Abstract
Machine learning has the potential to identify novel biological factors underlying successful antibody responses to influenza vaccines. The first attempts have revealed a high level of complexity in establishing influenza immunity, and many different cellular and molecular components are involved. Of note is that the previously identified correlates of protection fail to account for the majority of individual responses across different age groups and influenza seasons. Challenges remain from the small sample sizes in most studies and from often limited data sets, such as transcriptomic data. Here we report the creation of a unified database, FluPRINT, to enable large-scale studies exploring the cellular and molecular underpinnings of successful antibody responses to influenza vaccines. Over 3,000 parameters were considered, including serological responses to influenza strains, serum cytokines, cell phenotypes, and cytokine stimulations. FluPRINT, facilitates the application of machine learning algorithms for data mining. The data are publicly available and represent a resource to uncover new markers and mechanisms that are important for influenza vaccine immunogenicity.
Measurement(s) | immune response trait |
Technology Type(s) | digital curation |
Factor Type(s) | gender • race • visit_age • bmi • flu_vaccination_history • statin_use • influenza_infection_history • influenza_hospitalization • cmv_status • ebv_status |
Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.9902447
Similar content being viewed by others
Background & Summary
Influenza virus has a devastating societal impact, causing up to 650,000 deaths every year worldwide1. Vaccination with the seasonal mixture of strains is only partially effective, even among otherwise-healthy people, leading to serious pandemics. Vaccine efficacy is defined as the ability of a new seasonal influenza vaccine to prevent influenza-like illness compared to the placebo group, according to the US Food and Drug Administration (FDA) in their guideline for vaccine licensure2. Young children and elderly, due to high susceptibility to influenza infection3, are encouraged to be vaccinated annually making a placebo-controlled clinical efficacy study in this population an extremely costly and arduous undertaking. The alternative approach to correlate vaccine-mediated protection in these populations is based on immunogenicity endpoints, recommended by FDA. The appropriate immunogenicity endpoint is the influenza-specific antibody titer measured by a hemagglutination inhibition (HAI) assay to each viral strain included in the vaccine. Vaccine protection is then assessed based on seroconversion (4-fold increase in the HAI antibody titers after vaccination) and seroprotection (geometric mean HAI titer ≥40 after vaccination). The HAI titer ≥40 after vaccination is associated with a 50% reduction in risk of influenza infection or disease4.
Lack of pre-existing influenza immunity, especially T cells, has been identified as one of the major predispositions for failure to generate antibody response to vaccination5,6,7. However, exact phenotypes of CD4+ and CD8+ T cells, which are important for protective influenza immunity in general and to vaccination with live attenuated influenza vaccine (LAIV) in specific, remain elusive. The application of computational biology and machine learning to clinical datasets holds great promise for identifying immune cell populations and genes that mediate HAI antibody responses to influenza vaccines as a correlate of vaccine protection8,9,10,11,12,13,14,15. Unfortunately the correlates of protection identified are not consistent between cohorts and study years8,9,11,12. Some of the identified challenges leading to such discrepancy are small sample sizes and analysis of only one aspect of the biology, such as molecular correlates of protection by using transcriptome data16. Additionally, comparison of the results of different predictive models is hampered by the lack of a consensus regarding what defines the outcome of vaccination, i.e. high vs. low responders. For these reasons, it is necessary to generate a unified dataset that includes multiple measurements across age, gender and racially diverse populations, including different vaccine types. Specifically, it is of the utmost importance to include single-cell analysis at the protein level, such as mass cytometry combined with multiple high-dimensional biological measurements, since these have power to reveal heterogeneity of the immune system17,18,19,20,21.
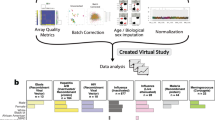
To accomplish that goal, we created FluPRINT, a dataset consisting of 13 data types in standardized tables on blood and serum samples taken from 740 individuals undergoing influenza vaccination with inactivated (IIV) or live attenuated seasonal influenza vaccines (LAIV) (Fig. 1). The FluPRINT dataset contains information on more than 3,000 parameters measured using mass cytometry (CyTOF), flow cytometry, phosphorylation-specific cytometry (phospho-flow), multiplex cytokine assays (multiplex ELISA), clinical lab tests (hormones and complete blood count), serological profiling (HAI assay) and virological tests. In the dataset, vaccine protection is measured using HAI assay, and following FDA guidelines individuals are marked as high or low responders depending on the HAI titers after vaccination. FluPRINT includes fully integrated and normalized immunology measurements from eight clinical studies conducted between 2007 to 2015 and assayed at the Human Immune Monitoring Center (HIMC) of Stanford University. Among those, one contains data from 135 donors enrolled in the 8-year long ongoing longitudinal study following immune responses to seasonal inactivated influenza vaccines. This is particularly interesting set of data that can deepen our understanding how repeated vaccination effects vaccine immunogenicity. The MySQL database containing this immense dataset is publicly available online (www.fluprint.com). The dataset represents a unique source in terms of value and scale, which will broaden our understanding of immunogenicity of the current influenza vaccines.
Overview of the FluPRINT dataset. The FluPRINT dataset consists of the 740 individuals from 8 clinical studies (SLVP015, SLVP017, SLVP018, SLVP021, SLVO024, SLVP028, SLVP029 and SLVP030) and 8 influenza seasons (from 2017 to 2015). (a) Pie chart shows distribution of donors across clinical studies. The dataset contains harmonized data from different assays, including mass and flow cytometry, phosphorylated cytometry (Phospho-flow), multiplex ELISA (Luminex assay), clinical lab tests, such as complete blood test, analysis of hormones and virological assays (CMV and EBV antibody titers) and serological profiling with hemagglutination inhibition assay, which was used to define high and low responders. (b) Distribution of assays across years available for each clinical study.
Methods
Clinical studies
All studies were approved by the Stanford Institutional Review Board and performed in accordance with guidelines on human cell research. All vaccines used were licensed in the US for use in the populations studied. Peripheral blood samples were obtained at the Clinical and Translational Research Unit at Stanford University after written informed consent/assent was obtained from participants. Samples were processed and cryopreserved by the Stanford HIMC BioBank according to the standard operating protocols available online at the HIMC website (https://iti.stanford.edu/himc/protocols.html).
Data collection
Data involving individuals enrolled in influenza vaccine studies at the Stanford-LPCH Vaccine Program was accessed from the Stanford Data Miner (SDM) which holds data processed by HIMC from 2007 up to date22. The FluPRINT cohort was assembled by filtering the SDM for assays available in studies involving influenza vaccination. This resulted in a dataset containing data from 740 healthy donors enrolled in influenza vaccine studies conducted by the Stanford-LPCH Vaccine Program from 2007 to 2015 in the following studies: SLVP015, SLVP017, SLVP018, SLVP021, SLVP024, SLVP028, SLVP029 and SLVP030. Online-only Table 1 provides a summary of all studies including information about clinical trial identification numbers on www.clinicaltrials.gov, clinical protocols, ImmPort accession numbers to access raw data and quality reports, and finally references to published works where data was used. ImmPort is a web portal that contains data from NIAID-funded immunology studies and clinical trials (https://immport.niaid.nih.gov/)23. All data contained in the FluPRINT dataset are made freely available through the Shared Data Portal on ImmPort repository. In all studies, except for study SLVP015, vaccine was administrated only once. The study SLVP015 was longitudinal study where 135 participants received vaccine in consecutive years from 2007–2015. In all studies, generally healthy participants were included, and in some studies (SLVP017 for the 2010, 2011 and 2013, SLVP021 and SLVP029) those that were vaccinated in the prior influenza season were excluded. A total of 121 CSV files containing processed data from various assays and studies were downloaded from SDM. The link to the 121 CSV files is provided on Zenodo24. Table 1 provides a summary of the demographic characteristics of the FluPRINT study population. The population spans a wide age range, from a 1-year-old to a 90-year-old, with a median age of 27 years. Among 740 individuals with available experimental data, 446 were females and 294 males. The majority (491) of the individuals were Caucasian (European American ancestry). The complete demographic information is available on the Zenodo25. Individuals were stratified into high and low responders, depending on their HAI antibody titers measured before and after vaccination, as described below. Figure 2 shows demographic information for the FluPRINT study population, including gender, ethnicity, cytomegalovirus (CMV) status, and age stratified by the outcome to vaccination. Out of 363 individuals with measured HAI responses, 111 were identified as high responders and 252 as low responders. Overall, no major differences in the gender, ethnicity distribution, or CMV status (Fig. 2a) or age (Fig. 2b) were observed between high and low responders.
Demographic characteristics for the FluPRINT study population stratified by the vaccination outcome. Distribution of individuals in the categories of high (red, n = 111) and low (grey, n = 252) responders regarding the (a) gender, ethnicity and CMV status (b) age distribution between high and low responders. Age is indicated in years.
Assays and data processing
All data used were analysed and processed at the HIMC26. The distribution of assays performed across clinical studies and years is illustrated in Fig. 1b. Overall, SLVP015 was the longest study, running from 2007 to 2014, spanning 135 unique individuals, while the majority of samples (249) came from the SLVP018 study (Fig. 1). Raw data, including report files, standards, controls, antibodies used are available at ImmPort (https://immport.niaid.nih.gov/) under identification numbers for each study provided in the Online-only Table 1. Online-only Table 2 provides information about all assays performed, protocols, validations used and references to the published manuscripts using the data. Protocols for all assays are available online at the HIMC website (https://iti.stanford.edu/himc/protocols.html).
Multiplex cytokine assay
Multiplex ELISA using Luminex was performed using either polystyrene bead (for 51/52-plex) or magnetic bead kits (62/63-plex) (eBioscience/Affymetrix). The processed Luminex data available in the FluPRINT is normalized at the plate level to mitigate batch and plate effects26. The two median fluorescence intensity (MFI) values for each sample for each analyte were averaged, and then log-base 2 transformed. Z-scores ((value–mean)/standard deviation) were computed, with means and standard deviations computed for each analyte for each plate. Thus, units of measurement were Zlog2 for serum Luminex. Some of the Luminex data was used in previous publications9,10,22,27,28. In 2009 and 2010, for SLVP015 and SLVP018 studies, serum analytes were analysed using MSD 4- and 9-plex kits (V-PLEX Human Proinflammatory Panel II, Mesoscale, Cat No. K15053D and Human ProInflammatory 9-Plex Ultra-Sensitive Kit, Mesoscale, Cat No. K15007C) as according to the manufacturer’s protocol. The assay named ‘Other Luminex’ was performed only for study SLVP015 in 2007 using the Human 42-Plex Polystyrene Kit (EMD Millipore, H42; MPXHCYTO060KPMX42) and data was processed in the same way as for the Luminex assays described above (measurement units reported were Zlog2)28.
Hemagglutination inhibition assay
Serum antibody titers before vaccination and day 28 after vaccination were measured using the standard HAI assay29 using strains of influenza contained in the vaccines9,10,27. Geometric mean titers (GMT) were calculated for all strains of the virus contained in the vaccine, while fold change is calculated as: GMT for all vaccine strains on day 28/GMT for all vaccine strains on day 0. High responders were determined as individuals that seroconverted (4-fold or greater rise in HAI titer) and were seroprotected (GMT HAI ≥ 40).
Virological assays
CMV and Epstein-Barr virus (EBV) analysis was performed using CMV IgG ELISA (Calbiotech, Cat No. CM027G) and EBV-VCA IgG ELISA (Calbiotech, Cat No. EVO10G), following manufacturer’s protocols10,27,30.
Immunophenotyping
Immunophenotyping was performed either with flow cytometry (Lyoplate)27,30 or mass cytometry (CyTOF)30,31,32. Data was analysed using FlowJo software using the standard templates. Gates were adjusted on a donor-specific basis, if necessary, to control for any differences in background or positive staining intensity. The statistics was exported for each gated population to a spreadsheet. The percentage of each cell type is determined and reported as a percent of the parent cell type.
Phosphorylation-specific cytometry
Phospho-flow assays were performed either using flow cytometry on PBMC (for studies SLVP015, SLVP018 and SLVP021 from 2007 to 2012)9,10,27,28,30 or mass cytometry on whole blood (for studies SLVP015, SLVP018 and SLVP021 in 2013)33,34. The percentage of each cell type is determined and reported as a percent of the parent cell type. Median values are reported to quantitate the level of phosphorylation of each protein in response to stimulation. For phospho-flow data acquired on flow cytometer a fold change value was computed as the stimulated readout divided by the unstimulated readout (e.g. 90th percentile of MFI of CD4+ pSTAT5 IFNα stimulated/90th percentile of CD4+ pSTAT5 unstimulated cells), while for data acquired using mass cytometry a fold change was calculated by subtracting the arcsinh (intensity) between stimulated and unstimulated (arsinh stim – arcsing unstim).
Automated importer and data harmonization
After collecting the data, a custom PHP script was generated to parse each of the 121 CSV files and to import data into the MySQL database. The source code for the script is available online at https://github.com/LogIN-/fluprint. The script optimizes the data harmonization process essential for combining data from different studies. Control and nonsense data were not imported, such as “CXCR3-FMO CD8+ T cells”, “nonNK-nonB-nonT-nonmonocyte-nonbasophils”, “viable”, etc. To standardize data, the original CSV entries were cleaned into the MySQL database readable format (e.g. quotes and parenthesis replaced with underscores, “+” with text “positive”, etc.). Additionally, classifications for ethnicity (Table 2), vaccine names (Table 3) and vaccination history (Table 4) were resolved into standard forms, while assays were numerated (Table 5). For example, “Fluzone single-dose syringe” and “Fluzone single-dose syringe 2009–2010” were mapped to “Fluzone” and given number 4 (Table 3). In all studies, vaccines were given intramuscularly for IIV and intranasally for LAIV, except for one study where a distinct licensed formulation of IIV was given intradermally and this was labelled as Fluzone Intradermal and given number 2. During data merging, we replaced text strings with binary values. For example, for the variable of gender, female and male were replaced with zero and one. To be able to distinguish between visits in consecutive years, a unique visit identification was calculated. For the original internal visit data, each visit in one year was labelled as V1 for day zero and V2 for day seven. However, if the same individual came in the consecutive year, day zero visit would again be labelled V1, and day seven as visit V2, causing repetition of values. To avoid such repetitions in the database, we generated a unique visit ID. Therefore, for the above example, first visit in the first year would be labelled V1 for day zero and V2 for day seven, but for the next year visits would be labelled as V3 for day zero and V4 for day seven. To distinguish between Luminex assays, the prefix L50 was given to each analyte analysed with the 51/52-plex Luminex kit. Finally, we imputed new values and calculated the vaccine outcome parameter using HAI antibody titers. High responders were determined as individuals that have HAI antibody titer for all vaccine strains ≥40 after vaccination and GMT HAI fold change ≥4, following FDA guidelines for evaluation of vaccine efficacy2. Vaccine outcome was expressed as a binary value: high responders were given value of one and low responders the value zero.
Generating tables
To build FluPRINT database, we generated four tables, as shown in Fig. 3. Table 6 depicts characteristics of the FluPRINT database. In the table donor, each row represents an individual given a unique encrypted identification number (study donor ID). Other fields provide information about the clinical study in which an individual was enrolled (study ID and study internal ID), gender and race. The second table, named donor_visits describes information about the donor’s age, CMV and EBV status, Body Mass Index (BMI) and vaccine received on each clinical visit. Each clinical visit was given a unique identification (visit ID) in addition to the internal visit ID (provided by the clinical study) to distinguish between visits in consecutive years. For each visit, we calculated vaccine response by measuring HAI antibody response. Information about vaccine outcome is available as geometric mean titers (geo_mean), difference in the geometric mean titers before and after vaccination (delta_geo_mean), and difference for each vaccine strain (delta_single). In the last field, each individual is classified as high and low responder (vaccine_resp). On each visit, samples were analysed and information about which assays were performed (assay field) and value of the measured analytes (units and data) are stored in the experimental_data table. Finally, the medical_history table describes information connected with each clinical visit about usage of statins (statin_use) and whether an influenza vaccine was received in the past (influenza vaccine history), if yes, how many times (total_vaccines_received, self reported). Also, we provide information which type of influenza vaccine was received in the previous years (1 to 5 years prior enrolment in the clinical study). Lastly, information about influenza infection (report of MD diagnosis) history and influenza-related hospitalization (participant report) is provided.
The FluPRINT database model. The diagram shows a schema of the FluPRINT database. Core tables, donors (red), donor_visits (yellow), experimental_data (blue) and medical_history (green) are interconnected. Tables experimental_data and medical_history are connected to the core table donor_visits. The data fields for each table are listed, including the name and the type of the data. CHAR and VARCHAR, string data as characters; INT, numeric data as integers; FLOAT, approximate numeric data values; DECIMAL, exact numeric data values; DATETIME, temporal data values; TINYINT, numeric data as integers (range 0–255); BOOLEAN, numeric data with Boolean values (zero/one). Maximal number of characters allowed in the data fields is denoted as number in parenthesis.
Data Records
The FluPRINT dataset described herein is available online for use by the research community and can be downloaded directly from a research data repository Zenodo35. Additionally, the dataset can be imported in the MySQL database for further manipulation and data extraction. The instructions how to import FluPRINT into the database are available at github (https://github.com/LogIN-/fluprint). The summary of the dataset, including the number of observations, fields and description for each table is provided in Table 6.
Technical Validation
The objective of the current study was to ensure that the FluPRINT dataset accurately reflects processed data available in SDM. Technical data validation was carried in previous published studies referred in the Online-only Table 2. Data was downloaded from the original source, and here we focused on ensuring that data records were accurately harmonized, merged and mapped in the unifying FluPRINT database.
The FluPRINT dataset was validated on two levels: (1) upon insertion and (2) after the data was inserted into the database. To validate data on insertion, we created loggers to monitor import of the CSV files into the database. This ensured easier and more effective troubleshooting of potential problems and contributed to the monitoring of the import process. Two different sets were used: (1) informative and (2) error loggers. Informative loggers provided information about which processing step has started or finished and how many samples have been processed in that particular step. This allowed us to monitor that correct number of samples was processed. Error loggers provided exact identification and name of the data which could not be imported into the database, usually caused by missing or incorrect user input, such as “… assay is missing. Skipping … ‘$row’”. This facilitated the process to identify erroneous data, which were then manually reviewed, corrected, and updated.
Once the database was built, a manual review of data was performed to ensure accuracy and integrity of the dataset. Several random individuals were chosen and the accuracy of data was evaluated by comparison with the raw data. Additionally, we evaluated total number of all donors, assays performed, clinical studies and years with the raw data available at the SDM.
Usage Notes
Recent advances in the computational biology and the development of novel machine learning algorithms, especially deep learning, make it possible to extract knowledge and identify patterns in an unbiased manner from large clinical datasets. Application of machine learning algorithms to clinical datasets can reveal biomarkers for different diseases, therapies36, including vaccinations8,9,12. The data from the FluPRINT study can be used to identify cellular and molecular baseline biomarkers that govern successful antibody response to influenza vaccines (IIV and LAIV) across different influenza seasons and a broad age population. The HAI antibody response to influenza vaccines is considered as an alternative way to compare immunogenicity of the vaccines in susceptible groups where placebo-controlled clinical efficacy study cannot be performed. Since FluPRINT dataset is provided as a database, this facilitates further analysis. Queries can be easily performed to obtain a single CSV file. For example, researchers interested in understanding which immune cells and chemokines can differentiate between high and low responders that received inactivated influenza vaccine could search the FluPRINT database. In the database, they can find all donors for which flow cytometry or mass cytometry were performed together with Luminex assays, for which donors the HAI response was measured, and all the donors who received inactivated influenza vaccine. The resulting CSV file can then easily be used for downstream analysis.
Major advantages of this dataset are the mapping of the vaccine outcome, classifying individuals as high or low responders, standardization of the data from different clinical studies, and from different assays. This data harmonization process allows for direct comparison of immune cell frequency, phenotype, and functionality and quantity of chemokines and cytokines shared between individuals before or after influenza vaccinations. By releasing the FluPRINT database and the source code, we provide users with the ability to continue building upon this resource and to update the database with their data and other databases.
Code availability
The source code for the PHP script and database schema are available from a public github repository (https://github.com/LogIN-/fluprint). Raw data files used to generate dataset are provided as single compressed file on Zenodo24. Full study population with demographic characteristics is provided as single CSV file25. Additionally, entire FluPRINT database export is available as CSV table and SQL file35. Database is also accessible at the project website https://fluprint.com.
References
Iuliano, A. D. et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300 (2018).
Food & Drug Administration. Clinical Data Needed to Support the Licensure of Seasonal Inactivated Influenza Vaccines, https://www.fda.gov/regulatory-information/search-fda-guidance-documents/clinical-data-needed-support-licensure-seasonal-inactivated-influenza-vaccines (2007).
Zhou, H. et al. Hospitalizations associated with influenza and respiratory syncytial virus in the United States, 1993–2008. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 54, 1427–1436 (2012).
de Jong, J. C. et al. Haemagglutination-inhibiting antibody to influenza virus. Developments in biologicals 115, 63–73 (2003).
Sridhar, S. et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 19, 1305–1312 (2013).
Bentebibel, S. E. et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Science translational medicine 5, 176ra132 (2013).
Trieu, M. C. et al. Long-term Maintenance of the Influenza-Specific Cross-Reactive Memory CD4+ T-Cell Responses Following Repeated Annual Influenza Vaccination. J Infect Dis 215, 740–749 (2017).
Furman, D. et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proceedings of the National Academy of Sciences of the United States of America 111, 869–874 (2014).
Furman, D. et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol 9, 659 (2013).
Furman, D. et al. Cytomegalovirus infection enhances the immune response to influenza. Science translational medicine 7, 281ra243 (2015).
Nakaya, H. I. et al. Systems Analysis of Immunity to Influenza Vaccination across Multiple Years and in Diverse Populations Reveals Shared Molecular Signatures. Immunity 43, 1186–1198 (2015).
Nakaya, H. I. et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 12, 786–795 (2011).
Sobolev, O. et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat Immunol 17, 204–213 (2016).
Nakaya, H. I. et al. Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proceedings of the National Academy of Sciences of the United States of America 113, 1853–1858 (2016).
Tsang, J. S. et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 157, 499–513 (2014).
Hagan, T., Nakaya, H. I., Subramaniam, S. & Pulendran, B. Systems vaccinology: Enabling rational vaccine design with systems biological approaches. Vaccine 33, 5294–5301 (2015).
Chattopadhyay, P. K., Gierahn, T. M., Roederer, M. & Love, J. C. Single-cell technologies for monitoring immune systems. Nat Immunol 15, 128–135 (2014).
Galli, E. et al. The end of omics? High dimensional single cell analysis in precision medicine. Eur J Immunol. https://doi.org/10.1002/eji.201847758 (2019).
Bendall, S. C., Nolan, G. P., Roederer, M. & Chattopadhyay, P. K. A deep profiler’s guide to cytometry. Trends Immunol 33, 323–332 (2012).
Simoni, Y., Chng, M. H. Y., Li, S., Fehlings, M. & Newell, E. W. Mass cytometry: a powerful tool for dissecting the immune landscape. Curr Opin Immunol 51, 187–196 (2018).
Newell, E. W., Sigal, N., Bendall, S. C., Nolan, G. P. & Davis, M. M. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36, 142–152 (2012).
Siebert, J. C., Munsil, W., Rosenberg-Hasson, Y., Davis, M. M. & Maecker, H. T. The Stanford Data Miner: a novel approach for integrating and exploring heterogeneous immunological data. J Transl Med 10, 62 (2012).
Bhattacharya, S. et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data 5, 180015 (2018).
Tomic, A. & Tomic, I. Raw data for the generation of the FluPRINT dataset. Zenodo. https://doi.org/10.5281/zenodo.3213899 (2019).
Tomic, A. & Tomic, I. Characteristics of the individuals included in the FluPRINT dataset. Zenodo. https://doi.org/10.5281/zenodo.3220934 (2019).
Whiting, C. C. et al. Large-Scale and Comprehensive Immune Profiling and Functional Analysis of Normal Human Aging. Plos One 10, e0133627 (2015).
Brodin, P. et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 160, 37–47 (2015).
Shen-Orr, S. S. et al. Defective Signaling in the JAK-STAT Pathway Tracks with Chronic Inflammation and Cardiovascular Risk in Aging Humans. Cell Syst 3, 374–384 e374 (2016).
Hirst, G. K. The Quantitative Determination of Influenza Virus and Antibodies by Means of Red Cell Agglutination. J Exp Med 75, 49–64 (1942).
Alpert, A. et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat Med 25, 487–495 (2019).
Leipold, M. D. & Maecker, H. T. Phenotyping of Live Human PBMC using CyTOF Mass Cytometry. Bio Protoc 5 (2), e1382. https://doi.org/10.21769/BioProtoc.1382 (2015).
Leipold, M. D. & Maecker, H. T. Mass cytometry: protocol for daily tuning and running cell samples on a CyTOF mass cytometer. J Vis Exp (69), 4398. https://doi.org/10.3791/4398 (2012).
Fernandez, R. & Maecker, H. Cytokine-stimulated Phosphoflow of PBMC Using CyTOF Mass Cytometry. Bio Protoc 5. https://doi.org/10.21769/BioProtoc.1496 (2015).
Fernandez, R. & Maecker, H. Cytokine-Stimulated Phosphoflow of Whole Blood Using CyTOF Mass Cytometry. Bio Protoc 5(11), e1495 (2015).
Tomic, A. & Tomic, I. The FluPRINT database. Zenodo. https://doi.org/10.5281/zenodo.3222451 (2019).
Krieg, C. et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med 24, 144–153 (2018).
Price, J. V. et al. Characterization of Influenza Vaccine Immunogenicity Using Influenza Antigen Microarrays. Plos One 8(5), e64555 (2013).
Wang, C. et al. Effects of Aging, Cytomegalovirus Infection, and EBV Infection on Human B Cell Repertoires. J Immunol 192, 603–611 (2014).
Jackson, K. J. L. et al. Human Responses to Influenza Vaccination Show Seroconversion Signatures and Convergent Antibody Rearrangements. Cell Host Microbe 16, 105–114 (2014).
Looney, T. J. et al. Human B-cell isotype switching origins of IgE. J Allergy Clin Immun 137, 579 (2016).
Haddon, D. J. et al. Mapping epitopes of U1-70K autoantibodies at single-amino acid resolution. Autoimmunity 48, 513–523 (2015).
Furman, D. et al. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat Med 23, 174–184 (2017).
de Bourcy, C. F. et al. Phylogenetic analysis of the human antibody repertoire reveals quantitative signatures of immune senescence and aging. Proceedings of the National Academy of Sciences of the United States of America 114, 1105–1110 (2017).
Kay, A. W. et al. Pregnancy Does Not Attenuate the Antibody or Plasmablast Response to Inactivated Influenza Vaccine. J Infect Dis 212, 861–870 (2015).
Roskin, K. M. et al. IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Science translational medicine 7, 302ra135 (2015).
Fang, F. Q. et al. Expression of CD39 on Activated T Cells Impairs their Survival in Older Individuals. Cell Rep 14, 1218–1231 (2016).
He, X. S. et al. Plasmablast-derived polyclonal antibody response after influenza vaccination. J Immunol Methods 365, 67–75 (2011).
Sasaki, S. et al. Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. J Clin Invest 121, 3109–3119 (2011).
He, X. S. et al. Heterovariant Cross-Reactive B-Cell Responses Induced by the 2009 Pandemic Influenza Virus A Subtype H1N1 Vaccine. J Infect Dis 207, 288–296 (2013).
Jiang, N. et al. Lineage Structure of the Human Antibody Repertoire in Response to Influenza Vaccination. Science translational medicine 5(171), 171ra19 (2013).
Horowitz, A. et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Science translational medicine 5, 208ra145 (2013).
Cheung, P. et al. Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell 173, 1385 (2018).
Kay, A. W. et al. Enhanced natural killer-cell and T-cell responses to influenza A virus during pregnancy. Proceedings of the National Academy of Sciences of the United States of America 111, 14506–14511 (2014).
Rubelt, F. et al. Individual heritable differences result in unique cell lymphocyte receptor repertoires of naive and antigen-experienced cells. Nat Commun 7, 1112 (2016).
Horns, F. et al. Lineage tracing of human B cells reveals the in vivo landscape of human antibody class switching. Elife 5, e16578 (2016).
de Bourcy, C. F. A., Dekker, C. L., Davis, M. M., Nicolls, M. R. & Quake, S. R. Dynamics of the human antibody repertoire after B cell depletion in systemic sclerosis. Sci Immunol 2(15), eaan8289 (2017).
O’Gorman, W. E. et al. The Split Virus Influenza Vaccine rapidly activates immune cells through Fc gamma receptors. Vaccine 32, 5989–5997 (2014).
Vollmers, C., Sit, R. V., Weinstein, J. A., Dekker, C. L. & Quake, S. R. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proceedings of the National Academy of Sciences of the United States of America 110, 13463–13468 (2013).
He, X. S. et al. Distinct Patterns of B-Cell Activation and Priming by Natural Influenza Virus Infection Versus Inactivated Influenza Vaccination. J Infect Dis 211, 1051–1059 (2015).
Le Gars, M. et al. Increased Proinflammatory Responses of Monocytes and Plasmacytoid Dendritic Cells to Influenza A Virus Infection During Pregnancy. J Infect Dis 214, 1666–1671 (2016).
Acknowledgements
We are grateful to all individuals that participated in the research studies. We appreciate helpful discussions from all members of the Davis and Y. Chien labs. We also thank all staff members from the HIMC (Yael Rosenberg-Hasson, Michael D. Leipold and Weiqi Wang) for data analysis, management and helpful discussions, HIMC Biobank (Rohit Gupta and Janine Bodea Sung) for sample processing and storage, Stanford-LPCH Vaccine Program (Sally Mackey, Alison Holzer and Savita Kamble) for management of clinical studies. The Clinical and Translational Research Unit at Stanford University was supported by an NIH/NCRR CTSA award UL1 RR025744. This work was supported by NIH grants (U19 AI090019, U19 AI057229) and the Howard Hughes Medical Institute to M.M.D., and by the EU’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant (FluPRINT, Project No. 796636) to A.T.
Author information
Authors and Affiliations
Contributions
A.T. downloaded data, coordinated the integration of the data into the FluPRINT database, advised on the database design and wrote the manuscript. I.T. built the MySQL database and wrote PHP script for the data import into the database, and contributed to writing the manuscript. H.T.M. managed the analysis, data collection and management of the SDM at the HIMC and advised during the manuscript preparation. C.L.D. was responsible for regulatory approvals, protocol design, study conduct, clinical data management and provided assistance during the manuscript preparation. M.M.D. conceived and supervised the clinical studies and advised during the analysis and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Tables
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Tomic, A., Tomic, I., Dekker, C.L. et al. The FluPRINT dataset, a multidimensional analysis of the influenza vaccine imprint on the immune system. Sci Data 6, 214 (2019). https://doi.org/10.1038/s41597-019-0213-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-019-0213-4
This article is cited by
-
Mucosal vaccines — fortifying the frontiers
Nature Reviews Immunology (2022)
-
Divergent trajectories of antiviral memory after SARS-CoV-2 infection
Nature Communications (2022)
-
treekoR: identifying cellular-to-phenotype associations by elucidating hierarchical relationships in high-dimensional cytometry data
Genome Biology (2021)