Abstract
T cells, as a crucial compartment of the tumour microenvironment, play vital roles in cancer immunotherapy. However, the basic properties of tumour-infiltrating T cells (TILs) such as the functional state, migratory capability and clonal expansion remain elusive. Here, using Smart-seq2 protocol, we have generated a RNA sequencing dataset of 11,138 T cells isolated from peripheral blood, adjacent normal and tumour tissues of 12 colorectal cancer (CRC) patients, including 4 with microsatellite instability (MSI). The dataset contained an expression profile of 10,805 T cells, as well as the full-length T cell receptor (TCR) sequences of 9,878 cells after quality control. To facilitate data mining of our T cell dataset, we developed a web-based application to deliver systematic interrogations and customizable functionalities (http://crctcell.cancer-pku.cn/). Functioning with our dataset, the web tool enables the characterization of TILs based on both transcriptome and assembled TCR sequences at the single cell level, which will help unleash the potential value of our CRC T cell data resource.
Design Type(s) | transcription profiling design • disease analysis objective |
Measurement Type(s) | transcription profiling assay |
Technology Type(s) | RNA sequencing |
Factor Type(s) | Microsatellite Instability • age • sex • experimental condition • tumor stage |
Sample Characteristic(s) | Homo sapiens • lymphocyte |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
CRC is among the common causes of cancer-related mortality worldwide1,2. While immune checkpoint blocking antibodies (ICBs) have shown impressive clinical benefits in cancers3,4,5,6, their benefits are highly uneven among CRC patients. Remarkably, only CRC patients with MSI showed pronounced responses to ICBs, while patients with microsatellite stability (MSS) derived no benefit7,8. The underlying mechanisms of such discrimination remain elusive. T cells play vital roles in killing malignant cells and are associated with responses to ICB-treatment9,10. It is thus important to understand the cellular underpinnings of TILs in CRC.
Single cell transcriptome analysis has become a compelling approach to decipher the properties of TILs, due to its ability to quantify gene expression and assemble TCR sequences simultaneously. In our recent Nature paper, we have performed single cell RNA sequencing of 11,138 T cells isolated from peripheral blood, adjacent normal and tumour tissues of 12 treatment-naïve CRC patients (Fig. 1a and Table 1), and developed STARTRAC (single T cell analysis by RNA sequencing and TCR tracking) indices to analyse the dynamic relationships among 20 identified T cell subsets11. Here, we provide the detailed description of our dataset and present a webserver to deliver comprehensive and customizable analyses.
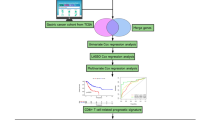
Schematic overview of the study design and analysis pipeline. (a) The experimental flowchart of this study. (b) The bioinformatics pipeline used for data analysis. Softwares used in each steps were labelled in blue. WES, whole exome sequencing; DEG, differentially expressed gene; dist, tissue distribution; expa, clonal expansion; migr, cross-tissue migration; tran, developmental transition.
The dataset contained an average of 1.25 million uniquely mapped read pairs per cell, with an average mapping rate of 96.6% (Online-only Table 1). After quality control, we obtained an expression profile of 12,547 genes for 10,805 cells, with an average of 3,182 genes detected per cell (Online-only Table 1). The expression data could be used to elucidate the expression distributions of genes including those currently pursued as immunotherapy targets in clinical trials (Fig. 2a), illuminating the potentially modulated T cell populations with different immunotherapies. Furthermore, the dataset can serve as a resource for further T cells exploration including the identification of novel regulatory mechanisms by depicting the specific expression patterns of transcription factors (Fig. 2b).
TCR sequences, composed of α- and β-chains, play major roles in the selection and activation of T cells12. Both α- and β-chains contribute to the determination of TCR antigen specificity, and different T cells with the same TCR could be functionally distinct13. To uncover information about T cell ancestry and clonality, we obtained full-length TCR sequences of 91.4% (9,878/10,805) cells with at least one pair of productive α-β chains after eliminating non-productive alleles or low-abundance TCRs (Fig. 3a and Supplementary File 1). Accordingly, T cells with identical TCRs were defined to be from the same clonotype, and a total of 7,274 clonotypes were obtained (Supplementary File 1). Indeed, a strong correlation was observed between the recurring frequencies of α-chains and that of β-chains, indicating a common ancestral cell of origin (Fig. 3b).
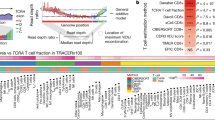
The TCR profile of single T cells. (a) The abundance distributions of TCR α- or β-chain. The gray lines represent the fitting values. (b) The relationship between the degrees of recurrent usage of various TCR α-chains with that of β-chains. Each dot represents a group of TCR α/β allele expressed in a given number of cells. Dot size represents the proportion of such group in all TCR chains detected. (c) TCR sharing patterns of different CD8+ T cell clusters enriched in different tissues.
The TCR sequences can be utilized to delineate TCR sharing patterns of both inter/intra-tissues and inter/intra-clusters (Fig. 3c), shedding light on the properties of T cells including clonal expansion, developmental transition and cross-tissue migration. Furthermore, TCR sequences, as well as the transcriptome data elucidating T cell functions, could serve as a data resource for the discovery of antigen specificity in therapeutic applications14.
In our related work, we have revealed important insights of the T cell biology based on STARTRAC indices11. For instance, tumour-resident CD8+ effector memory and dysfunctional T cells showed mutually exclusive developmental transition patterns, suggesting a TCR-based cell fate decision. In addition, we found that a special subset of IFNG+ TH1-like T cells with CXCL13+BHLHE40+ were preferentially enriched in MSI tumours, which might contribute to the favourable responses of MSI patients to ICBs.
While some discoveries have been made, the unprecedented data resource of CRC T cells is still attractive to many biologists. To facilitate data mining of our T cell dataset, we developed iSTARTRAC (the interactive platform of STARTRAC), a web server to deliver customizable functionalities for further T cell investigation. iSTARTRAC provides key functions including cluster visualization, gene expression demonstration, differential expression analysis, TCR sharing illustration and discrimination of differences between MSI and MSS patients (Fig. 4).
Schema describing the key functionalities of the iSTARTRAC web server. iSTARTRAC provides six functional modules including cluster atlas, gene expression, DEG analysis, TCR-based analysis, STRATRAC indices and MSI versus MSS. Each module implements several customizable analyses for user input samples.
The comprehensive and customizable analyses with simple clicking through iSTARTRAC could greatly facilitate data reuse in the field of cancer immunology, and the accompanying scientific discussion will further expedite the process of therapeutic discovery and understanding the mechanism of immunotherapies with respect to T cell functions.
Methods
These methods are expanded version of descriptions in our related work11, which provided detailed descriptions of experimental procedures including human specimens, single cell collection, cell sorting, reverse transcription, amplification and sequencing, and those of computational processing including quality control, data processing, TCR assembly, unsupervised clustering and definition of STARTRAC indices11. While most part of the methods described here was cited from that report, we specifically aim to emphasize the samples and the methods used to generate the single cell RNA-seq data.
Clinical human specimens
Twelve patients with CRC were enrolled and pathologically diagnosed with colorectal adenocarcinoma at Peking University People’s Hospital. All patients in this study provided written informed consent for sample collection and data analyses. This study was approved by the Research and Ethical Committee of Peking University People’s Hospital and complied with all relevant ethical regulations.
The patients included eight with MSS (P0701, P1012, P1207, P1212, P1228, P0215, P0411 and P0309) and four with MSI (P0123, P0909, P0825 and P0413) status. Among these 4 MSI patients, 3 had positive lymph nodes (P0123, P0413 and P0909), two of them had poorly-differentiated disease (P0825 and P0909), and none of them had distal metastasis. There were eight females and four males, and the median age of diagnosis was 67, ranging from 35 to 82. Among these 12 patients, one was diagnosed at stage I, five at stage II, five at stage III, and one at stage IV, which was classified according to the guidance of AJCC version 8. None of them were treated with chemotherapy or radiation prior to tumour resection. The available clinical characteristics are summarized in Table 1.
Sample collection and preparation
Fresh tumour and adjacent normal tissue samples (at least 2 cm from matched tumour tissues) were surgically resected from the above-described patients. Patients P0701, P0909, P1212, P1228, P0215, P0411, P0413, P0825, P0123 and P0309 had peripheral blood and paired tumour and adjacent normal tissues, whereas patients P1012 and P1207 had only fresh tumour tissue and matched peripheral blood.
Tumours and adjacent normal tissues were cut into approximately 1-mm3 pieces in the RPMI-1640 medium (Invitrogen) with 10% fetal bovine serum (FBS; Sciencell), and enzymatically digested with MACS Tumour Dissociation Kit (Miltenyi Biotec) for 30 min on a rotor at 37 °C, according to the manufacturer’s instruction. The dissociated cells were subsequently passed through a 40-µm cell-strainer (BD) and centrifuged at 400 g for 10 min. After the supernatant was removed, the pelleted cells were suspended in red blood cell lysis buffer (Solarbio) and incubated on ice for 2 min to lyse red blood cells. After washing twice with PBS (Invitrogen), the cell pellets were re-suspended in sorting buffer (PBS supplemented with 1% FBS). PBMCs were isolated using HISTOPAQUE-1077 (Sigma-Aldrich) solution as previously described15. In brief, 3 ml of fresh peripheral blood was collected before surgery in EDTA anticoagulant tubes and subsequently layered onto HISTOPAQUE-1077. After centrifugation, lymphocyte cells remained at the plasma–HISTOPAQUE-1077 interface and were carefully transferred to a new tube and washed twice with PBS. Red blood cells were removed via the same procedure described above. These lymphocytes were re-suspended in sorting buffer.
Single-cell sorting, reverse transcription, amplification and sequencing
Single-cell suspensions were stained with antibodies against CD3, CD4, CD8 and CD25 (anti-human CD3, UCHT1; anti-human CD4, OKT4; anti-human CD8, OKT8; anti-human CD25, BC96; eBioscience) for fluorescence-activated cell sorting (FACS), performed on a BD Aria III instrument. Single cells of different subtypes including cytotoxic T (TC) cells, T helper (TH) cells and regulatory T (Treg) cells were enriched by gating 7AAD−CD3+CD8+, 7AAD−CD3+CD4+CD25−/+ and 7AAD-CD3+CD4+ CD25++ T cells, respectively, and sorted into 96-well plates (Axygen) chilled to 4 °C, prepared with lysis buffer with 1 µl 10 mM dNTP mix (Invitrogen), 1 µl 10 µM Oligo dT primer, 1.9 µl 1% Triton X-100 (Sigma), and 0.1 µl 40 U µl-1 RNase Inhibitor (Takara). The single-cell lysates were sealed and stored frozen at −80 °C immediately. Single-cell transcriptome amplifications were performed according to the Smart-Seq2 protocol15,16. The External RNA Controls Consortium (ERCC; Ambion; 1:4,000,000) was added into each well as the exogenous spike-in control before the reverse transcription. The amplified cDNA products were purified with 1× Agencourt XP DNA beads (Beckman). A procedure of quality control was performed following the first round of purification, which included the detection of CD3D by qPCR (forward primer, 5′-TCATTGCCACTCTGCTCC-3′; reverse primer, 5 primer, 5′-TCATTGCCACT) and fragment analysis by analyser AATI. For those single-cell samples with high quality after quality control (cycle threshold <30), the DNA products were further purified with 0.5× Agencourt XP DNA beads, and the concentration of each sample was quantified by Qubit HsDNA kits (Invitrogen). Multiplex (384-plex) libraries were constructed and amplified using the TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme Biotech). The libraries were then purified with Agencourt XP DNA beads and pooled for quality assessment by fragment analyser. For all the 12 patients, purified libraries were analysed by an Illumina Hiseq 4000 sequencer with 150-bp pair-end reads. For patient P1207, only CD8+ T cells were collected due to the temporary lack of CD4 antibody.
Bulk DNA isolation and sequencing
Genomic DNA of peripheral blood and tissue samples of patients with CRC were extracted using the QIAamp DNA Mini Kit (QIAGEN) according to the manufacturer’s specification. The concentrations of DNA were quantified using the Qubit HsDNA Kits (Invitrogen) and the qualities of DNA were evaluated with agarose gel electrophoresis. Exon libraries were constructed using the SureSelectXT Human All Exon V5 capture library (Agilent). Samples were sequenced on the Illumina Hiseq 4000 sequencer with 150-bp paired-end reads.
Multi-colour immunohistochemistry
OpalTM multi-colour immunohistochemistry (IHC) staining were performed with antibodies of rabbit anti-human CD3 (Abcam, clone SP7, 1:400), mouse anti-human CD8 (Abcam, clone 144B, 1:500), rabbit anti-human CD4 (Abcam, clone EPR6855, 1:400) and mouse anti-human FOXP3 (Abcam, clone mAbcam22510, 1:500) to validate the existence of infiltrating TC, TH and Treg cells in tumour tissues. The specimens were collected and prepared for the formalin-fixed paraffin-embedded tissues sections as previously mentioned15. Antigen was retrieved by AR9 buffer (pH 6.0, PerkinElmer) and boiled in the oven for 15 min. After a pre-incubation with blocking buffer at room temperature for 10 min, the sections were incubated at room temperature for 1 h with aforementioned antibodies. A secondary horseradish peroxidase-conjugated antibody (PerkinElmer) were added and incubated at room temperature for 10 min. Signal amplification was performed using TSA working solution diluted at 1:100 in 1× amplification diluent (PerkinElmer) and incubated at room temperature for 10 min. The multispectral imaging was collected by Mantra Quantitative Pathology Workstation (PerkinElmer, CLS140089) at 20× magnification and analysed by InForm Advanced Image Analysis Software (PerkinElmer) version 2.3. For each patient, a total of 8–15 high-power fields were taken based on their tumour sizes.
Microsatellite instability testing
DNA purified from tumour tissues using QIAamp DNA Mini Kit (QIAGEN) was subjected to multiplex fluorescent PCRbased assay (Promega) by amplifying seven loci including five mononucleotide repeats (NR21, BAT26, BAT25, NR24 and Mono27) and two pentanucleotide repeats (PentaC and PentaD) and was compared with DNA extracted from matched adjacent normal tissues. Multiplex PCR products were analysed by ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Quality control and preprocessing of single cell RNA-seq data
Low-quality read pairs of single-cell RNA sequencing (scRNA-seq) data were filtered out if at least one end of the read pair met one of the following criteria: (1) ‘N’ bases account for ≥10% of the read length; (2) bases with quality <5 account for ≥50% of the read length; and (3) the read contains adaptor sequence. The filtered read pairs were processed using HTSeqGenie pipeline (R package version 4.8) to obtain the gene expression table. Specially, read pairs were then mapped to human ribosomal RNA (rRNA) sequences (download from RFam database) and the read pairs with both ends unmapped were kept for downstream analysis. Read pairs passing this filter for rRNA were aligned to human reference sequence (hg19) using GSNAP17, with parameters ‘–novelsplicing 1 -n 10 -i 1 -M 2’. To calculate the expression levels of genes, the gene model file ‘knownGene.txt’ (30 June 2013 version), downloaded from UCSC, was used. The R function findOverlaps was used to count the number of uniquely mapped read pairs located in each gene and the count table tabulated as genes by cells was used for downstream analysis. The transcripts per million (TPM) table was derived from the count table and the TPM value was calculated by
where Cij is the count value of gene i in cell j. It should be noticed that the TPM here is a simplified version based on the hypothesis that all mapped reads are approximate the same length.
Low-quality cells were filtered if the library size or the number of expressed genes (counts larger than 0) was smaller than predefined thresholds. Both thresholds were defined as the medians of all cells minus 3× the median absolute deviation. Furthermore, if the proportion of mitochondrial gene counts was larger than 10%, these cells were discarded. Only cells with the average TPM of CD3D, CD3E and CD3G larger than 10 were kept for subsequent analysis. We further identified CD4+, CD8+, CD4−CD8− (double negative) and CD4+CD8+ (double positive) T cells based on the gene expression data. Given the average TPM of CD8A and CD8B, one cell was considered as CD8 positive or negative if the value was larger than 30 or less than 3, respectively; given the TPM of CD4, one cell was considered as CD4 positive or negative if the value was larger than 30 or less than 3, respectively. Hence, the cells can be in silico classified as CD4+CD8−, CD4−CD8+, CD4+CD8+, CD4−CD8− and other cells that cannot be clearly defined.
While TPM is an intuitive and popular measurement to standardize the total number of transcripts between cells, it is insufficient and could bias downstream analysis because TPM can be dominated by a handful of highly expressed genes. Therefore, we mainly used TPM for preliminary data processing and gene expression visualization. Recently, methods for normalizing scRNA-seq data including scran18 have been proposed to implement robust and effective normalization, and thus we used the size-factor normalized read count for main analyses in our study including dimensionality reduction, clustering and finding markers for each cluster.
After discarding genes with average counts of fewer than or equal to 1, the count table of the cells passing the above filtering was normalized by a pooling strategy. We applied the R package scran18 in Bioconductor to perform the normalization process. Specifically, cells were pre-clustered using the ‘quickCluster’ function with the parameter ‘method = hclust’. Size factors were calculated using ‘computeSumFactors’ function with the parameter ‘sizes = seq (20,100,by = 20)’ which indicates the number of cells per pool. Raw counts of each cell were divided by their size factors, and the resulting normalized counts were then scaled to log2 space and used for batch correction.
Scran utilizes a pooling strategy implemented in ‘computeSumFactors’ function, in which size factors for individual cells were deconvoluted from size factors of pools. To avoid violating the assumption that most genes were not differentially expressed, hierarchical clustering based on Spearman’s rank correlation was performed with ‘quickCluster’ function first, then normalization was performed in each resulting cluster separately. The size factor of each cluster was further re-scaled to enable comparison between clusters.
To remove the possible effects of different donors on expression, the normalized table was further centred by patient. Thus, in the centred expression table, the mean values of the cells for each patient were zero. A total of 12,548 genes and 10,805 cells were retained in the final expression table. If not explicitly stated, ‘normalized read count’ or ‘normalized expression’ in this study refers to the normalized and centred count data for simplicity.
Unsupervised clustering analysis of CRC single T cell RNA-seq dataset
The cell clusters used here were the same as defined in our related Nature paper11. The expression tables of CD8+CD4− T cells and CD8−CD4+ T cells as defined by the aforementioned in silico classification but excluding MAIT cells and iNKT cells, were fed into an iteratively unsupervised clustering pipeline separately. Specifically, given expression table, the top n genes with the largest variance were selected, and then the expression data of the n genes were analysed by single-cell consensus clustering (SC3)19. n was tested from 500, 1000, 1500, 2000, 2500 and 3000. In SC3, the distance matrices were calculated based on Spearman correlation and then transformed by calculating the eigenvectors of the graph Laplacian. Then the k-means algorithm was applied to the first d eigenvectors multiple times where d was chosen from 4% to 7% of the total number of input cells. Finally, hierarchical clustering with complete agglomeration was performed on the SC3 consensus matrix and k clusters were inferred. The SC3 parameters k, which was used in the k-means and hierarchical clustering, was tried from 2 to 10. For each SC3 run, the silhouette values were calculated, the consensus matrix was plotted, and cluster specific genes were identified. Such information was used to determine the optimal k and n. Once the stable clusters were determined, the above procedure was iteratively applied to each of these clusters to reveal the sub-clusters. After obtained the stable clusters by SC3, we further redefined the cluster labels of indeterminate cells with the silouatte values less than zero by R package XGBoost20. The training datasets were composed of cells with the silouatte >0, while cells to be reclassified with the silouatte <0 were then redefined to clusters with the largest predicting score. The in silico classified CD8+CD4− MAIT cells had distinct gene expression patterns compared with other CD8+CD4− T cells, and were defined as cluster “CD8_C08-SLC4A10”.
When the clustering results were obtained, one-way ANOVA implemented by R function aov was performed to identify the differentially expressed genes among the clusters. R function TukeyHSD was used to identify which cluster pairs showed a significant difference. A gene was defined as being significantly differentially expressed based on the following criteria: 1) adjusted P-value (Benjamini-Hochberg method) of F test less than 0.05; 2) the absolute difference of any one significant cluster pair (P-value of Tukey’s ‘Honest Significant Difference’ method less than 0.01) larger than 1. The significantly differentially expressed genes were categorized in the cluster that showed the highest expression.
The t-SNE method implemented in R package Rtsne was used for clustering visualization. To visualize the cell density on the t-SNE plot, kernel density estimation was performed using R function kde (ks package), and the contour lines encompassing the top 10%, 20%, …90% cells with highest densities were shown. A total of 8,530 T cells, including 3,628 CD8+CD4− and 4,902 CD8−CD4+ T cells with clustering definitions, were used in the t-SNE projection. Other cells such as CD8+CD4+ and CD8−CD4− T cells were not included in this visualization.
Analysis pipelines of bulk exome sequencing data
The bulk exome sequencing data were cleaned following the same procedure for the scRNA-seq data processing. The cleaned read pairs were then processed according to the BWA-Picard/ Genome Analysis Toolkit (GATK)-Strelka pipeline. In brief, the cleaned read pairs were aligned to human genome reference version b37 (downloaded from ftp://ftp.broadinstitute.org:/bundle) by the BWA-MEM algorithm21. The alignments were then sorted and de-duplicated by Picard (Broad Institute). GATK22 was used to realign multiple reads around putative INDEL by Smith–Waterman alignment algorithm and re-calibrate base quality. The analysis-ready bam files were input into the GATK UnifiedGenotyper module to call SNP/INDEL and into Strelka23 to call somatic SNV/INDEL and into ADTEx24 (version 1.0.4) to call somatic copy number alterations. The mutations were annotated with ANNOVAR25.
TCR assembly
TraCeR26 was used to deduce the TCR sequences of each cell. The outputs of TraCeR include the assembled nucleotide sequences for both α and β chains, the coding potential of the nucleotide sequences (that is, productive or not), the translated amino acid sequence, the CDR3 sequences and the estimated TPM value of α or β chains. Only cells with TPM values larger than 10 for the α chain and larger than 15 for the β chain were kept. For cells with two or more α or β chains assembled, the α–β pair that was productive and of the highest expression level was defined as the dominant α–β pair in the corresponding cell. If two cells had identical dominant α–β pairs, the dominant α–β pair was identified as clonal TCRs.
To integrate with the gene expression data, the TCR-based analysis was performed only for cells that passed the aforementioned quality control pipeline (total 10,805). Thus, 9,878 cells with TCR information were used in the integrative analysis27 (Supplementary File 1). If one cell had an α chain composed of V segment TRAV1-2 and one of the following J segments (TRAJ33, TRAJ20 and TRAJ12), the cell was classified as a MAIT cell28. If the α chain of one cell was rearranged by V segment TRAV10 and J segment TRAJ18, the cell was classified as an invariant natural killer T cell29. In the 9,878 cells with at least one pair of productive α and β chains, only 3 cells were identified as invariant natural killer T cells, and 102 cells were identified as MAIT cells, including 71 CD8+CD4− T cells classified in silico.
Definition of STARTRAC indices
We present STRATRAC as a framework, defined by four indices, to analyse different aspects of T cells based on paired single cell transcriptomes and TCR sequences. The first index, named as STARTRAC-dist (STARTRAC-distribution), utilizes the ratio of observed over expected cell numbers in tissues to measure the enrichment of T cell clusters across different tissues. Given a contingency table of T cell clusters by tissues, we first apply Chi-squared test to evaluate whether the distribution of T cell clusters across tissues significantly deviates from random expectations. We then calculate the STARTRAC-dist index for each combination of T cell clusters and tissues according the following formula:
where Ro/e is the ratio of observed cell number over the expected cell number of a given combination of T cell cluster and tissue. The expected cell number for each combination of T cell clusters and tissues are obtained from the Chi-squared test. \({I}_{dist}^{STARTRAC}\) can indicate whether cells of a certain cluster are enriched (Ro/e > 1) or depleted (Ro/e < 1) in a specific tissue.
The other three STARTRAC indices, STARTRAC-expa (STARTRAC-expansion), STARTRAC-migr (STARTRAC-migration) and STARTRAC-tran (STARTRAC-transition), are designed to measure the degree of clonal expansion, tissue migration, and state transitions of T cell clusters upon TCR tracking, respectively. The MAIT cells were not included in these types of analyses because they have distinct TCRs. For STARTRAC-expa, which uses the standard TCR clonality measurement30 but is specifically applied to different T cell clusters in our analyses, we first adopt the normalized Shannon entropy to calculate the evenness of the TCR repertoire of the given T cell cluster and then define the STARTRAC-expa index as 1-evenness. Mathematically, the STARTRAC-expa index of a specific cluster with N clonotypes is defined by the following formula:
where pi is the cell frequency of clonotype i in the cluster, and a clonotype is defined by identical, full-length, paired α and β TCR chains. STARTRAC-expa ranges from 0 to 1, with 0 indicating no clonal expansion for each clonotype while 1 indicating that the cluster is composed of only one clonally expanded clonotype, with high STARTRAC-expa indicating high clonality.
For T cells with identical TCR clonotypes, even if they are present in different tissues or in different development states, logically they could be likely derived from a single naïve T cell, clonally expanded initially at one location and migrated across tissues or have undergone state transitions. Based on this principle, we define STARTRAC-migr and STARTRAC-tran to evaluate the extent of tissue migration and state transition of each clonotype, respectively. For each clonotype, given its distribution across tissues (peripheral blood, adjacent normal mucosa and tumour), we define its STARTRAC-migr index \({I}_{migr}^{t}\) as:
where \({p}_{j}^{t}\) is the ratio of the number of cells with TCR clonotype t in tissue j to the total number of cells with TCR clonotype t and \({\sum }_{j=1}^{J}{p}_{j}^{t}=1\). For two T cell clusters with similar clonal expansion and clonal size, the one with clonal cells broadly distributed in various tissues would likely be more mobile. Similarly, its STARTRAC-tran index \({I}_{tran}^{t}\) can be defined as:
where \({p}_{k}^{t}\) is the ratio of the number of cells with TCR clonotype t in cluster k to the total number of cells with TCR clonotype t, \({\sum }_{k=1}^{K}{p}_{k}^{t}=1\), and K is the total number of cell clusters. The input of STARTRAC-migr is the observed cell frequency across tissues of a certain clonotype, while the input of STARTRAC-tran is the observed cell frequency across cell clusters of a certain clonotype. By contrast, the input of STARTRAC-expa is the observed cell frequency across clonotypes of a certain cell cluster, and the input for the traditional TCR clonality measure is the observed sequence frequency across a TCR repertoire of a given sample.
After the extent of tissue migration of each clonotype is quantified by STARTRAC-migr, given a cluster with total T clonotypes, the STARTRAC-migr index at the cluster level \({I}_{migr}^{STARTRAC}\) can be defined as the weighted average of all TCR clonotype migration indices contained in the cluster:
where \({p}_{cls}^{t}\) is the ratio of the number of cells with clonotype t in cluster cls to the total number of cells in cluster cls.
Similarly, when the extent of state transition of each clonotype is quantified by STARTRAC-tran, given a cluster with total T clonotypes, the STARTRAC-tran index at the cluster level can be defined as the weighted average of all TCR clonotypes state transition indices contained in the cluster:
where \({p}_{cls}^{t}\) is the ratio of the number of cells with clonotype t in cluster cls to the total number of cells in cluster cls.
Besides the overall evaluation of the extents of migration and state transitions by STARTRAC-migr and STARTRAC-tran, we also define pairwise STARTRAC-migr (pSTARTRAC-migr) and STARTRAC-tran (pSTARTRAC-tran) indices for precise quantification. For example, given a clonotype t and two tissue types (e.g., blood and tumour), the pSTARTRAC-migr index \({p}^{{I}_{migr}^{t}}\) is calculated by the following formula:
where \({p}_{j}^{t}\) is the ratio of the number of cells with TCR clonotype t in tissue j to the total number of cells with TCR clonotype t in tissues 1 and 2 (i.e., blood and tumour), and \({\sum }_{j=1}^{2}{p}_{j}^{t}=1\). In other words, pSTARTRAC-migr uses the same formula as STARTRAC-migr but limits the number of tissues to two and the frequencies of cells between two specified tissues are re-calculated. Likewise, given a clonotype t and two T cell clusters (e.g., TEM and TEX), the pSTARTRAC-tran index \({p}^{{I}_{tran}^{t}}\) is calculated by the following formula:
where \({p}_{k}^{t}\) is the ratio of the number of cells with TCR clonotype t in cluster k to the total number of cells with TCR clonotype t in clusters 1 and 2 (i.e., TEM and TEX), and \({\sum }_{k=1}^{2}{p}_{k}^{t}=1\). Thus, pSTARTRAC-tran uses the same formula as STARTRAC-tran but limits the number of clusters to two and the frequencies of cells between the two specified clusters are re-calculated. Once pairwise STARTRAC-migr and STARTRAC-tran for clonotypes are obtained, the corresponding indices for clusters are calculated via weighted average according to their clonotype compositions.
Summary of scRNA-seq data and bioinformatics workflow used for data processing
For all the 12 patients, a total of 35.5 G raw reads and 5.4 T raw bases were obtained after sequencing. After preprocessing, we obtained 32.5 G high-quality reads with an average high-quality rate of 91.3% (Online-only Table 1). Accordingly, we summarized the data processing procedures and tools used in each step in a flowchart, consisting of quality control filtering, TCRs assembly, expression quantification, data normalization and downstream analyses (Fig. 1b).
Data Records
As described in our related research paper11, the raw sequencing data have been deposited in the European Genome-phenome Archive database under study accession id EGAS00001002791 and dataset accession id EGAD0000100391031, which are available in FASTQ file format upon request and approval. The DATA ACCESS AGREEMENT is provided at https://github.com/zhangyybio/single-T-cell-data-access. Applicants can request access to the data by directly downloading it or by sending an email to cancerpku@pku.edu.cn. The process that is used to approve an application includes verifying the institution, participants and research purposes of the application, and the authorization by EGA. In general this process will take about two weeks. In principal, any academic research institutions complying with the laws and bioethic regulation policies of China will be approved. The publication moratorium described in the Data Access Agreement officially expires concurrent with publication of this Data Descriptor. The processed gene expression data were deposited in the Gene Expression Omnibus database under accession id GSE10898932. The clinical data recording available clinical characteristics of the collected 12 CRC patients are summarized in Table 1 and the genomic features are summarized in Table 2 and Online-only Table 2. Online-only Table 3 lists the DNA fragment sizes of short tandem repeat loci from tested patients in microsatellite instability testing experiment. Basic statistics of single cell sequencing data are provided in Online-only Table 1. The cluster information and TCR typing data are presented in Supplementary File 1, which has also been uploaded to Figshare27.
Technical Validation
Validating the presence of tumour-infiltrating lymphocytes
OpalTM multi-colour IHC staining were performed with anti-CD3, CD8,CD4, and FOXP3 antibodies to validate the existence of infiltrating TC, TH and Treg cells in tumour tissues (Fig. 5a).
Quality assessment of single cell RNA-seq data. (a) OpalTM multi-colour IHC staining to validate the existence of T cells in CRC tumours (exemplified by P0215 and P1212). (b) One representative example of cDNA size distribution derived from tumour of P0309. (c) One representative fragmentation profile of sequencing library after tagmentation prepared from pooled amplicons produced by PCR amplification of cDNA from samples of P0413. (d) The densities of GC content per sequence for two representative samples of P1212 and P1228. (e) Heatmaps demonstrating the expression levels of classic marker in each T cell subtypes. The right-sided barplots showed the percentages of cell with the expression of corresponding genes (TPM > 0). RFU, relative fluorescence unit.
Validating the genomic features of CRC patients
Exome sequencing of bulk tumours from 12 patients showed that four patients harboured mutations in TP53 and five patients harboured mutations in APC/FBXW7. These genomic alterations were consistent with the characteristics of colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) from The Cancer Genome Atlas (TCGA)33. Summarized tables were provided for the statistics of somatic mutations (Table 2) and selected cancer-associated somatic mutations (Online-only Table 2) that were detected in these patients.
Validating the genomic alterations of MSI patients
Among the 12 CRC patients, 4 patients (P0123, P0909, P0825 and P0413) showed deficient in DNA mismatch repair based on IHC testing of four markers (MLH1, MSH2, MSH6, and PMS2)11, which was also supported by the much higher mutation load (Table 2). To further confirm the MSI status of these patients, we performed microsatellite instability testing by multiplex fluorescent PCR-based assay. Indeed, we found that 4 tumours from MSI patients were characterized by MSI-H phenotypes with two or more mononucleotide loci showing instability (Online-only Table 3).
Validation of RNA samples & RNA-seq libraries
Quality control procedure was performed following the first round of purification of amplified cDNA products, including the detection of CD3D by qPCR and fragment analysis. For single cell samples with high quality (cycle threshold <30), the DNA products were further purified and the concentration of each sample was quantified (Fig. 5b). The constructed multiplex libraries were purified and pooled for quality assessment (Fig. 5c).
Validating the quality of scRNA-seq data
Quality control analyses revealed that the raw sequence data were of high quality, with an average high-quality rate of 91.3% (Online-only Table 1). We assessed the qualities of clean data by statistics of per sequence quality scores and per sequence GC contents. For each sequence, an average of 87.9% bases have a quality score higher than phred quality 30 (Q30), and 94.5% bases have a quality score higher than phred quality 20 (Q20) (Online-only Table 1). In addition, the GC contents of each sample showed a similar normal distribution, with a mean value of 46.2% (Fig. 5d and Online-only Table 1). These statistics indicated that high-quality RNA-seq reads were obtained for downstream analysis.
Validating cell types by marker genes
To evaluate the accuracy of FACS, we examined the expression of conventional marker genes of T cell subsets, including CD3D, CD3E, CD3G, CD8A, CD8B, CD4, IL2RA and FOXP3 (Fig. 5e). While dropout event is prevalent and challenging in single cell RNA-seq data, the gene expression levels of classical T cell markers were consistent with protein levels measured by FACS. Specifically, all T cells were characterized by high expression of CD3 genes (CD3D, CD3E and CD3G). Most TC cells expressed high-level of CD8 (CD8A, CD8B) but low-level of CD4, whereas TH cells and Tregs exhibited the opposite pattern. Tregs showed high expressions of IL2RA encoding transmembrane protein CD25 and regulatory transcription factor FOXP3 compared with TH cells (Fig. 5e). Therefore, the expression patterns of classic T cell markers confirmed the reliability of T cell subtypes.
Usage Notes
To facilitate reuse of our T cell dataset and broaden the user community, we developed a web server and will use the following sections to elaborate the design and functionalities provided by iSTARTRAC. iSTRATRAC is available at http://crctcell.cancer-pku.cn/.
Design and implementation
Although we have provided an online portal at http://crc.cancer-pku.cn to depict gene expressions, only limited functionalities were presented, hindering the wide usage of our data. Here, to facilitate further exploration of our T cell data, we have developed a much enhanced web server iSTARTRAC to enable the comprehensive and customizable analyses.
The iSTARTRAC website is deployed on server with 64GB RAM and CPU Gold 6149 × 16 cores running the Ubuntu (version 16.04.4) Linux (version 4.4.0) operating system. The interface is constructed using the Shiny web application framework (version 1.2.0) in R (version 3.5.0) running on the Shiny-server (version 1.5.6.875).
iSTARTRAC is freely available to all users with no login requirement, and can be accessed by most web browsers including Google Chrome, Mozilla Firefox, Safari and Internet Explorer. The website automatically adjusts the look and feel according to different browsers and devices, but Google Chrome is recommended to achieve the best visualization.
Sample options panel
In each module of iSTARTRAC, four categories of basic options are available for modulating the input samples of interest, including Cluster, Cell Type, Tissue Type and Patient. The Cluster icon consists of 20 clusters including 8 for CD8+ T cells and 12 for CD4+ T cells, and the Cell Type icon is composed of five cell types including CD8+ T cells, CD4+ T cells, CD4+ CD25− T cells, CD4+ CD25+ T cells and CD4+ CD25++ T cells defined by FACS. Peripheral blood (P), adjacent normal (N) and tumour infiltrating (T) are included in the Tissue Type icon. The Patient icon contains eight MSS patients, as well as four MSI patients.
Moreover, iSTARTRAC presents interactive sliders that can be adjusted to change the dot sizes and line widths to achieve optimal visualization of the plots. Plots are regenerated on-the-fly as the user changes sliders or samples, providing an interactive experience that makes it possible to perform customizable analyses.
Functionalities
iSTARTRAC provides key interactive and customizable functions including cluster visualization, gene expression demonstration, differential expression analyses between clusters or cell types, TCR sharing illustration, customizable analysis of STARTRAC indices and discrimination of differences between MSI and MSS patients (Fig. 4).
Cluster atlas
iSTARTRAC dynamically demonstrates the tSNE plot of cell clusters for user-defined T cells derived from given cell clusters, tissue origins, cell types and patients (in the ‘tSNE Plot’ tab). In addition, an annotation table of basic information of T cells is shown and users are allowed to download the table by clicking the DOWNLOAD button (in the ‘Table’ tab).
Gene expression
In this module, iSTARTRAC interactively plots expression distribution of a given gene in different clusters according to user-defined sample selections. The results can be presented in tSNE plot (in the ‘tSNE Plot’ tab), violin plots (in the ‘Violin Plot’ tab), or box plots (in the ‘Box Plot’ tab).
Differential expression analysis
iSTARTRAC performs differential expression (DE) analyses and identifies differentially expressed genes (DEGs) between any two given clusters (in ‘Cluster DEG’ tab) or cell types (in ‘Cell Type DEG’ tab), illustrating the results in volcano plots. Single cell transcriptome data is exceptionally appropriate for dissecting the intrinsic cellular heterogeneity. In addition to the commonly used unsupervised clustering, pairwise gene expression distribution, a simple and effective approach similar to FACS with proteins, can also be utilized to detect cell subpopulations. Accordingly, iSTARTRAC allows users to input a pair of genes to dynamically compartmentalize cell subpopulations and performs differential expression analysis for any two subdivided populations (in ‘in silico FACS’ tab). Users can adjust the thresholds of low/high-expression, as well as the significance thresholds of fold change and p-values after multiple testing adjustments. Furthermore, summary tables of signature gene for CD8+ and CD4+ T cells are provided and can be downloaded (in ‘Table’ tab).
TCR-based analysis
For any user-defined frequency of clonal cells, iSTARTRAC provides a tSNE plot to illustrate the distribution of clonal cells in each cluster, with non-clonal cells (cells harbouring TCRs with a frequency below the defined threshold) coloured in grey as background (in ‘tSNE Plot’ tab). The enormous TCR repertoire, which is essential for recognising foreign antigens and tumour neoantigens, could serve as tags to track T cell lineages. Accordingly, iSTARTRAC plots a heatmap to depict the TCR sharing patterns of various clusters enriched in different tissues (in ‘TCR Sharing’ tab), providing the clues of cross-tissue migration and state transition. In addition, iSTARTRAC presents bar plots to show the clonotype statistics of user-defined samples (in ‘Clonotype Statistics’ tab). A summary table of TCR typing is displayed and can be downloaded, which contains the information of TCR sequences and corresponding samples (in ‘Table’ tab).
STRATRAC indices
For given samples, iSTARTRAC dynamically illustrates the STRATRAC-dist indices to dissect the tissue preference of T cell clusters, yielding a discrete enrichment table decorated with colours (in ‘STARTRAC-dist’ tab). Users are allowed to adjust the thresholds for discretizing enrichment levels quantified by Ro/e (the ratio of observed over expected cell numbers in tissues to measure the enrichment of T cell clusters across different tissues). To reveal dynamic relationships of T cell subsets with respect to clonal expansion, migration and development transition, iSTARTRAC plots STRATRAC-expa/migr/tran indices for samples of user interest (in ‘STRATRAC-expa/migr/tran’ tab). Furthermore, pairwise STRATRAC-migr (in ‘pSTRATRAC-migr’ tab) and pairwise STRATRAC-tran (in ‘pSTRATRAC-tran’ tab) could also be dynamically illustrated according to user defined sample selections.
MSI versus MSS
With this module, users can delineate differences in term of cell compositions (in ‘Cell Percentage’ tab), STARTRAC indices (in ‘STARTRAC-expa/migr/tran’ tab) and gene expressions (in ‘DEG Analysis’ tab) between MSI and MSS patients for user-specified dataset of interest.
Summary of scRNA-seq data application
The compendium dataset provided here, was produced primarily to illustrate the dynamic relationships of tumour-infiltrating lymphocytes in CRC, including functional states, clonal expansions, migrations and developmental transitions11.
The dataset can be further utilized to detect the transcript isoforms, non-coding transcripts and the potential splice variants. The differential isoform usages of T cell subtypes will shed new light on the underlying regulatory mechanisms of phenotypic differentiation and will provide opportunities for immuno-oncology modulation by determining the subtype specific expression of known and novel isoforms in TILs.
In addition, our dataset could serve as a resource for the comparison of different library preparation methods such as Smart-seq2 protocol and 10X platform, providing specific features of RNA-seq data produced with Smart-seq2 protocol.
The interactive platform, iSTARTRAC, could be explored by experimental biologists to dissect regulatory mechanisms of T cell differentiation, identify novel targets of immunotherapy, as well as to compare the differences of T cell compositions, gene expressions and STARTRAC indices between MSI and MSS patients. The comprehensive and customizable analyses with simple clicking through iSTARTRAC will facilitate data mining in cancer immunology community and help unleash the potential value of our CRC T cell data resource.
Code Availability
Sequencing data were processed using SAMtools (version 0.1.19), Picard (version 2.18.9) and GATK (version 3.8-1-0). Clean reads were aligned to human reference genome (hg19) using GSNAP (version 2014-10-22). TraCeR (version 2015-10-21) was used to assemble the TCR sequences of single T cells.
All downstream analyses were performed using open source R (version 3.5.0). A series of R package were utilized for data analyses including HTSeqGenie (version 4.8.0) for expression quantification, single-cell consensus clustering (SC3, version 1.7.2) for unsupervised clustering and Rtsne (version 0.13) for dimension reduction.
Static visualizations of iSTARTRAC are rendered as Portable Document Format (PDF). Tables are generated with R package DT (version 0.5), which provides R interface to the JavaScript library DataTables and allows for data querying, selection and download.
Other R packages used by iSTARTRAC includes ggplot2 (version 3.1.0) for plotting box plots, violin plots and volcano plots, ComplexHeatmap (version 1.18.1) for plotting heatmaps, limma (version 3.36.5) for detecting DEGs, ks (version 1.11.3) for plotting cell densities, Startrac (version 0.1.0) for obtaining indices of STARTRAC, RColorBrewer (version 1.1-2) for colour palettes and org.Hs.eg.db (version 3.6.0) for converting gene names etc.
Code for preliminary data processing including size-factor normalization, dimensional reduction and clustering is available on Figshare (https://doi.org/10.6084/m9.figshare.8204624.v1), and code for STARTRAC is available on GitHub (https://github.com/Japrin/STARTRAC).
References
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–386 (2015).
Arnold, M. et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 66, 683–691 (2017).
McDermott, D. F. et al. Survival, Durable Response, and Long-Term Safety in Patients With Previously Treated Advanced Renal Cell Carcinoma Receiving Nivolumab. J. Clin. Oncol. 33, 2013–2020 (2015).
Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science 348, 56–61 (2015).
Reck, M. et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 375, 1823–1833 (2016).
Khalil, D. N., Smith, E. L., Brentjens, R. J. & Wolchok, J. D. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 13, 394 (2016).
Le, D. T. et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Kalyan, A., Kircher, S., Shah, H., Mulcahy, M. & Benson, A. Updates on immunotherapy for colorectal cancer. J. Gastrointest. Oncol 9, 160–169 (2018).
Mellman, I., Coukos, G. & Dranoff, G. Cancer immunotherapy comes of age. Nature 480, 480–489 (2011).
Farhood, B., Najafi, M. & Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 234, 8509–8521 (2019).
Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018).
Coulie, P. G., Van den Eynde, B. J., van der Bruggen, P. & Boon, T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat. Rev. Cancer 14, 135–46 (2014).
Han, A., Glanville, J., Hansmann, L. & Davis, M. M. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014).
Pasetto, A. et al. Tumor- and Neoantigen-Reactive T-cell Receptors Can Be Identified Based on Their Frequency in Fresh Tumor. Cancer Immunol. Res. 4, 734–743 (2016).
Zheng, C. et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 169, 1342–1356 (2017).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Wu, T. D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
Lun, A. T., Bach, K. & Marioni, J. C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17, 75 (2016).
Kiselev, V. Y. et al. SC3: consensus clustering of single-cell RNA-seq data. Nat. Methods 14, 483–486 (2017).
Chen, T. & Guestrin C. XGBoost: A Scalable Tree Boosting System. KDD ‘16 Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 785–794 (2016).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Saunders, C. T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012).
Amarasinghe, K. C. et al. Inferring copy number and genotype in tumour exome data. BMC genomics 15, 732 (2014).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Stubbington, M. J. T. et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods 13, 329–332 (2016).
Zhang, Y., Zheng, L. T., Zhang, L. & Zhang, Z. Expression profile and TCR typing information of single T cells derived from 12 treatment-naive colorectal cancer patients. figshare, https://doi.org/10.6084/m9.figshare.7634762.v1 (2019).
Wilgenburg, V. B. et al. MAIT cells are activated during human viral infections. Nat. Commun. 7, 11653 (2016).
Godfrey, D. I., Stankovic, S. & Baxter, A. G. Raising the NKT cell family. Nat. Immunol. 11, 197–206 (2010).
Kirsch, I., Vignali, M. & Robins, H. T-cell receptor profling in cancer. Mol. Oncol 9, 2063–2070 (2015).
European Genome-phenome Archive, https://identifiers.org/ega.dataset:EGAD00001003910 (2018).
Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Gene Expression Omnibus, https://identifiers.org/geo:GSE108989 (2018).
Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Acknowledgements
We thank C.X. Ye for sample preparation and F. Wang, X. Zhang and J.S. Li for assistance with FACS. We thank Dr. Z. Tang for assistance with website construction. We thank the Computing Platform of the CLS (Peking University). This project was supported by Beijing Advanced Innovation Centre for Genomics at Peking University, Key Technologies R&D Program (2016YFC0900100), National Natural Science Foundation of China (81573022, 31530036, 91742203 and 81672375) and Amgen Corporation (USA). L.Z. was supported by the Postdoctoral Foundation of CLS.
Author information
Authors and Affiliations
Contributions
Z.Z. and Y.Z. designed experiments. L.Z. performed the experiments. Y.Z., L.T.Z., X.R. and X.H. analysed sequencing data. Y.Z. constructed the website. Y.Z. and Z.Z. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online-only Tables
ISA-Tab metadata file
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Zhang, Y., Zheng, L., Zhang, L. et al. Deep single-cell RNA sequencing data of individual T cells from treatment-naïve colorectal cancer patients. Sci Data 6, 131 (2019). https://doi.org/10.1038/s41597-019-0131-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-019-0131-5
This article is cited by
-
Construction of an immune predictive model and identification of TRIP6 as a prognostic marker and therapeutic target of CRC by integration of single-cell and bulk RNA-seq data
Cancer Immunology, Immunotherapy (2024)
-
Drug conjugates for targeting regulatory T cells in the tumor microenvironment: guided missiles for cancer treatment
Experimental & Molecular Medicine (2023)
-
T cells in health and disease
Signal Transduction and Targeted Therapy (2023)
-
Intestinal cellular heterogeneity and disease development revealed by single-cell technology
Cell Regeneration (2022)
-
Single-cell RNA sequencing coupled to TCR profiling of large granular lymphocyte leukemia T cells
Nature Communications (2022)