control of Guiana Space Center (www.cnes-csg.fr). Other sampling sites were chosen because they represented several typical French Guiana’s habitat types, as defined by Guitet et al.24: margin of inselbergs, white-sands forests, terra-firme forests, seasonally flooded forests. The plots are at altitudes ranging from 35 to 800 m.
The 126 plots were assigned to one of three topographies according to the classification of Ferry et al.26: plateau if the plot is situated on upper part of hill with vertical water drainage; slope if the plot is situated along a slope and exhibited a superficial lateral drainage, and seasonally flooded if the plot is situated in a bottomland regularly inundated during rainy season with a water table always observed above 60 cm depth and present at the surface soil for at least two consecutive months26. Two main types of soils were selected. First, clay-rich soils or terra-firme are sand-silt-clay mixture of soils very commonly found in French Guiana27,28. Second, white-sand soils are soils derived from podzols as well as quartzites and weathered granite on the margin of the inselbergs according to the definition given by Baraloto et al.28.
Sampling description
We developed an easily and reproducible field experimental procedure to collect and identify fruiting bodies. Each sampling site coordinates were recorded and associated with the World Geodetic System 1984 (WGS 1984) and UTM 21-22N for map projection. We took advantage of pre-existing 1-ha botanical plots to carry out inventories and proceeded as follows: we randomly positioned three sub-plots of 20 × 20 m in each main 1 ha-plot where two collectors exhaustively sampled all visible sporocarps, for a period of 1.5 h maximum per sub-plot. Hypogeous fungi were not targeted during these inventories. All visible sporocarps were photographed, numbered and dried using a field drier the same day and a ~0.5 cm2 tissue sample of each sporocarp was stored for DNA in CTAB (2% Cetyltrimethylammonium bromide).
Taxonomic identification
The dataset of 5219 specimens gathers 245 genera belonging to 75 families. Species names of the closest morphospecies were assigned by M. Roy in the field based on existing literature9,11,14,16,29. Then, more precise taxonomic identification of all fungi collected was done in collaboration with R. Courtecuisse, C. Decock, T. Henkel, P.-A. Moreau, M. Roy, S. Welti, G. Grühn, J. Fournier, C. Lechat in the field or later by examination of vouchered specimens by A. Verbeken, F. Wartchow and B. Buyck, and using existing literature29. Homogeneity and consistency of all taxonomic names were controlled afterward. All dry voucher specimens were deposited at one of the following herbaria: LIP herbarium (Lille, Université de Lille, Département de Botanique); PC herbarium (Mycological herbarium of the Paris Natural History Museum, Paris); MUCL, Catholic University of Louvain; HSC, Humboldt State University.
Barcoding
Among the collection, 771 specimens were barcoded as followed. DNA was extracted using the CTAB method30, the internal transcribed spacer (ITS1f-ITS1r primers from Taberlet et al.31) was amplified by PCR and sequenced using Illumina Miseq technology (2 × 250 bp) by Fasteris (Plan-les-Ouates, Switzerland) or at the Genotoul platform (www.genotoul.fr). We used tagged primers to distinguish sequences from each specimen. Raw data of the Illumina sequencing were analyzed with the OBITools package32 as well as scripts in R33. Briefly, we first conducted paired-end read assembly, read assignment to samples and read dereplication. Low-quality sequences, i.e. those shorter than expected (under 80 bp), containing ambiguous nucleotides, corresponding to singletons and displaying low score paired-end alignments were excluded from the analysis. Scores of pairwise alignments were calculated with Sumatra package (www.metabarcoding.org) which uses the same clustering algorithm as UCLUST and CD-HIT. This algorithm is mainly useful to detect the ‘erroneous’ sequences created during amplification and sequencing protocols, deriving from ‘true’ sequences. For each sample, sequences having pairwise alignments with a score below 97% of similarity were removed and considered as erroneous. Last, for each specimen, the most abundant sequence was kept as representative of the specimen. The last UNITE dataset (https://unite.ut.ee/) was used as reference for the taxonomic assignment of specimen target sequence. The molecular assignation was then compared to the morphological one to confirm the identification of the sequence. In case of discrepancy between the morphological and the molecular identification, the sequence was discarded. At the end, 642 sequences (140 to 256 bp length) were submitted to GenBank. The barcoding of remaining specimens is ongoing with the aim to sequence at least one specimen of each species or genus.
Data Records
The dataset contains a record for each sample. Each record contains a unique code identifying the specimen in the collection; a code attributed by the herbarium where it is deposited; a name corresponding to the most precise identification by one of the mycologists involved in this work; the name of the specialist who identified it; the complete description of the sampling plot (city, site, plot, geographical coordinates, altitude, habitat, topography, soil type, substratum and host, name of the collector; collection date), the barcode name (ITS1), the obtained sequence for this barcode and the GenBank accession number of the barcode.
The dataset is managed locally in a shared database and is accessible publicly in the GBIF repository (www.gbif.org) under the https://doi.org/10.15468/ymvlrp 34. Updates of the online dataset are planned when major changes will occur. All unique ITS1 barcodes (642) were submitted to GenBank under accession numbers MF03888735 to MK54705636.
Technical Validation
Homogeneity and consistency of all taxonomic names were controlled afterward thanks to MycoBank (http://www.mycobank.org) and Index Fungorum (http://www.indexfungorum.org).
The dataset described in this work was gathered thanks to a field experimental procedure to collect Basidiomycota fruiting bodies exhaustively across several typical French Guiana’s habitat types. We analyse the efficiency of this sampling method by building accumulation curves (Fig. 3). These curves show that we sampled the majority of Basidiomycota families present in French Guiana. But these curves also indicate that, despite our sampling effort, we probably missed some Basidiomycota genera. This underlies the crucial necessity to continue this collection.
Sampling accumulation curves. (A) At the Family level and (B) at the Genus level. (C) Represents rarefaction curves for each sampling site. In (A) the flattened curve shows that we sampled the majority of Basidiomycota families present in French Guiana. On the contrary, curve in (B) does not really flatten, indicating that, despite our sampling effort, we probably missed some Basidiomycota genera, and underlying the crucial necessity to continue this collection. (C) Shows an unbalanced sampling effort by sites, probably indicating differences in species richness across contrasting sites but also differences in sampling effort. Accumulation curves were performed using Coleman method.
References
Roy, M. et al. Diversity and Distribution of Ectomycorrhizal Fungi from Amazonian Lowland White-sand Forests in Brazil and French Guiana. Biotropica 48, 90–100, https://doi.org/10.1111/btp.12297 (2016).
Sulzbacher, M. A., Grebenc, T., Giachini, A. J., Baseia, I. G. & Nouhra, E. R. Hypogeous sequestrate fungi in South America – how well do we know them? Symbiosis 71, 9–17, https://doi.org/10.1007/s13199-016-0461-4 (2017).
Osmundson, T. W. et al. Filling Gaps in Biodiversity Knowledge for Macrofungi: Contributions and Assessment of an Herbarium Collection DNA Barcode Sequencing Project. PLoS One 8, e62419, https://doi.org/10.1371/journal.pone.0062419 (2013).
Montagne, C. J. F. Cryptogamia Guyanensis seu plantarum cellularium in Guyana Gallica: annis 1835-1849, a Cl. Leprieur collectarum enumeratio universalis. Vol. 3 (1855).
Berkeley, M. J. Decades of fungi. Decades LXI–LXII. Rio Negro fungi. Hooker’s Journal of Botany and Kew Garden Miscellany 8, 272–280 (1856).
Berkeley, M. J. & Cooke, M. C. The Fungi of Brazil, including those collected by J. W. H. Trail, Esq., M.A., in 1874. Botanical Journal of the Linnean Society 15, 363–398, https://doi.org/10.1111/j.1095-8339.1876.tb00248.x (2009).
Perkins, J. Paul Hennings. Botanical Gazette 47, 239–241, https://doi.org/10.1086/329853 (1909).
Kirk, P. M., Cannon, P. F., Minter, D. W. & Stalpers, J. A. Ainsworth and Bisby’s Dictionary of the Fungi (10th edition). Vol. 23 (Cab International, 2008).
Singer, R. The Agaricales in modern taxonomy. (Koeltz Scientific Books, 1986).
Singer, R., Araujo, I. & Ivory, M. H. The Ectotrophically Mycorrhizal Fungi of the Neotropical Lowlands, Especially Central Amazonia. (Lubrecht & Cramer Limited, 1983).
Trierveiler-Pereira, L. & Baseia, I. G. A checklist of the Brazilian gasteroid fungi (Basidiomycota). Mycotaxon 108, 441–444, https://doi.org/10.5248/108.441 (2009).
Sulzbacher, M. A., Grebenc, T., Jacques, R. J. S. & Antoniolli, Z. I. Ectomycorrhizal fungi from southern Brazil - a literature-based review, their origin and potential hosts. Mycosphere 1, 61–94, https://doi.org/10.5943/mycosphere/4/1/5 (2013).
Ryvarden, L. Neotropical polypores. Vol. 34–36 (Fungiflora, 2016).
Henkel, T. W. et al. Ectomycorrhizal fungal sporocarp diversity and discovery of new taxa in Dicymbe monodominant forests of the Guiana Shield. Biodiversity and Conservation 21, 2195–2220, https://doi.org/10.1007/s10531-011-0166-1 (2012).
Henkel, T. W. et al. Cantharellaceae of Guyana II: New species of Craterellus, new South American distribution records for Cantharellus guyanensis and Craterellus excelsus, and a key to the Neotropical taxa. Mycologia 106, 307–324, https://doi.org/10.3852/106.2.307 (2014).
Vasco-Palacios, A. M. & Franco Molano, A. E. Diversity of Colombian macrofungi (Ascomycota - Basidiomycota). Vol. 121 (2013).
Vasco-Palacios, A. M., Hernandez, J., Peñuela-Mora, M. C., Franco-Molano, A. E. & Boekhout, T. Ectomycorrhizal fungi diversity in a white sand forest in western Amazonia. Fungal Ecology 31, 9–18, https://doi.org/10.1016/j.funeco.2017.10.003 (2018).
López-Quintero, C. A., Straatsma, G., Franco-Molano, A. E. & Boekhout, T. Macrofungal diversity in Colombian Amazon forests varies with regions and regimes of disturbance. Biodiversity and Conservation 21, 2221–2243, https://doi.org/10.1007/s10531-012-0280-8 (2012).
Courtecuisse, R. et al. Check-List of Fungi from French Guiana. Mycotaxon 57, 1–86 (1996).
Geml, J. et al. Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Molecular Ecology 23, 2452–2472, https://doi.org/10.1111/mec.12765 (2014).
Truong, C. et al. How to know the fungi: combining field inventories and DNA-barcoding to document fungal diversity. New Phytologist 214, 913–919, https://doi.org/10.1111/nph.14509 (2017).
Roy, M. et al. The (re)discovery of ectomycorrhizal symbioses in Neotropical ecosystems sketched in Florianópolis. New Phytologist 214, 920–923, https://doi.org/10.1111/nph.14531 (2017).
Gargominy, O. et al. TAXREFv12, référentiel taxonomique pour la France: méthodologie, mise en œuvre et diffusion - Rapport Patrinat 2018–117. 156, https://doi.org/10.15468/vqueam (2018).
Guitet, S. et al. Landform and landscape mapping, French Guiana (South America). Journal of Maps 9, 325–335, https://doi.org/10.1080/17445647.2013.785371 (2013).
Gourlet-Fleury, S., Guehl, J.-M. & Laroussinie, O. Ecology and management of a neotropical rainforest. Lessons drawn from Paracou, a long-term experimental research site in French Guiana (2004).
Ferry, B., Morneau, F., Bontemps, J.-D., Blanc, L. & Freycon, V. Higher treefall rates on slopes and waterlogged soils result in lower stand biomass and productivity in a tropical rain forest. Journal of Ecology 98, 106–116, https://doi.org/10.1111/j.1365-2745.2009.01604.x (2010).
ter Steege, H. et al. An analysis of the floristic composition and diversity of Amazonian forests including those of the Guiana Shield. Journal of Tropical Ecology 16, 801–828, https://doi.org/10.1017/S0266467400001735 (2000).
Baraloto, C. et al. Disentangling stand and environmental correlates of aboveground biomass in Amazonian forests. Global Change Biology 17, 2677–2688, https://doi.org/10.1111/j.1365-2486.2011.02432.x (2011).
Pegler, D. N. Agaric Flora of the Lesser Antilles. H.S.M.O., London, Kew Bulletin Additional Series IX. Royal Botanic Gardens. 668p. (1983).
Doyle, J. J. Isolation of plant DNA from fresh tissue. Focus 12, 13–15 (1990).
Taberlet, P., Bonin, A., Zinger, L. & Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring. (Oxford University Press, 2018).
Boyer, F. et al. Obitools: a unix-inspired software package for DNA metabarcoding. Molecular Ecology Resources 16, 176–182, https://doi.org/10.1111/1755-0998.12428 (2016).
R Development Core Team. R: A Language and Environment for Statistical Computing (2013).
Schimann, H., Roy, M. & Jaouen, G. Fungi of French Guiana. The Global Biodiversity Information Facility. https://doi.org/10.15468/ymvlrp (2019).
Schimann, H. et al. GenBank, https://identifiers.org/ncbi/insdc:MF038887 (2017).
Jaouen, G. et al. GenBank, https://identifiers.org/ncbi/insdc:MK547056 (2019).
Tedersoo, L. et al. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Diversity 90, 135–159, https://doi.org/10.1007/s13225-018-0401-0 (2018).
Acknowledgements
Authors are grateful to Régis Courtecuisse, Stéphane Welti and Pierre-Arthur Moreau (University of Lille, France), Cony Decock (Catholic University of Louvain, Belgium), Christian Lechat (www.ascofrance.fr), Jacques Fournier, Gerald Grühn (ONF, Mende, France), Terry Henkel (Humbolt State University, California, USA), Bart Buyck (Department of Systematic and Evolution, Museum National d’Histoire Naturelle, Paris), Annemieke Verbeken (Department of Biology, Gent University, Belgium), Felipe Wartchow (Department of Systematics and Ecology, Universidade Federal da Paraiba, Joao Pessoa, Brazil) for identification of specimens. Authors thank all people who have contributed to complete this database by adding specimens found opportunistically: Amaia Pelozuelo, Hadrien Lalagüe, Axel Touchard, Bertrand Goguillon, Christopher Baraloto, Daniel Sabatier, Emeric Auffret, Eddy Poirier, Thomas Denis, Gunther Fleck, Jérôme Barbut, Jocelyn Cazal, Jennifer Devillechabrolle, Jan Hacker, Marie Fleury, Olivier Bruneaux, Olivier Gargomini, Olivier Tostaing, Pascal Petronelli, Sébastien Brosse, Serge Fernandez, Sophie Gonzalez, Thibaud Decaens, Thierry Munnier, Xavier Desmier, Yannick Estevez, Yves Gaert. Authors thank the Centre Spatial Guyanais, the Reserve Naturelle de l’Amana (http://www.reserveamana.com/), the Reserve Naturelle de la Trinité (http://www.reserve-trinite.fr/), the Parc Amazonien de Guyane (http://www.parc-amazonien-guyane.fr/), the Réserve Naturelle Régionale Trésor (http://www.reserve-tresor.fr/), the Reserve Naturelle des Nouragues (http://www.nouragues.fr/) for autorisations and access to the sites. Part of the material was collected during the “Our Planet Reviewed” Guyane-2015 expedition in the Mitaraka range, in the core area of the French Guiana Amazonian Park, organized by the MNHN and Pro-Natura international MNHN/PNI Guyane 2015 (APA-973-1). The expedition was funded by the European Regional Development Fund (ERDF), the Conseil régional de Guyane, the Conseil général de Guyane, the Direction de l’Environnement, de l’Aménagement et du Logement and by the Ministère de l'Éducation nationale, de l’Enseignement supérieur et de la Recherche. It was realized in collaboration with the Parc amazonien de Guyane. Authors thank the ‘Investissement d’avenir’ grant from the Agence Nationale de la Recherche (CEBA, ref. ANR-10- LABX-25-01). Data have been collected from access to genetic resources in French Guiana, that has come through a declarative process with non-commercial uses at the competent administrative authority, in accordance with article L.421-7 of the environmental code (ABSCH-IRCC-FR-246832-1 and TREL1820249A/51).
Author information
Authors and Affiliations
Contributions
C.B. coordinated the project. H.S., C.B. and M.R. developed the sampling design. C.B., H.S., M.R., E.L., S.M., A.S., C.D. and T.H. collected the specimens in the field. G.J. curated and managed the data, and conceived the database. E.L. and S.M. extracted DNA and managed the barcoding of the specimens. A.S. conceived and developed the pipeline of analysis for molecular data. H.S. and G.J. wrote the first draft of the manuscript and all authors contributed to revisions.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Jaouen, G., Sagne, A., Buyck, B. et al. Fungi of French Guiana gathered in a taxonomic, environmental and molecular dataset. Sci Data 6, 206 (2019). https://doi.org/10.1038/s41597-019-0218-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-019-0218-z
This article is cited by
-
Citizen science helps in the study of fungal diversity in New Jersey
Scientific Data (2023)
-
Allophlebia, a new genus to accomodate Phlebia ludoviciana (Agaricomycetes, Polyporales)
Mycological Progress (2022)